Abstract
The human β-globin locus control region (LCR) harbors both strong chromatin opening and enhancer activity when assayed in transgenic mice. To understand the contribution of individual DNase I hypersensitive sites (HS) to the function of the human β-globin LCR, we have mutated the core elements within the context of a yeast artificial chromosome (YAC) carrying the entire locus and then analyzed the effect of these mutations on the formation of LCR HS elements and expression of the genes in transgenic mice. In the present study, we examined the consequences of two different HS2 mutations. We first generated seven YAC transgenic lines bearing a deletion of the 375-bp core enhancer of HS2. Single-copy HS2 deletion mutants exhibited severely depressed HS site formation and expression of all of the human β-globin genes at every developmental stage, confirming that HS2 is a vital, integral component of the LCR. We also analyzed four transgenic lines in which the core element of HS2 was replaced by that of HS3 and found that while HS3 is able to restore the chromatin-opening activity of the LCR, it is not able to functionally replace HS2 in mediating high-level globin gene transcription. These results continue to support the hypothesis that HS2, HS3, and HS4 act as a single, integral unit to regulate human globin gene transcription as a holocomplex, but they can also be interpreted to say that formation of a DNase I hypersensitive holocomplex alone is not sufficient for mediating high-level globin gene transcription. We therefore propose that the core elements must productively interact with one another to generate a unique subdomain within the nucleoprotein holocomplex that interacts in a stage-specific manner with individual globin gene promoters.
Locus control regions (LCRs) are highly specialized tissue-specific DNA regulatory elements that are able to confer position-independent and copy number-dependent expression of cis-linked genes when examined in transgenic mice. Since the discovery of the human β-globin LCR (13, 20), a growing number of genes or loci have been found to be regulated by LCR-like activities. Most LCRs appear to be composite elements and, perhaps not coincidentally, contain several DNase I hypersensitive sites (HS). The examples of genes regulated by such elements include the human β-globin (45), the T-cell-specific CD2 (18), the T-cell receptor α/δ (41), and the chicken lysozyme loci (2). Higgs et al. (23) and Montoliu et al. (35) have shown that single HS located upstream of the α-globin or tyrosinase genes also bear multiple activities normally attributed to an LCR.
The human β-globin LCR, located from approximately 8 to 22 kbp upstream of the ɛ-globin gene (13, 14, 47, 48) is composed of four erythroid cell-specific (HS1 to HS4) and one ubiquitous (HS5) DNase I HS. This region mediates chromatin opening over the whole β-globin gene locus and also is responsible for stimulating high-level expression of the globin genes throughout erythroid cell development (12). Perhaps most remarkably, the LCR enhances transcription over a considerable distance, more than 60 kbp in the adult human β-globin gene. Characteristically, it is able to confer position-independent and copy number-dependent expression to linked globin genes in transgenic mice. In addition, the human β-globin LCR is required for altered timing of DNA replication of the locus, which initiates in erythroid cells from an origin located between the adult δ- and β-globin genes (1, 27).
The mechanism(s) by which the LCR elicits its multiple activities is unclear. One model has proposed that the LCR acts only indirectly, by providing an open, accessible chromatin environment and that gene-proximal regulatory elements mediate the developmental switches as well as high levels of globin gene expression (32). In a second, mutually exclusive model, it was proposed that the LCR, after inducing chromatin opening, directly interacts by DNA looping to stimulate high-level, stage-specific expression of individual globin genes (10, 49).
The contribution of individual HS elements to the overall function of the LCR is also somewhat controversial. Previous studies showed that human β-globin LCR element HS2 is a potent enhancer when analyzed in both transient- and stable-transfection assays, showing that HS2 functions as a classical enhancer in the absence or presence of chromatin (7, 46, 48). In contrast, HS3 and HS4 were shown to function as potent activators only when the constructs were integrated into chromosomal DNA, indicating that for these two elements, chromatin environment plays an important role in their activity (7, 26, 36). Subsequent studies revealed that HS2, HS3, and HS4 act as strong individual enhancers in transgenic mice when linked to cosmid constructs bearing the human γ- and β-globin genes (16). More importantly, Fraser et al. (16) showed that individual HS elements exhibit stage-specific preferences for enhancing particular globin genes in the various hematopoietic compartments: only HS2 appeared to be equally active at the embryonic, fetal, and adult stages. It was originally concluded that HS2 alone was able to provide copy number-dependent and position-independent transcription to linked genes (5, 15), but this view was challenged when it was shown that HS2 was able to protect from position-of-integration effects only in multiply integrated transgene copies (8).
The entire transcriptional stimulatory activity of LCR HS2 was originally mapped to a 375-bp HindIII-XbaI restriction fragment (46). This region contains a cluster of binding sites for ubiquitous as well as tissue-specific transcription factors. Several of these sites within the HS2 core element are highly conserved among different species (22). It was shown that one of these highly conserved elements is a 60-bp motif encompassing a tandem maf-responsive element (MARE) (37; also referred to as an NF-E2 binding site). This element was shown to harbor most of the enhancer activity of HS2, and a similar configuration of tandem MAREs is not found in other LCR HS sites (39).
Here we report the analysis of seven transgenic lines containing intact human β-globin yeast artificial chromosomes (YACs) bearing a deletion of the 375-bp core enhancer of HS2. The results show that deletion of HS2 results in a severe reduction of ɛ-globin gene expression in the embryonic yolk sac and that expression of the adult β-globin gene is significantly reduced in fetal liver and adult spleen. Moreover, in three of the lines bearing single-copy transgenes, expression of all of the genes is much more severely affected than in the context of multiple tandemly integrated copies. We also analyzed four transgenic lines carrying intact human β-globin YACs in which the 220-bp core of HS3 was substituted for the 375-bp HS2 core enhancer. These results show that HS3 is unable to functionally compensate for the loss of HS2 and demonstrate that both HS2 and HS3 provide unique contributions to LCR activity. Moreover, the substitution mutants are expressed in a far more consistent manner from line to line than are the deletion mutants, suggesting that although transcription is impaired, the substitution mutations protect the transgenes from position-of-integration effects.
These experiments extend our earlier observations showing that deletion of single core elements for HS3 or HS4 from intact, single-copy transgenic YACs severely diminishes LCR-mediated transcriptional activation (4, 29). Together, the data show that deletion of any single-core HS element (HS2, HS3, or HS4) abrogates the activity of the entire LCR. Deletion of any of these core elements also causes variable DNase I hypersensitivity within the LCR that is dependent on the site of integration into the mouse genome. We interpret these results as being most compatible with a model in which an LCR holocomplex provides a unique chromatin architecture that protects transgenes from position-of-integration effects. This architectural complex includes both the core elements and their flanking sequences, and it can be partially restored if the core element plus flanking sequences of any single LCR HS site are deleted, if one HS core element is used to replace another, or, indeed, even if multiple copies of a particular transgene have integrated in tandem into the mouse genome. Additionally, the core HS elements appear to be required to cooperatively form a subdomain, or active site, within the superstructure provided by the holocomplex, and it is this active site within the holocomplex that is required specifically for transcriptional stimulation.
MATERIALS AND METHODS
Generation of HS2 mutant human β-globin YACs by homologous recombination in yeast.
