Abstract
Gain-of-function polymorphisms in the transcription factor IFN regulatory factor 5 (IRF5) are associated with an increased risk of developing systemic lupus erythematosus. However, the IRF5-expressing cell type(s) responsible for lupus pathogenesis in vivo is not known. We now show that monoallelic IRF5 deficiency in B cells markedly reduced disease in a murine lupus model. In contrast, similar reduction of IRF5 expression in macrophages, monocytes, and neutrophils did not reduce disease severity. B cell receptor and TLR7 signaling synergized to promote IRF5 phosphorylation and increase IRF5 protein expression, with these processes being independently regulated. This synergy increased B cell–intrinsic IL-6 and TNF-α production, both key requirements for germinal center (GC) responses, with IL-6 and TNF-α production in vitro and in vivo being substantially lower with loss of 1 allele of IRF5. Mechanistically, TLR7-dependent IRF5 nuclear translocation was reduced in B cells from IRF5-heterozygous mice. In addition, we show in multiple lupus models that IRF5 expression was dynamically regulated in vivo with increased expression in GC B cells compared with non-GC B cells and with further sequential increases during progression to plasmablasts and long-lived plasma cells. Overall, a critical threshold level of IRF5 in B cells was required to promote disease in murine lupus.
Keywords: Autoimmunity
Keywords: Autoimmune diseases
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by autoantibody production, inflammation, and tissue damage in multiple organs resulting from an overactivation of the immune system through various mechanisms (1, 2). Disease development is due to a combination of genetic and environmental factors, among which polymorphisms in IFN regulatory factor 5 (IRF5) have been strongly associated with an increased risk of developing SLE (3, 4). Although the exact mechanism of how these polymorphisms in IRF5 lead to an increased risk of developing SLE is incompletely understood, it is thought that they lead to increased levels of IRF5 protein expression and/or functional change (3, 4). Notably, global homozygous or heterozygous deficiency of IRF5 has conferred protection in many mouse models of lupus (5–10). Thus, there is both human and mouse genetic evidence that suggests that IRF5 expression levels contribute to disease pathogenesis. However, the previous studies of global heterozygous and homozygous gene deficiencies (5–10) do not address how cell type–specific expression of IRF5 impacts disease and how IRF5 expression and activation are modulated in the pathogenic cell type(s) during disease development.
IRF5 is a transcription factor that plays an important role in regulating immune responses downstream of TLRs, specifically, TLR-3, TLR-4, TLR-7, and TLR-9, and other innate immune receptors, including nucleotide-binding oligomerization domain 2, retinoic acid inducible gene I, and dectin-1 (11–15). Of these, TLR7 has been most closely associated with disease pathogenesis in mouse lupus models (16). Similar to other IRF family members, IRF5 resides predominantly in the cytosol in latent form as a monomer in unstimulated cells. Upon activation, specific serine residues in the C-terminal region are phosphorylated, which causes IRF5 to homodimerize or heterodimerize and undergo nuclear translocation. The IRF5 dimer then assembles with transcriptional coactivators such as CREB-binding protein or p300 in the nucleus, which enables promoter binding and the induction of gene transcription (17, 18).
Although IRF5 is also expressed outside the immune system, it is constitutively expressed in B cells, myeloid DCs (mDCs), plasmacytoid DCs (pDCs), and monocytes/macrophages (19, 20) and plays a key role in the induction of a number of proinflammatory cytokines and chemokines (11, 21). The extent of involvement of IRF5 in cytokine production is both cell type dependent and stimulus dependent (3, 20). Beyond cytokine production, IRF5 has also been linked to promoting M1 macrophage polarization (22), IgG2a/c class-switching (6), and upregulation of costimulatory molecules and Blimp-1 in B cells (23, 24). Because B cells, mDCs, pDCs, and monocytes/macrophages have all been implicated in lupus pathogenesis, it is plausible that IRF5 in any 1 or more of these cell types may be required for disease pathogenesis.