Generation of mutant human β-globin YACs by homologous recombination in yeast was carried out as described previously (4). A KpnI-BglII fragment containing the core and flanking sequences of HS2 (from +7764 to +9218; numbering within the locus as found at the Globin Gene Server Web Site [17a]). was isolated from λ DNA and ligated to the KpnI-BamHI sites of pGEM7. A PCR-fragment was generated from the pGEM7/HS2 plasmid corresponding to the HS2 5′-flanking region, incorporating unique restriction enzyme sites for EcoRI and XbaI (from EcoRI at +7771 to XbaI at +8490). A similar PCR fragment was generated corresponding to the 3′-flanking region of HS2, incorporating restriction sites for XhoI and BamHI (from XhoI at +8861 to BamHI at +9220). The PCR fragments were digested with XbaI (5′ flank) or XhoI (3′ flank), the ends were filled with Klenow to generate blunt ends, and the fragments were subsequently digested with EcoRI (5′) or BamHI (3′) and then ligated into EcoRI-BamHI-digested pRS306 (4). The targeting vector for replacing the 375-bp HS2 core enhancer with the 220-bp HS3 element was generated by ligating the two HS2-flanking sequences (EcoRI-XbaI and XhoI-BamHI fragments), together with the XbaI-XhoI HS3 core enhancer (4), into the EcoRI-BamHI sites of pRS306.
The vectors were linearized with AvaI and used to transform yeast cells containing the human β-globin YAC (A201F4.3 [46a]). Transformed yeast cells were plated onto agarose plates lacking uracil. DNA was prepared from cells growing on uracil-deficient medium and analyzed by Southern blotting for correct integration of the HS2 deletion or substitution target plasmid. Clones that had the plasmid integrated into the homologous site of the β-globin YAC were grown in uracil-containing medium and plated onto agarose plates containing 5′-fluoroorotic acid (FOA). Growth on FOA-containing plates indicates removal of the URA3 gene by homologous excision within the YAC, generating cells with either wild-type or the desired mutant β-globin locus structure. DNA was prepared from these cells and analyzed by Southern blotting for the absence of the core enhancer of HS2 or for the duplication of HS3. Yeast clones bearing the mutant β-globin YACs were embedded in agarose plugs for pulsed-field gel electrophoretic analyses or DNA isolation.
Generation of human β-globin YAC transgenic mice.
YAC DNA was isolated from pulsed-field gels as previously described (4) and injected into fertilized CD1 oocytes at 0.5 to 1 ng/μl. The offspring were screened by PCR for the presence of the left and right YAC vector arms. Animals containing both vector arms were bred to generate F1 mice.
Structural analyses of human β-globin YAC transgenic mice.
For analysis of the copy number of the transgene, we first used an end-fragment assay. DNA isolated from the tail of transgenic F1 founders was digested with PstI and electrophoresed in 1% agarose gels. After being blotted to nylon membranes, the DNA was hybridized to radiolabeled DNA probes corresponding to either the left (723-bp PstI-AlwNI fragment) or the right (639-bp PvuII-AvaI fragment) vector arm of the YAC (30). This assay distinguishes single-copy from multiple-copy transgenes and further allows the determination of the arrangement of multiple-copy transgenes (head to tail, tail to tail, head to head) by the appearance of characteristic PstI junction fragments (30).
To analyze the overall integrity of the transgenes, thymus cells isolated from each transgenic line were embedded in agarose, digested with proteinase K for 48 h, and digested with SfiI. After pulsed-field gel electrophoresis, the DNA was blotted to nylon membranes and probed with various fragments spanning the β-globin gene locus (4, 17). SfiI releases a 100-kbp fragment containing a segment of the β-globin YAC from a site between HS3 and HS4 to the right vector arm, into which an SfiI site was introduced by homologous recombination (46a).
RT-PCR analysis of globin gene expression in human β-globin YAC transgenic mice.
RNA was isolated from erythroid cells at various stages of development (embryonic yolk sac at 9.5 days postcoitum (d.p.c.) fetal liver at 14 d.p.c., and adult spleen) and analyzed by reverse transcription-PCR (RT-PCR) as described previously (4).
DNase I HS mapping.
DNase I HS mapping was carried out as described previously (12, 29). The probe used to detect the human β-globin LCR HS was a 914-bp HindIII-XbaI restriction fragment derived from the 3′-flanking sequence of HS4. The probe for analyzing the mouse βmajor-globin gene promoter HS site was a 498-bp PstI-SacI fragment from the 5′ region of the gene (28).
RESULTS
Generation of HS2-mutant human β-globin YACs.
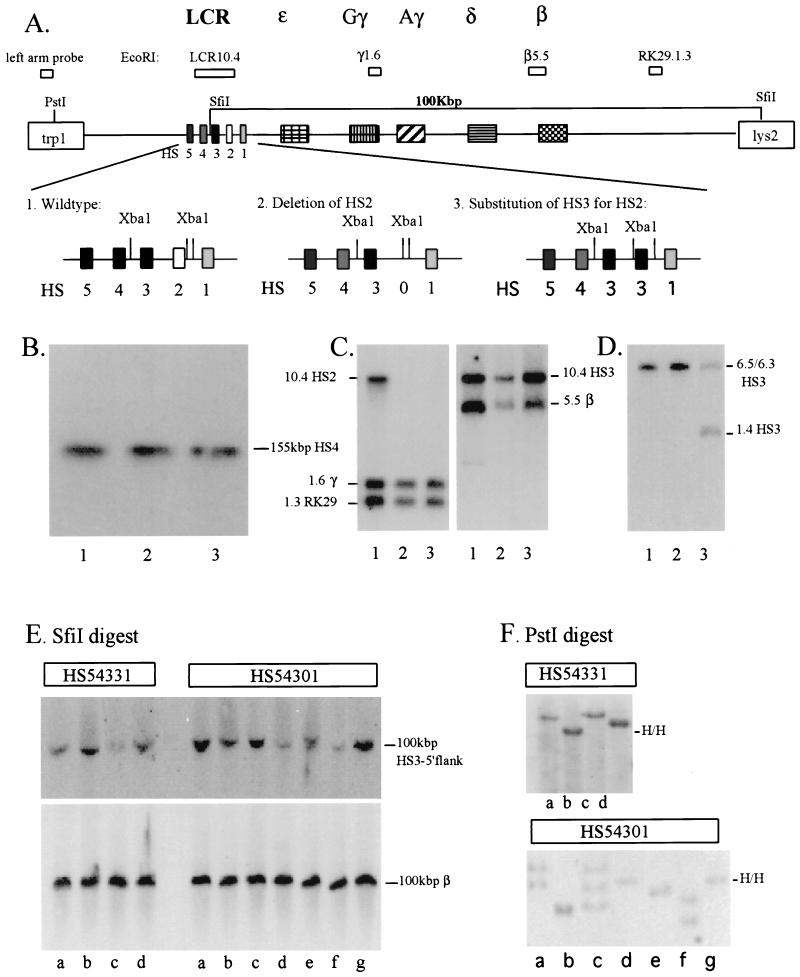
The enhancer activity of LCR HS2 has previously been mapped to a 375-bp DNA fragment, referred to as the HS2 core enhancer (46). We deleted this 375-bp fragment from a human β-globin YAC by using homologous recombination in yeast (Fig. 1A) (4, 30, 31). For this study, we also modified the original 155-kbp human β-globin YAC (called A201F4 [17]) by inserting the LYS2 gene, as well as a restriction enzyme site for SfiI, into the YAC right vector arm, thus disrupting the URA3 gene (46a). Disruption of URA3 facilitated subsequent manipulations since it can be used for both positive and negative selection of homologous recombinants in yeast. Additionally, the introduction of a new SfiI site enhanced our ability to analyze the structure of the locus. Digestion of intact transgenes with SfiI is predicted to yield a 100-kbp DNA fragment that contains sequences from 5′ to HS3 through to the YAC right vector arm.
FIG. 1.
Generation and structural analysis of HS2-mutant human β-globin YACs in yeast and transgenic mice. (A) Replacement of the 375-bp core enhancer of HS2 by the 220-bp core enhancer of HS3. Yeast cells were transformed with the targeting vectors (pRS306, linearized with AvaI, with or without HS3 embedded in HS2-flanking sequences). Yeast clones containing the vector integrated at the homologous site were grown in the presence of FOA, which selects for cells excising the URA3 marker together with either the mutant copy, thus restoring the wild-type configuration (1, HS54321), or together with the wild-type HS2 core element, generating a mutant LCR in which either HS2 is deleted (2, HS54301) or replaced by HS3 (3, HS54331), respectively. (B) Pulsed-field gel electrophoretic and Southern blot analysis of wild-type (lane 1) and mutant human β-globin YACs (HS54301 [lane 2] and HS54331 [lane 3]). Yeast chromosomes, embedded in agarose plugs, were loaded on a pulsed-field gel. After electrophoresis, the DNA was transferred to nylon membranes and hybridized to a radiolabeled 290-bp HS4 core enhancer fragment. (C and D) Southern blot analysis of yeast DNA carrying either wild-type (lane 1) or mutant YACs (HS54301 [lane 2] and HS54331 [lane 3]). Yeast DNA was isolated and digested with EcoRI (C) or XbaI (D). After electrophoresis the DNA was transferred to nylon membranes and hybridized with various radiolabeled probes derived from throughout the human β-globin gene locus (as indicated in panel A) (4). (E) Pulsed field gel electrophoretic and Southern blot analysis of the integrity of mutant human β-globin YAC transgenic mice (mutant lines HS54331a to HS54331d and HS54301a to HS54301g, as indicated). After electrophoresis, the DNA was transferred to nylon membranes and hybridized to radiolabeled probes derived from the 5′-flanking region of HS3 (top) or to a fragment corresponding to the second intron of the adult β-globin gene (bottom). (F) Analysis of the copy number of the mutant human β-globin YAC transgenes (same numbering as in panel E). Mouse genomic DNA, isolated from the tails of transgenic mice, was digested with PstI. After electrophoresis and transfer to nylon membranes, the DNA was hybridized to a 723-bp PstI-AwlI fragment derived from the left YAC vector arm (H/H indicates the presence of two left-arm fragments integrated into a head-to-head configuration in HS54331b, HS54301e, and HS54301g).
The strategy for deleting HS2 or replacing it with HS3 is outlined in Fig. 1A. The targeting vectors were linearized and used to transform yeast cells carrying the modified human β-globin YAC (called A201F4.3). After homologous integration and excision in yeast, the clones were analyzed for integrity by conventional and pulsed-field gel Southern blots. These studies show that no gross deletions or rearrangements occurred during the targeting or excision steps (Fig. 1B). Figure 1C demonstrates that all markers of the β-globin gene locus are present in the mutant YACs and that HS2 was successfully deleted or replaced with HS3 (Fig. 1C and D). The mutant YACs (called HS54301 and HS54331, according to our previous nomenclature [4]) were isolated and then microinjected to generate transgenic mice.
Structural analysis of transgenic mice carrying human β-globin YACs with HS2 deleted or replaced by HS3.
We generated and analyzed seven human β-globin YAC transgenic lines bearing the deletion of the HS2 core enhancer (called lines HS54301a to HS54301g) and four transgenic lines in which the HS3 core replaced HS2 (HS54331a to HS54331d). We performed four different experiments (three of which are discussed here) to analyze the structural integrity and copy number of the YAC transgenic lines.
First, we assayed transgenic-mouse genomic DNA for the presence of characteristic restriction fragments derived from the human β-globin gene locus (Fig. 1A) from 5′ of the LCR to far downstream of the β-globin gene (4). Animals in which specific globin locus fragments were missing or were not of the correct size were discarded from further analysis (results not shown).
Next, we analyzed the integrity of the integrated β-globin YACs by pulsed-field gel electrophoresis (Fig. 1E). Mouse genomic DNA from thymus cells was embedded in agarose, digested with SfiI, and separated on 1% pulsed-field agarose gels. The DNA transferred to nylon membranes was then hybridized to probes derived from the 5′-flanking region of HS3 (4) or from the human β-globin gene (indicated in Fig. 1A). Figure 1E shows that all of the lines analyzed here exhibit an SfiI restriction fragment of 100 kbp that hybridizes to both 5′ and 3′ markers, indicating that all of the β-globin YACs are integrated intact. Transgenic line HS54301e has an additional hybridizing fragment which is slightly larger than 100 kbp and which hybridizes only to the HS3 5′ probe, indicating that this line carries at least one intact and one broken YAC (see below).
Finally, we determined the copy number of the transgenes by end-fragment analyses for both left and right vector arms (30). As shown in Fig. 1F for the left-arm analysis, three of the HS54301 transgenic lines bear one copy of the YAC (lines b, d, and e), three of the lines harbor two copies (lines a, f, and g), and one of the lines carries three copies (line c). The end-fragment analysis for the right arm was consistent with the results of the left-arm analysis except for transgenic line HS54301e (above), which revealed a single copy bearing the right arm (results not shown). The four lines in which HS3 replaced HS2 (HS54331) bear one (lines a, c, and d) or two (line b) transgenic YAC copies.
Taken together with the data from the pulsed-field analysis, these experiments demonstrate that transgenic line HS54301e carries one intact copy and one broken copy of the human β-globin YAC arranged in a 5′-to-5′ joining configuration. Further analysis of this line with internal fragments showed that the second copy of the YAC extends from the left YAC vector arm to within the LCR just 3′ to HS3, indicating that this transgenic line is also single copy with respect to the globin genes (data not shown). Three of the four substitution lines (HS54331a, HS54331c, and HS54331d) harbor intact, single copies of the mutant human β-globin YAC. One line (HS54331b) has an additional left-vector-arm fragment, which is characteristic for a 5′-to-5′ configuration of the transgenes (Fig. 1F). Since this line displays only two hybridizing fragments in the right-arm analysis (results not shown), we conclude that this line carries two intact copies of the mutant YAC. The copy numbers of the YAC transgenes, as determined in the end-fragment assay, were confirmed in independent experiments by quantifying the intensities of internal fragments in the YAC in comparison to the intensity of a band from the mouse GATA-2 locus (reference 29 and data not shown).