B lineage cells are thought to be central to the pathogenesis of lupus, although the optimal therapeutic intervention targeting this lineage is not yet established (25). Clinical trials of B cell depletion in lupus have shown variable efficacy, leading to a shift in focus to developing more effective B cell–depleting strategies and also targeting long-lived plasma cell populations (26). In mouse models of lupus, B cell deficiency is protective (16). A role for B cell–intrinsic TLR signaling was demonstrated in bone marrow chimera studies, showing that B cell–specific expression of the TLR adaptor protein MyD88 is required for disease development (27). Given that B cells and their expression of key molecules in the TLR signaling pathways are important in disease development, we hypothesized that the function of IRF5 in B cells might be a central component of pathogenesis. Furthermore, the fact that heterozygous global deletion of IRF5 markedly reduces disease in lupus models suggested that loss of 1 allele of IRF5 in B cells might confer protection (5, 10, 28, 29).
To examine the role of B cell–specific IRF5, we conditionally deleted 1 allele of IRF5 in the FcγRIIB−/−Yaa mouse lupus model, the model we previously used to demonstrate the effect of global homozygous and heterozygous IRF5 deletion (5). We found that heterozygous deletion of IRF5 in B cells led to a marked reduction in disease severity. In contrast, a similar reduction of IRF5 expression in macrophages, monocytes, and neutrophils did not impact disease. Thus, there was a critical threshold level of IRF5 expression in B cells that was necessary to induce disease, and this suggests that future therapeutic approaches targeting IRF5 in SLE will need to effectively reduce IRF5 expression or activity in B cells.
Results
IRF5 expression in bone marrow–derived cells is required for the development of autoimmune disease.
IRF5 is expressed in both immune cells and non–bone marrow–derived cells (19), and previous reports have suggested a role for IRF5 in non–bone marrow–derived cells in lupus pathogenesis (30). To evaluate the relative contribution of IRF5 expression in bone marrow–derived cells to disease pathogenesis in the FcγRIIB−/−Yaa model, we performed bone marrow chimera studies. Bone marrow from FcγRIIB−/− Yaa IRF5+/+ or FcγRIIB−/− Yaa IRF5–/– mice was transferred into irradiated C57BL/6 (B6) recipients. Disease parameters were evaluated 28 weeks after transfer. Mice that received the FcγRIIB−/− Yaa IRF5+/+ bone marrow developed splenomegaly, antinuclear autoantibodies (ANAs), and kidney disease, whereas mice that received the FcγRIIB−/− Yaa IRF5–/– bone marrow did not. These studies demonstrate that IRF5 expression in bone marrow–derived cells was sufficient to drive the lupus-like disease observed in FcγRIIB−/−Yaa mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.141395DS1).
Generation of FcγRIIB−/−Yaa mice with a B cell–specific deletion of IRF5.
To study the role of IRF5 expression specifically in B cells, we first generated an IRF5-floxed allele on a C57BL/6 genetic background by introducing loxP sites flanking exons 3–6 including a neo cassette flanked by Frt sites to the 5′ end of exon 3 (Supplemental Figure 2). Mice were then bred to the Flpe deleter mice to remove the neo cassette. The resulting IRF5fl/fl mice were intercrossed with Mb1cre/wt and FcγRIIB−/−Yaa mice, generating FcγRIIB−/−Yaa mice with a heterozygous deletion of IRF5 in B cells (Mb1cre/wt IRF5F/+ mice; termed IRF5ΔB) and littermate control FcγRIIB−/−Yaa mice with no deletion of IRF5 in B cells (Mb1wt/wt IRF5F/+ mice; termed IRF5F/+). We chose to focus on the heterozygous deletion of IRF5 in B cells given our previous finding that global heterozygous IRF5 deletion protected mice from disease to a similar extent as homozygous deletion, a finding that has been confirmed in other mouse lupus models (5, 10, 28, 29).
In Mb1cre-expressing animals, the cre transgene is driven by the CD79α promoter and results in the deletion of the floxed allele in 95% of B cells (31). In some cases, the Mb1cre transgene has been reported to induce ectopic, germ line deletion of floxed genes (32). Therefore, it was important to ensure that the deletion mediated by Mb1cre in our system was restricted to the B cell lineage.