Expression in human β-globin YAC transgenic mice with a 375-bp deletion of the core enhancer of HS2.
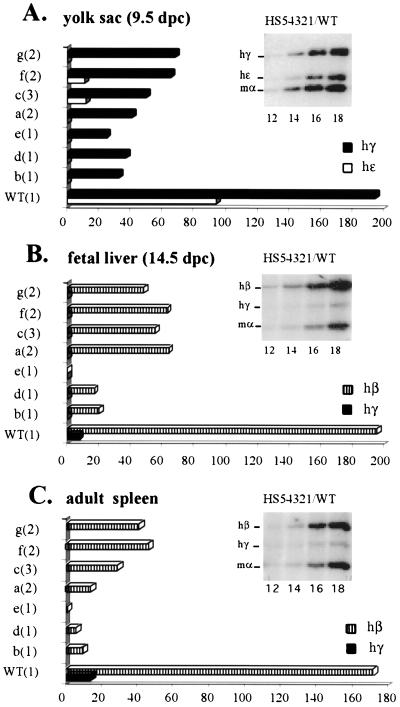
To analyze the expression of individual human globin genes in transgenic mice, we performed a multiplex RT-PCR assay which amplifies simultaneously the transcripts of the human ɛ-, γ- and β-globin genes as well as those of the endogenous mouse α-globin gene, which serves as the internal control (4). The PCR products, taken from the linear range of amplification as determined by cycle number titration (insets in Fig. 2), were analyzed by phosporimaging, and the expression levels of individual human globin genes were presented as expression per gene copy. Two samples derived from transgenic littermates were taken for each line at each developmental stage (9.5-d.p.c. yolk sac, 14.5-d.p.c. fetal liver, or adult spleen).
FIG. 2.
Analysis of globin gene expression in YAC transgenic mice bearing a deletion of the 375-bp core enhancer of HS2. RNA was extracted from yolk sac (at 9.5 d.p.c.), fetal liver (14.5 d.p.c.), or adult spleen (4 to 6 weeks) and subjected to RT-PCR analysis was performed with a mixture of globin primers (amplifying human ɛ-, γ-, and β-as well as mouse α-globin genes) and a radiolabeled nucleotide. After electrophoresis, samples (amplifying within the linear range) were analyzed and quantified by phosphorimaging, as described previously (4). The insert in each panel shows an autoradiograph depicting characteristic PCR products obtained from RNA extracted from the yolk sac (A), fetal liver (B), or anemic spleen (C) of transgenic embryos or mice carrying the wild-type β-globin YAC HS54321 (cycle numbers are indicated below the lanes). (A) Expression profile of human globin genes in the embryonic yolk sac taken at 9.5 d.p.c. (B) Expression profile of human globin genes in the fetal liver taken at 14.5 d.p.c. (C) Expression profile of human β-globin genes in the adult spleen taken at from 4- to 6-week-old anemic mice. The lines analyzed in each columns are indicated on the left of each panel with the transgene copy numbers shown in parentheses (WT, wild-type HS54321; a to g, HS54301 mutant lines). Levels of individual globin gene transcription are expressed as a percentage of mouse α-globin gene expression (which was set at 100%) per gene copy.
The results of the expression analysis for the HS2 core deletion mutants are summarized in Fig. 2. These data show that deletion of HS2 severely impairs the expression of the human ɛ-globin gene in the embryonic yolk sac, which was reduced by approximately 10-fold in multicopy lines (HS54301a, HS54301c, HS54301f, and HS54301g) or to undetectable levels in single-copy transgenes (HS54301b, HS54301d, and HS54301e). Expression of the γ-globin genes in the 9.5-d.p.c. embryonic yolk sacs was reduced by three- to sixfold. In the fetal-liver and adult-spleen stages, expression of the β-globin gene was diminished, again by 3- to 5-fold in multiple copy lines but by more than 10-fold in single copy lines. Expression of the γ-globin genes in the fetal liver or adult spleen was undetectable in all of the HS2 deletion mutant lines.
It is important to note that the effect of deleting the HS2 core element was always more dramatic in single-copy than in multicopy transgenic-animal lines (see Discussion). In these lines, expression of all the human globin genes is significantly reduced and comparable to what we previously found when analyzing single-copy β-globin YAC transgenic mice with deletion of either core element HS3 or HS4 (4). This result supports our previous conclusion that individual LCR HS sites must cooperate to generate the active LCR and identifies one more component that is crucial for this cooperativity.
The effect of deleting HS2 is less variable in multiple-copy transgenic lines than in the single-copy lines. We interpret this to mean that deletion of HS2 impairs the function of the LCR in two different ways. The variation in expression levels of the human globin genes probably results from the conclusion of Milot et al. (34) that deleting individual HS elements renders an enfeebled LCR unable to provide position-independent expression to integrated LCR mutant transgenes. However, in addition to this factor, these data strongly imply that deletion of the HS2 core directly impairs the ability of the mutant LCR to enhance the expression of individual globin genes. This contention is best illustrated by the observation that expression of the ɛ-globin gene in all seven deletion mutant lines never exceeds 10% of the expression level in the wild-type line. We conclude that this impairment of the enhancement effect is independent of the site of transgene integration effects and thus reveals the true potential of LCR activity in these mutants (see Discussion).
Expression analysis of mutant human β-globin YACs containing HS3 substituted for the core enhancer of HS2.
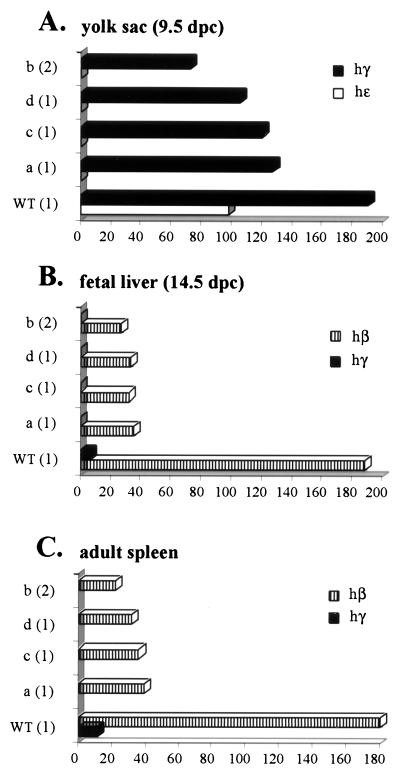
Expression of the human β-globin genes in the four mutant transgenic lines carrying a replacement of HS2 with HS3 (HS54331a to HS54331d) was analyzed in the same manner as for the HS2 deletion mutants. Figure 3 summarizes these results. Transcription of the human ɛ-globin gene in the embryonic yolk sac is undetectable in all four lines (HS54331a through HS54331d), showing that replacing HS2 with HS3 did not rescue the expression of the ɛ-globin gene. Expression of the γ-globin genes in the embryonic yolk sac was reduced 1.5- to 2-fold compared to expression of these genes in the wild-type line. The replacement of HS2 with HS3 therefore partially rescues the expression of the γ-globin genes in the yolk sac stage. These results together suggest that the LCR containing HS3 in place of HS2 has a preference for enhancing transcription of the γ-globin genes over the ɛ-globin gene in the embryonic yolk sac environment.