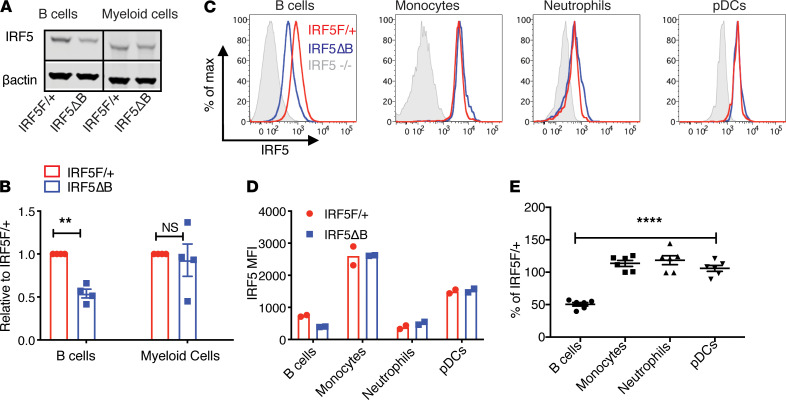
To evaluate IRF5 deletion, we first used flow cytometry to sort mature B cells (B220+CD93–) and myeloid cells (B220–CD3–CD11b+Ly6G–) from IRF5F/+ mice expressing or not expressing Mb1cre and performed Western blot on the B cell and myeloid cell populations. Mb1cre-mediated deletion of IRF5F/+ mice resulted in an approximately 50% reduction in IRF5 expression in B cells (Figure 1, A and B). This is consistent with the reduction we observed in the B cells of IRF5 germ line heterozygous mice (Supplemental Figure 3, A and B). Importantly there was no difference in IRF5 protein expression in myeloid cells from the IRF5ΔB and IRF5F/+ mice. To corroborate the Western blot studies and extend the analysis to specific IRF5-expressing immune cell types not captured in the myeloid gating strategy, we measured IRF5 expression by intracellular flow cytometry (Figure 1, C–E). This confirmed that IRF5 reduction in IRF5F/+ FcγRIIB−/−Yaa mice expressing Mb1cre was restricted to B cells as the MFI of IRF5 was reduced by approximately 50% in B cells but was not reduced in monocytes, neutrophils, or pDCs (Figure 1, C–E). The gating strategy used to define each immune cell population is shown in Supplemental Figure 3C. Figure 1D represents MFI values from 2 separate mice of each genotype from 1 experiment. Figure 1E is a compilation of MFI values from 6 IRF5ΔB mice normalized to the IRF5F/+ littermate controls from the same experiment. The intracellular staining for IRF5 was specific, because cells from IRF5 germ line knockout mice stained with the anti-IRF5 antibody had a similar staining profile as the isotype control antibody (Supplemental Figure 3D). Taken together, these data indicate that we successfully generated FcγRIIB−/−Yaa mice with B cell–specific deletion of 1 allele of IRF5.
Figure 1. B cell–specific reduction of IRF5 expression in IRF5ΔB mice.
All analyses were done in 8- to 10-week-old FcγRIIB−/−Yaa mice. (A) Representative Western blot of IRF5 protein expression in sorted splenic B cells (CD19+) and myeloid cells (CD11b+Ly6G–) from IRF5F/+ and IRF5ΔB mice. All lanes were run on the same gel but were noncontiguous. (B) Expression of IRF5 in B cells and myeloid cells from IRF5ΔB mice normalized to IRF5F/+ (n = 4). Data were analyzed using 2-tailed, unpaired Welch’s t test; **P < 0.01. (C) Representative flow cytometry plots of intracellular IRF5 expression in CD19+ B cells, CD11b+Ly6C+ monocytes, CD11b+Ly6G+ neutrophils, and CD11b–PDCA1+Ly6C+ pDCs from IRF5F/+, IRF5ΔB, and IRF5–/– global knockout mice. (D) MFI values of IRF5 in B cells, monocytes, neutrophils, and pDCs from IRF5F/+ and IRF5ΔB mice (representative experiment of 3 individual experiments, n = 2 for each genotype). (E) IRF5 expression in B cells, monocytes, neutrophils, and pDCs from IRF5ΔB mice normalized to the IRF5F/+ littermate control in each experiment (n = 6). Data are shown as mean ± SEM and were analyzed using 1-way ANOVA with Tukey’s post hoc test; ****P < 0.0001. IRF5, IFN regulatory factor 5; pDCs, plasmacytoid DCs.