FIG. 3.
Analysis of YAC transgenic mice in which the 375-bp core enhancer of HS2 is replaced by the 220-bp core element of HS3. RNA was analyzed at the various stages of mouse hematopoiesis as described in the legend to Fig. 2. Shown are the expression profiles of the human globin genes in the embryonic yolk sac (9.5 d.p.c.) (A), fetal liver (14.5 d.p.c.) (B), and adult spleen (C). Indicated on the left of each panel is the specific line analyzed in each column (WT, transgene carrying wild-type YAC HS54331; a to d, HS54331 mutant lines). Numbers in parentheses are the copy numbers of the YACs transgenes. Expression is based on levels of mouse α-globin (set at 100%) calculated per integrated gene copy.
Transcription of the γ-globin genes in the fetal liver and adult spleen was virtually undetectable in the four substitution mutant lines, whereas expression of the adult β-globin gene was reduced four- to fivefold compared to that in the wild type. Once again, replacing HS2 with HS3 did not rescue transcription of the adult β-globin gene. In fact, it appears that the transcription of the β-globin gene in the fetal liver is less efficient in the substitution mutants than in the lines bearing a deletion of HS2. Since expression of the γ-globin genes was also undetectable at this stage in the substitution mutant lines, we conclude that an LCR in which HS2 is replaced by HS3 is only partially active in fetal liver erythroid cells. Taken together, the results show that HS3 is unable to functionally replace HS2, and we conclude that these two elements play different roles in the regulation of the human β-globin genes. It should be noted, however, that the expression of the human β-globin genes in the substitution mutant lines is far more consistent from line to line than in the deletion mutants, suggesting that the replacement of HS2 with HS3 may still allow integration position-independent expression of the locus.
Analysis of DNase I hypersensitivity in the LCR of HS2 mutant human β-globin YAC transgenic mice.
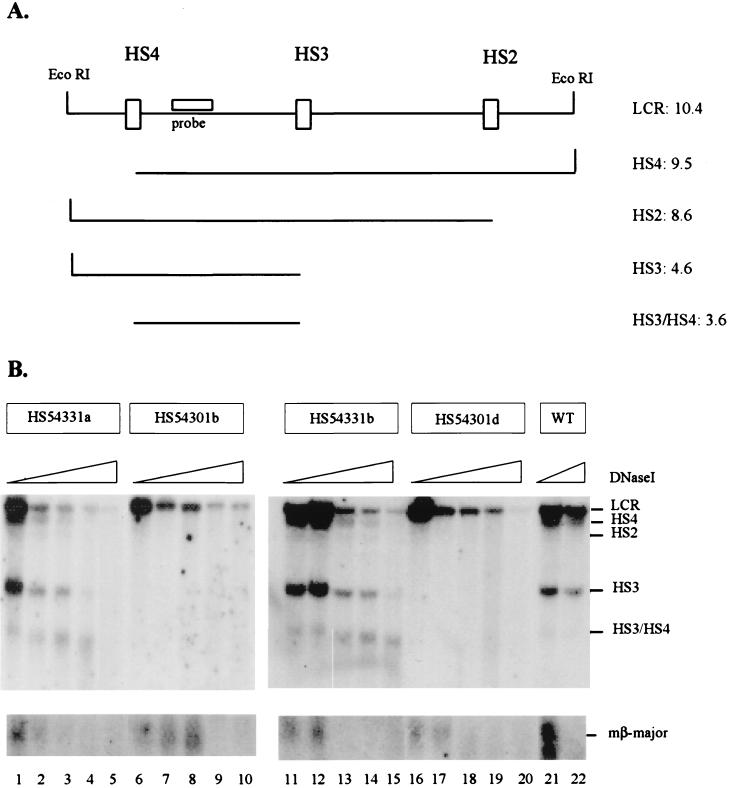
To examine whether the inability of HS3 to functionally replace HS2 is because it cannot reform HS2 hypersensitivity, we performed DNase I HS-mapping experiments on wild-type, HS2 deletion mutant, and HS2-HS3 substitution mutant β-globin YAC transgenic lines. Figure 4A is a diagrammatic representation of the LCR region analyzed in these experiments. The results show that in single-copy wild-type transgenes the LCR elements HS2, HS3, and HS4 are formed efficiently (Fig. 4B). The same is true for the two lines in which HS2 was replaced by HS3 (HS54331a and HS54331b). This result demonstrates that the HS3 core element can reform hypersensitivity within the HS2 region and that the other HS sites are formed with similar efficiency to those detected in the wild-type lines. We therefore conclude that the inability of HS3 to functionally replace HS2 is not due to its ineffectiveness in reconstituting an HS site within the HS2 region or to any negative effect it might exert on the formation of the other LCR HS sites.
FIG. 4.
DNase I HS mapping of the human β-globin LCR in HS2 mutant and wild-type human β-globin YAC transgenic mice. Nuclei were isolated from spleens of anemic transgenic mice bearing wild-type or HS2 mutant human β-globin YACs. The nuclei were incubated with increasing concentrations of DNase I, and the DNA was isolated and digested with EcoRI. After gel electrophoresis the DNA was hybridized to radioactively labeled probes corresponding to the 3′-flanking region of HS4 (as indicated in panel A) or to the 5′ promoter region of the mouse βmajor-globin gene. (A) Diagrammatic representation of the fragments generated by DNase I and EcoRI in the human β-globin LCR (indicated on the right are the sizes, in kilobase pairs of the various fragments). (B) HS formation in the wild-type (WT) or HS2 mutant human β-globin YAC transgenic mice. Two lines representing each mutant were analyzed for HS formation within the LCR, the substitution lines HS54331a and HS54331b (lanes 1 to 5 and 11 to 15, respectively) and two deletion mutants HS54301b and HS54301d (lanes 6 to 10 and 16 to 20, respectively). Lanes 21 and 22 show hypersensitivity in the single-copy transgene carrying the wild-type human β-globin YAC (HS54321a) (4). As a control for the DNase I digest, the nylon membranes were rehybridized to a probe detecting a characteristic HS site in the mouse βmajor-globin gene promoter (shown at the bottom of the picture).
We also analyzed two single-copy lines bearing β-globin YACs with a deletion of HS2 (HS54301b and HS54301d [Fig. 4B]). The results show that in both of these lines, all of the LCR HS are formed poorly compared with those detectable in the wild-type or substitution mutant lines. The control experiment shows that although the LCR HS form weakly, the HS in the mouse βmajor promoter in these two HS2 deletion mutant lines form with similar efficiencies to those detected in the wild-type or substitution mutant lines (Fig. 4B). This result supports our previous conclusion that deletion of individual core HS elements dramatically affects HS formation in single-copy transgenes (29).
DISCUSSION
In this study, we have analyzed the consequence of deleting HS2 or of replacing it by HS3 on the formation of DNase I HS and on expression of the human β-globin genes in mutant β-globin YAC transgenic mice. Analysis was restricted to 11 lines bearing at least one intact copy of the mutant YACs. The results show that the HS2 core element is required for both chromatin opening and optimal expression of all human β-globin genes at all developmental stages. HS2 replacement by HS3 restores the chromatin-opening activity but not the transcription-stimulatory activity of the LCR. These results support our previous conclusion that individual HS elements synergize to generate a fully potential LCR and that specific HS elements must play distinct roles in the process of globin gene switching (4).