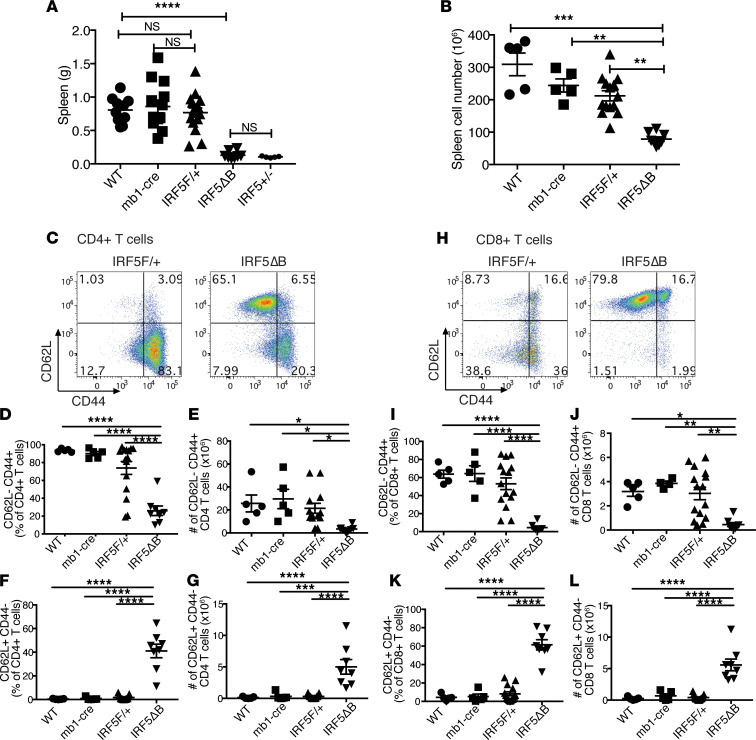
Monoallelic deletion of IRF5 in B cells reduces splenomegaly and T cell activation in FcγRIIB−/−Yaa mice.
Splenomegaly is seen in 10%–45% of patients with SLE and in multiple mouse models of lupus-like disease (33). Therefore, we evaluated the effect of heterozygous deletion of IRF5 in B cells on splenomegaly and found that it was markedly reduced and to the same extent as that seen in global IRF5-heterozygous FcγRIIB−/−Yaa mice (Figure 2A). No reduction in splenomegaly was seen in FcγRIIB−/−Yaa mice expressing Mb1cre alone or the IRF5-floxed allele alone. The reduction in spleen weight also correlated with a reduction in overall spleen cell number (Figure 2B). There was no significant difference in the number of CD3+ T cells and CD19+ B cells between the genotypes, indicating that the splenomegaly in this model was due to the expansion of a non–B cell, non–T cell population (Supplemental Figure 4).
Figure 2. Splenomegaly and T cell activation are reduced in IRF5ΔB mice.
All analyses were done in 5-month-old FcγRIIB−/−Yaa mice. (A) Spleen weights from WT (n = 10), mb1cre (n = 13), IRF5F/+ (n = 16), IRF5ΔB (n = 8), and IRF5+/– (global heterozygous deletion, n = 5) mice. (B) Splenic cell counts from WT (n = 5), mb1cre (n = 5), IRF5F/+ (n = 14), and IRF5ΔB (n = 8) mice. (C) Representative flow cytometry plots of CD4+CD62L–CD44+ (effector/memory) and CD62L+CD44– (naive) T cells from spleen of IRF5F/+ and IRF5ΔB mice. (D and E) Percentage and number of CD62L–CD44+ CD4+ T cells from WT (n = 5), mb1cre (n = 5), IRF5F/+ (n = 15), and IRF5ΔB (n = 8) mice. (F and G) Percentage and number of CD62L+CD44–CD4+ T cells. (H) Representative flow cytometry plots of CD8+CD62L–CD44+ (effector/memory) and CD62L+CD44– (naive) T cells from spleen of IRF5F/+ and IRF5ΔB mice. (I and J) Percentage and number of CD62L–CD44+CD8+ T cells from WT (n = 5), mb1cre (n = 5), IRF5F/+ (n = 15), and IRF5ΔB (n = 8) mice. (K and L) Percentage and number of CD62L+CD44–CD8+ T cells. Data are shown as mean ± SEM and were analyzed using 1-way ANOVA with Tukey’s post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. IRF5, IFN regulatory factor 5.
We next evaluated the effect of heterozygous deletion of IRF5 in B cells on T cell activation using flow cytometry, to assess T cell activation based on CD44 and CD62L expression. In both the CD4+ (Figure 2, C–E) and CD8+ (Figure 2, H–J) T cell populations, we found a dramatic reduction in the percentage and number of effector/memory (CD62L– CD44+) T cells in the IRF5ΔB mice compared with the FcγRIIB−/−Yaa mice and the littermate FcγRIIB−/−Yaa mice expressing Mb1cre alone or the IRF5-floxed allele alone. Notably, as with splenomegaly, no impact on T cell activation was seen in FcγRIIB−/−Yaa mice expressing Mb1cre alone or the IRF5-floxed allele alone compared with FcγRIIB−/−Yaa mice. Conversely, the percentage and number of naive (CD62L+CD44–) T cells in the IRF5ΔB mice was greatly increased compared with the other experimental groups for both the CD4+ (Figure 2, F and G) and CD8+ (Figure 2, K and L) populations. These data indicate that heterozygous deletion of IRF5 in B cells markedly reduced splenomegaly and prevented T cell activation in FcγRIIB−/−Yaa mice and that the Mb1cre allele or the IRF5-floxed allele alone had no impact on these measurements.
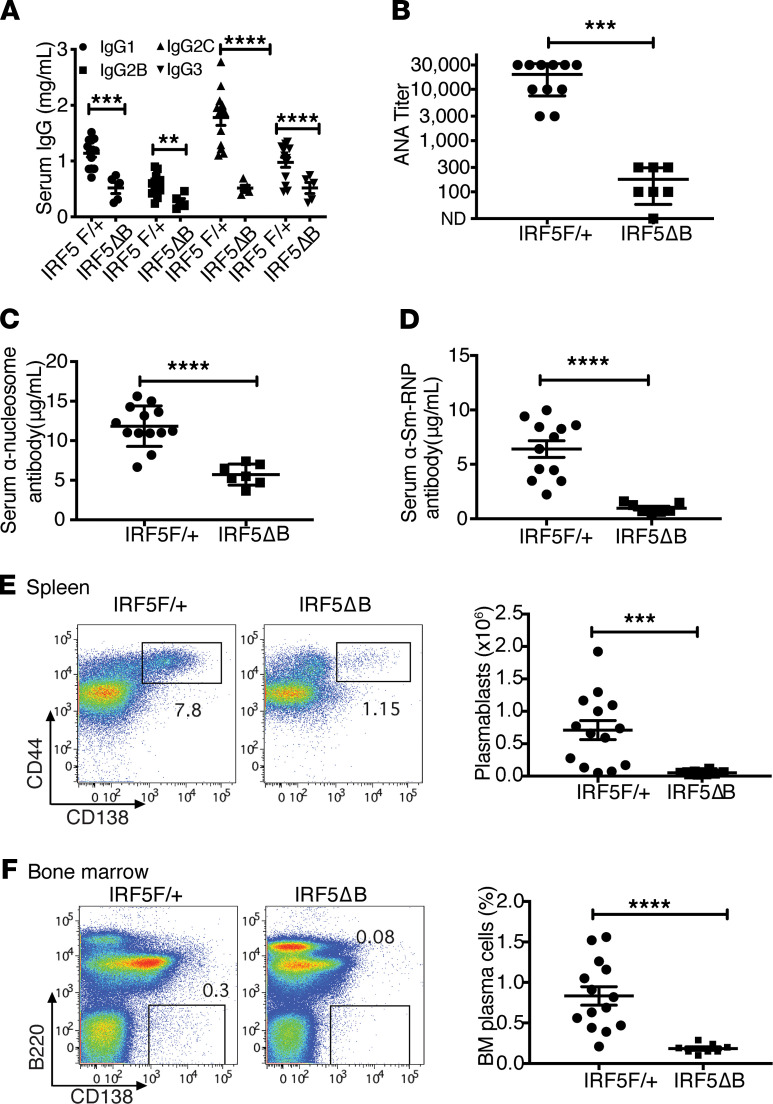
Monoallelic deletion of IRF5 in B cells reduces serum autoantibodies, splenic plasmablasts, and bone marrow plasma cells.