HS2 is an integral component of the human β-globin LCR.
We have deleted the 375-bp HS2 core enhancer fragment from a human β-globin YAC by homologous recombination in yeast. We found, after analyzing globin gene expression in seven transgenic lines containing the mutated YAC, that deletion of the core enhancer of HS2 affects the expression of all the human globin genes. As is also true for deletion of the core enhancers of HS3 and HS4 (4), deletion of HS2 had a more severe effect on expression of all the human β-globin genes in single-copy YAC transgenic lines than in lines bearing multiple copies (Fig. 2). According to Milot et al. (34), who found that deletion of HS elements impairs the ability of the LCR to provide position-independent expression, we interpret these results to mean that when multiple copies of the mutated YACs integrate into the genome, this leads to more effective protection from any negative effect of surrounding chromatin at the site of transgene integration.
Although deletion of HS2 leads to a decrease in expression of all the human β-globin genes, this mutation most severely affected transcription of the embryonic ɛ-globin gene in the yolk sac (reduced to less than 10% of wild-type levels) and that of the adult β-globin gene (reduced to less than 30% of wild-type levels).
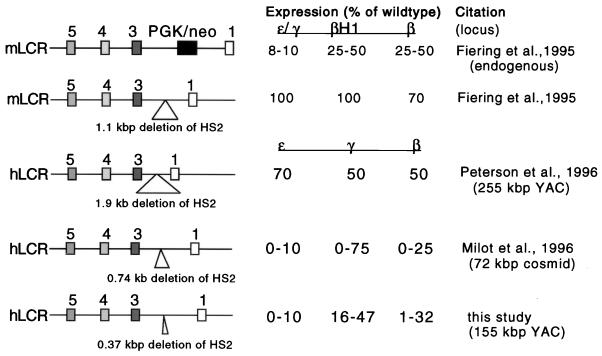
Several other studies have previously examined the effect of deleting HS2 from either the endogenous mouse locus or from YAC or cosmid human β-globin transgenes. As shown in Fig. 5, which summarizes the data in these reports, the results presented here are most consistent with those of Milot et al. (34). In summary, the combined data indicate that deletion of the HS2 core element affects the function of the LCR in two different ways. First, deletion of the HS2 core no longer allows the LCR to protect human β-globin gene expression from position of integration effects. This conjecture is supported by the observation that HS2 deletion drastically impairs LCR hypersensitivity in single-copy transgenes (Fig. 4). This effect is not HS2 specific, since it is also observed in other core HS element deletion mutants (4, 29, 34). This indicates that the LCR requires (at least) HS2, HS3, and HS4 to be able to counteract the influence of repressive activities located in the vicinity of transgene integration (e.g., heterochromatin or other strong repressive elements).
FIG. 5.
Comparison of results obtained from studies examining deletion of HS2 from the endogenous mouse or from the human β-globin gene loci. Expression levels of individual globin genes were calculated as percentages of expression levels of particular genes expressed from the wild-type loci (set at 100%).
Second, deletion of the HS2 core element also impairs the potential of the LCR to enhance the transcription of individual globin genes. We conclude that this impaired transcriptional activation function of the LCR is independent of the site of transgene integration effect, for two reasons. First, the dramatic reduction in expression of human ɛ- and β-globin genes is consistent in all seven lines analyzed here as well as in the four deletion mutant lines analyzed by Milot et al. (34). Second, a very similar effect on the expression of these genes was found in the four lines in which HS2 was replaced by HS3.
Other studies examining the effects of larger deletions around HS2 observed milder phenotypes than the data reported here and by Milot et al. (34). Fiering et al. (11) deleted a 1.1-kbp fragment encompassing HS2 from the endogenous mouse locus. Similarly, Peterson et al. (44) analyzed a human β-globin YAC transgenic line in which HS2 was deleted as a 1.94-kbp fragment, removing both the core element and substantial flanking sequence from the LCR. Both of these deletions resulted in only mild expression phenotypes.
Several explanations could account for the different HS2 deletion effects on expression of the globin genes. First, additional DNA sequences present in the endogenous mouse locus or in the larger, 248-kbp YAC could compensate for the loss of HS2 from a smaller, 155-kbp YAC (examined here) or from the 72-kbp cosmid examined by Milot et al. (34). We have detected such (apparent) compensation previously in the human β-globin locus, when we found that deletion of the Aγ-globin 3′ “enhancer” from the 150-kbp YAC had no consequences on gene expression (31). A second possibility is that sequences flanking the core elements harbor potentially repressive activities (25, 26). Finally, it is possible that the spacing between HS elements is important for the function of the LCR. Peterson et al. (44) deleted 1.94 kbp of HS2, thereby bringing HS1 significantly closer to HS3, which could potentially partially restore LCR function. The same explanation could account for the relatively mild phenotype observed after deleting the 1.1-kbp HS2 fragment from the endogenous mouse locus (11). Deleting only the core element, or replacing mouse HS2 with a 2-kbp selection cassette (Fig. 5), leaves a larger gap between HS3 and HS1, which might then cause a more severe phenotype.
Importantly, it should be noted that these results, obtained from the analysis of HS2 deletion mutants, represent the third example in which the removal of LCR core HS elements had a far more severe effect on expression of the globin genes than did larger deletions encompassing flanking sequences. Deletion of the 220-bp HS3 core enhancer dramatically affects the expression of the human globin genes (4, 38), whereas deletion of a larger fragment (2.3 kbp) results in a far milder phenotype (44). Similarly, deletion of the 290-bp core element of HS4 also results in severely diminished levels of globin gene expression (4) while deletion of a larger HS4-containing fragment (875 bp) (34) does not significantly alter globin gene expression. Similar mechanisms to those discussed above could account for these differences.
HS3 is unable to functionally substitute for HS2.
We have previously shown that HS3 is able to functionally replace HS4, indicating that HS3 carries all of the activities attributable to HS4 (4). The reverse, however, was not true: HS4 could not replace HS3, demonstrating that HS3 harbors unique activities not present in HS4. Here we analyzed four YAC transgenic lines in which the core element of HS3 was substituted for the core element of HS2 and showed that HS3 was not able to functionally replace HS2. This result indicates that these two LCR elements are not equivalent but harbor specific functions that are both critical and unique for human β-globin gene expression.
Two observations may be important. First, it appears that globin gene expression in the substitution lines is more quantitatively consistent than in lines bearing deletions of the same HS sites. This is especially obvious when one compares expression among single-copy lines (compare Fig. 2, HS54301 lines b, d, and e, with Fig. 3, HS54331 lines a, c, and d). This observation suggests that the replacement of HS2 by HS3 restores integration site position independence and may be due to the contention that HS3 harbors intrinsic chromatin-opening activity (9). In support of this possibility, our analysis of HS site formation in single-copy substitution and deletion mutant lines showed that all LCR HS sites are formed in single-copy substitution mutant lines but not in single-copy lines bearing a deletion of HS2 (Fig. 4). This result, combined with our previous data (29), indicates that several combinations of β-locus HS elements, other than the resident ones, can mediate the chromatin opening activity of the LCR.
Expression of the embryonic ɛ-globin gene and that of the adult β-globin gene appear to be more severely affected in the substitution than in the deletion mutant lines, whereas expression of the γ-globin genes is less affected in the substitution mutant lines. These results suggest that an LCR in which HS3 replaces HS2 has a preference for enhancing the expression of the γ-globin genes and does not seem to be functional in mediating efficient transcription of the ɛ- or β-globin genes. In this context, it should be noted that we observed a similar phenotype in the analysis of YAC transgenic mice in which HS3 replaced HS4. In these lines, both ɛ- and β-globin gene transcription was severely reduced whereas transcription of the γ-globin genes was less affected. These results, together with the data from the four substitution lines presented here, suggest that efficient transcription of the ɛ- and β-globin genes, but not expression of the γ-globin genes, requires activities present in both HS2 and HS3. In support of this interpretation, it was found that HS2 and HS3 synergize in activating the embryonic ɛ-globin gene in cell transfection studies (26). The synergy between these two elements is also required for regulation of the globin genes over long distances, which appears to be particularly important for activating the adult β-globin gene (3).
Taken together, the data suggest that HS2 contributes to two discrete activities attributable to the human β-globin LCR. First, HS2 is required as part of the integral holocomplex, to open the chromatin in the β-globin gene locus (Fig. 4). Second, HS2 is separately required to stimulate high-level ɛ- and β-globin gene transcription during erythroid-cell development. The chromatin-opening activity of the LCR appears to be almost fully restored when HS2 is replaced by HS3, indicating that HS2 does not uniquely specify this property. Nonetheless, HS3 is unable to substitute for HS2 in stimulating high levels of globin gene transcription, demonstrating that HS2 contributes uniquely to the LCR transcription-stimulatory function.
The LCR is organized as a holocomplex, and the core enhancer elements generate a subdomain required for stimulating globin gene transcription.
The results presented in this study, taken together with the data from several earlier reports, are consistent with a model in which individual HS elements interact with one another to form a cooperative, higher-order chromatin architecture, termed the LCR holocomplex. This complex mediates chromatin opening over the whole β-globin gene locus and enhances the transcription of individual globin genes during erythroid-cell development. The concept that the LCR might function as a single unit was first invoked by Grosveld and colleagues (19, 21) and the data presented in our previous study (4) and here provide direct experimental support for this hypothesis. Milot et al. (34) also confirmed many of these ideas by direct mutagenesis of the LCR. Given the numerous studies on the transcriptional effects of specific mutations within the LCR, there are now ample data to suggest how the individual HS elements might act to form a unique chromatin architectural unit. Furthermore, other data suggest how formation of this unique structure could be mediated by proteins binding to the core HS element regions as well as to their flanking sequences (24, 33, 40, 42, 43).
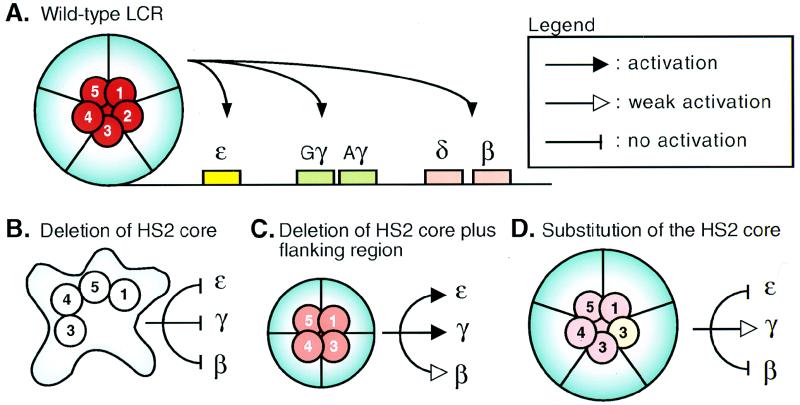
To explain how different deletions of the individual HS elements differentially affect the two aspects of LCR activity (chromatin opening and enhancement), we propose that the individual HS core elements aggregate into a precise substructure within the overall architecture of the holocomplex to create a single, cooperative “active site.” Thus, the holocomplex structure provides a necessary but not sufficient condition for stimulating globin gene transcription. Viewed from the perspective of this more detailed hypothesis, a critical feature of the holocomplex is that it provides the structural framework for formation of this subdomain or single active site. Interpreted in light of this model, the present data indicate that the holocomplex is required to provide position-independent chromatin opening and that such a complex can be formed with either the resident or (some) substituted core HS elements. A subdomain within the holocomplex, adopting different conformations at different erythroid-cell developmental stages, would then be responsible for enhancing competitive stimulatory interactions with the proteins bound at individual globin gene promoters by DNA looping (6).
The model proposed here is consistent with many previous observations and thereby provides coherence to seemingly disparate experimental results. First, deletion of core elements from the wild-type LCR structure (Fig. 6A), containing little if any flanking sequence, has a much more severe consequence (see above) (4, 34, 38) (Fig. 6B) on transcription-stimulatory activity than does deletion of any of the core-plus-flanking sequences (11, 44) (Fig. 6C). The reason for this may be that a reasonable semblance of the LCR holocomplex architecture can re-form if the entire HS plus flanking sequences are removed (Fig. 6C). Finally, replacement of one HS core element with another could lead to partial restoration of the active-site subdomain, allowing transcription of some genes at near normal abundance from within the essentially wild-type holocomplex (see above) (4) (Fig. 6D). As is true of any useful model, this hypothesis is subject to direct experimental tests, and we are pursuing strategies to determine the veracity of the holocomplex hypothesis.
FIG. 6.
HS core elements form a synergistic active subdomain within the LCR holocomplex. The figure depicts a cartoon of a model consistent with our data. (A) The LCR in the wild-type configuration. The core elements, together with the flanking regions, generate the holocomplex (light blue), with the core elements folded together to establish the specific configuration required for the active site (red). (B) Deletion of the HS2 core enhancer results in collapse of the LCR holocomplex and active site and renders the LCR unable to confer high-level or transgene integration position-independent transcription to the globin genes. (C) Deletion of the HS2 core element together with its flanking sequences allows the formation of an imperfect, alternative structure for the holocomplex, which retains transgene integration site-independent transcription but is impaired in its activity provided by the subdomain (light red). (D) Replacement of the HS2 by the HS3 core element allows the formation of an essentially wild-type holocomplex (thus retaining position-independent transcription of the integrated transgenes), but the active site is again unable to stimulate all the different globin genes at each developmental stage because it (light red) is not perfectly re-formed at each stage.
ACKNOWLEDGMENTS
We acknowledge the outstanding technical assistance of Jie Fan, Wei Min Song, and Yoko Tanimoto. We also thank Kazuhiko Igarashi and Jordan Shavit for encouraging discussions.
This work was supported by a senior research fellowship from the Chicago Chapter of the American Heart Association (to J.B.) and by the NIH (grants HL 24415 to J.D.E. and DK 52356 to J.B.).