Antinuclear autoantibodies are a hallmark of SLE and are thought to contribute to disease pathogenesis (34). We first evaluated the effect of B cell–specific heterozygous deletion of IRF5 on serum IgG isotype levels and found that this significantly reduced the levels of all 4 IgG isotypes, IgG1, IgG2b, IgG2c, and IgG3 (Figure 3A). We next measured serum ANA and, as expected, found that IRF5F/+ mice had a high ANA titer as measured by immunofluorescence on HEp2 cells (Figure 3B). However, there was a marked reduction of ANA titer in the IRF5ΔB mice (Figure 3B). This reduction was seen in both autoantibodies directed against DNA-containing autoantigens (antinucleosome; Figure 3C) and autoantibodies directed against RNA-containing autoantigens (anti-Sm/RNP; Figure 3D).
Figure 3. Serum IgG, autoantibodies, and plasma cells are reduced in IRF5ΔB mice.
All analyses were done in 5-month-old FcγRIIB−/−Yaa mice. (A–D) IRF5F/+ (n = 11) and IRF5ΔB mice (n = 5–7) were analyzed. (A) Serum IgG isotype concentrations. (B) Serum antinuclear autoantibody titers. (C) Serum anti-nucleosome IgG concentration. (D) Serum anti-Sm/RNP IgG concentrations. (E) Representative flow cytometry plots and total numbers of splenic plasmablasts in IRF5F/+ (n = 15) and IRF5ΔB (n = 8) mice. (F) Representative flow cytometry plots and percentages of bone marrow plasma cells in IRF5F/+ (n = 15) and IRF5ΔB (n = 8) mice. Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test; **P < 0.01, ***P < 0.001, ****P < 0.0001. IRF5, IFN regulatory factor 5.
To determine how B cell–specific heterozygous deletion of IRF5 might cause the observed reduction in serum IgG and autoantibody levels, we evaluated plasmablast numbers in the spleen and plasma cell percentages in the bone marrow. It has been recently reported that B cell–intrinsic IRF5 expression is necessary for human B cell differentiation into plasmablasts in vitro (35), and IRF5-deficient mice have fewer plasma cells (23). We found that there was an appreciable population of plasmablasts in the spleens of IRF5F/+ mice at 5 months of age and that this plasmablast population was greatly reduced in the IRF5ΔB mice (Figure 3E). Similarly, there was a large reduction in plasma cells in the bone marrow compartment of the IRF5ΔB mice at 5 months of age compared with the IRF5F/+ mice at 5 months of age (Figure 3F). These data explain the reduction of IgG and autoantibodies observed with the heterozygous deletion of IRF5 in B cells in the FcγRIIB−/−Yaa mice and demonstrate that B cell–intrinsic IRF5 was necessary for plasmablast and plasma cell development in a mouse model of lupus.
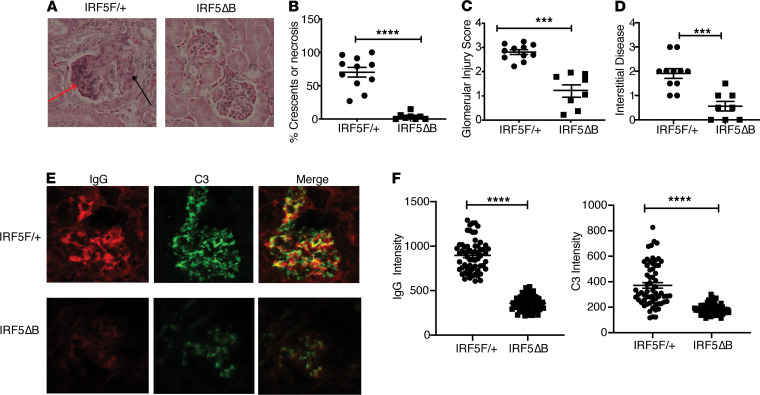
Monoallelic deletion of IRF5 in B cells reduces disease manifestations in the kidney.
The renal disease in lupus patients and in many mouse lupus models is characterized by immune complex deposition, complement activation, and glomerular inflammation with resultant glomerular crescent formation and cellular injury, often accompanied by tubulointerstitial disease (36). To assess whether the deletion of 1 allele of IRF5 in B cells might impact renal disease, we measured glomerular inflammation and injury as well as interstitial disease in kidneys from 4- to 5-month-old IRF5F/+ and IRF5ΔB mice. The IRF5F/+ mice all had glomerular crescents and/or necrosis, whereas these features were essentially absent in the IRF5B mice (Figure 4, A and B). The more severe renal disease in the IRF5F/+ mice was also evident as assessed by the overall glomerular injury score and interstitial disease (Figure 4, C and D) and by the extent of immune complex and complement deposition (Figure 4, E and F).