REFERENCES
- 1.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human β-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 2.Bonifer C, Vidal M, Grosveld F, Sippel A E. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990;9:2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnick E, Tze L. Synergism between hypersensitive sites confers long-range gene activation by the β-globin locus control region. Proc Natl Acad Sci USA. 1997;94:4566–4571. doi: 10.1073/pnas.94.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bungert J, Dave U, Lieuw K E, Lim K-C, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 5.Caterina J, Ryan T M, Pawlik R D, Brinster R L, Behringer R R, Townes T M. Human β-globin locus control region: analysis of the 5′ DNase I hypersensitive site HS 2 in transgenic mice. Proc Natl Acad Sci USA. 1991;88:1626–1630. doi: 10.1073/pnas.88.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi O-R, Engel J D. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 7.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis J, Talbot D, Dillon N, Grosveld F. Synthetic human β-globin 5′ HS2 constructs function as locus control regions only in multicopy transgene concatemers. EMBO J. 1993;12:127–134. doi: 10.1002/j.1460-2075.1993.tb05638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J, Tan U K, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 10.Engel J D. Developmental regulation of human β-globin gene transcription: a switch of loyalties? Trends Genet. 1993;9:304–309. doi: 10.1016/0168-9525(93)90248-g. [DOI] [PubMed] [Google Scholar]
- 11.Fiering S, Epner E E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-like globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 12.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 13.Forrester W C, Takegawa S, Papayannopoulou T, Stammatoyannopoulos G, Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser P, Hurst J, Collis P, Grosveld F. DNase1 hypersensitive sites 1, 2 and 3 of the human β-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser P, Pruzina S, Grosveld F. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression to the globin genes. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 17.Gaensler K M L, Burmeister M, Brownstein B H, Taillon-Miller P, Myers R M. Physical mapping of yeast artificial chromosomes containing sequences from the human β-globin gene region. Genomics. 1991;10:976–984. doi: 10.1016/0888-7543(91)90188-k. [DOI] [PubMed] [Google Scholar]
- 17a.Globin Gene Server Website. 15 May 1994, posting date. [Online.] Pennsylvania State University. http://globin.cse.psu.edu. [4 December 1998, last date accessed.]
- 18.Greaves D R, Wilson F D, Lang G, Kioussis D. Human CD2 3′-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 19.Grosveld F, Antoniou M, Berry M, deBoer E, Dillon N, Ellis J, Fraser P, Hanscombe O, Hurst J, Imam A, Lindenbaum M, Philipsen S, Pruzina S, Strouboulis S, Raguz-Bolognesi S, Talbot D. The regulation of human globin gene switching. Philos Trans R Soc Lond Ser B. 1993;339:183–191. doi: 10.1098/rstb.1993.0015. [DOI] [PubMed] [Google Scholar]
- 20.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 21.Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Importance of globin gene order for correct developmental expression. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- 22.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 23.Higgs D R, Wood W G, Jarman A P, Sharpe J, Lida J, Pretorius I-M, Ayyub H. A major positive regulatory region located far upstream of the human α-globin gene locus. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M. Multivalent DNA binding complex generated by small maf and bach1 as a possible biochemical basis for β-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 25.Jackson J D, Miller W, Hardison R C. Sequences within and flanking hypersensitive sites 3 and 2 of the β-globin locus control region required for synergistic versus additive interaction with the ɛ-globin gene promoter. Nucleic Acids Res. 1996;24:4327–4335. doi: 10.1093/nar/24.21.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson J D, Petrykowska H, Philipsen S, Miller W, Hardison R. Role of DNA sequences outside the cores of DNase hypersensitive sites (HSs) in functions of the β-globin locus control region: domain opening and synergism between HS2 and HS3. J Biol Chem. 1996;271:11871–11878. doi: 10.1074/jbc.271.20.11871. [DOI] [PubMed] [Google Scholar]
- 27.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 28.Konkel D A, Tilghman S M, Leder P. The sequence of the chromosomal mouse beta-globin major gene: homologies in capping, splicing and poly(A) sites. Cell. 1978;15:1125–1132. doi: 10.1016/0092-8674(78)90040-5. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Lim K C, Engel J D, Bungert J. Individual LCR hypersensitive sites cooperate to generate an open chromatin domain spanning the human β-globin locus. Genes Cells. 1998;3:415–430. doi: 10.1046/j.1365-2443.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Bungert J, Engel J D. Mutation of gene-proximal regulatory elements disrupts human ɛ-, γ- and β-globin expression in YAC transgenic mice. Proc Natl Acad Sci USA. 1997;94:169–174. doi: 10.1073/pnas.94.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, Tanimoto K, Bungert J, Engel J D. The Aγ-globin 3′ element provides no unique function(s) for β-globin locus gene regulation. Proc Natl Acad Sci USA. 1998;95:9944–9949. doi: 10.1073/pnas.95.17.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin D I K, Fiering S, Groudine M. Regulation of β-globin gene expression: straightening out the locus. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 33.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milot E, Strouboulis J, Trimborn T, Wijgerde M, deBoer E, An Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 35.Montoliu L, Umland T, Schütz G. A locus control region at −12 kb of the tyrosinase gene. EMBO J. 1996;14:6026–6034. [PMC free article] [PubMed] [Google Scholar]
- 36.Moon A M, Ley T J. Functional properties of the beta-globin locus control region in K562 erythroleukemia cells. Blood. 1991;77:2272–2284. [PubMed] [Google Scholar]
- 37.Motohashi H, Shavit J A, Igarashi K, Yamamoto M, Engel J D. The world according to maf. Nucleic Acids Res. 1997;25:2953–2960. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navas P A, Peterson K R, Li Q, Skarpidi E, Rohde A, Shaw S E, Clegg C H, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol. 1998;17:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ney P A, Sorrentino B P, McDonagh K T, Nienhuis A W. Tandem AP-1-binding sites within the human β-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev. 1990;4:993–1006. doi: 10.1101/gad.4.6.993. [DOI] [PubMed] [Google Scholar]
- 40.Orkin S. Regulation of globin gene expression in erythroid cells. Eur J Biochem. 1995;231:271–281. doi: 10.1111/j.1432-1033.1995.tb20697.x. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz B D, Cado D, Chen V, Diaz P W, Winoto A. Adjacent DNA elements dominantly restrict ubiquitous activity of a novel chromatin-opening region to specific tissues. EMBO J. 1997;16:5037–5045. doi: 10.1093/emboj/16.16.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc finger protein GATA1. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with mafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson K R, Clegg C H, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatoyannopoulos G, Neinhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Neinhuis A W, Majerus P, Varmus H, editors. The molecular basis of blood diseases. 2nd ed. Philadelphia, Pa: The W. B. Saunders Co.; 1994. pp. 107–155. [Google Scholar]
- 46.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 1990;9:2169–2178. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Tanimoto, K., Q. Liu, J. Bungert, and J. D. Engel. Submitted for publication.
- 47.Tuan D, Solomon W, Li Q, London I M. The ‘β-like globin’ gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuan D Y, Solomon W B, London I M, Lee D P. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “beta-like globin” genes. Proc Natl Acad Sci USA. 1989;86:2554–8. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]