Figure 4. Decreased renal disease in IRF5ΔB mice.
All analyses were done in 5-month-old FcγRIIB−/−Yaa mice. (A) Representative renal histology of IRF5F/+ and IRF5ΔB mice. Red arrow depicts necrotic cells; black arrow depicts cellular crescent (original magnification, ×20). (B–D) Quantification of renal disease by (B) percentage of glomeruli with crescents or necrosis, (C) glomerular injury score, and (D) interstitial disease. IRF5F/+ (n = 11) and IRF5B (n = 8). (E) Representative examples and (F) quantitation of glomerular IgG and complement C3 deposition measured by fluorescence intensity in 11–14 glomeruli per mouse from 5 mice per group. All scored glomeruli are shown (original magnification, ×20). Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test; ***P < 0.001, ****P < 0.0001. IRF5, IFN regulatory factor 5.
The deletion of IRF5 in macrophages, monocytes, and neutrophils did not reduce disease severity in FcγRIIB−/−Yaa mice.
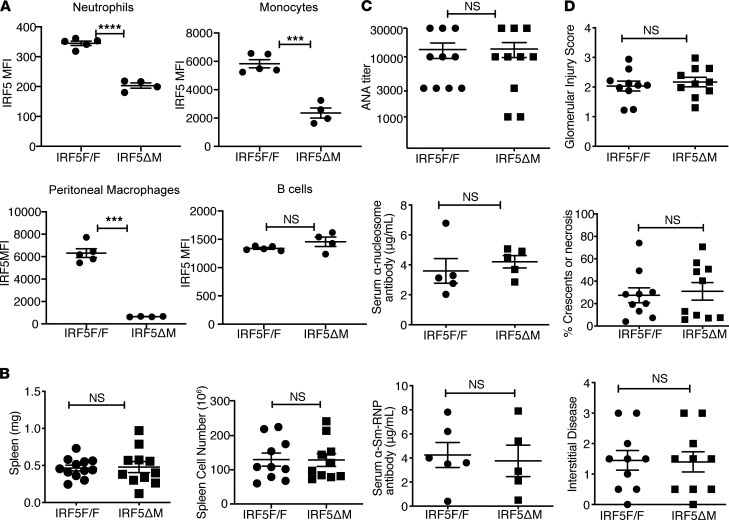
Although our data demonstrated that IRF5 expression in B cells was required for disease pathogenesis, this did not exclude the possibility that IRF5 expression in other bone marrow–derived cells might also be required. To address this possibility, we deleted IRF5 in macrophages, monocytes, and neutrophils in the FcγRIIB−/−Yaa model using the LysMcre system (37, 38). We found substantial reductions in IRF5 expression in peritoneal macrophages and splenic monocytes and neutrophils in the IRF5F/F FcγRIIB−/−Yaa mice expressing LysMcre (termed IRF5ΔM) compared with littermates not expressing LysMcre (termed IRF5fl/fl), but we found no difference in IRF5 expression in B cells (Figure 5A). However, we found no difference in spleen size, spleen cell number, autoantibody levels, or renal disease between the IRF5fl/fl mice and their IRF5ΔM littermates at 5 months of age (Figure 5, B–D).
Figure 5. LysMcre-mediated deletion of IRF5 does not reduce disease in FcγRIIB−/−Yaa mice.
All analyses were done in FcγRIIB−/−Yaa littermates that either did not express LysMcre (IRF5fl/fl) or did express LysMcre (IRF5ΔM). (A) Flow cytometry quantitation of intracellular IRF5 expression (MFI values) in neutrophils (CD11b+Ly6G+), monocytes (CD11b+Ly6C+), and B cells (CD19+) from spleen, and in peritoneal macrophages (F/480+CD11b+), in 8- to 10-week-old FcγRIIB−/−Yaa littermates (n = 4 or n = 5). Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test; ***P < 0.001, ****P < 0.0001. (B–D) All analyses were done in 5-month-old FcγRIIB−/−Yaa littermates. (B) Spleen weight and cell counts from IRF5fl/fl (n = 10–12) and IRF5ΔM mice (n = 10–12). (C) Serum antinuclear autoantibody titers, serum anti-nucleosome IgG concentration, and serum anti-Sm/RNP IgG concentration (n = 5–10). (D) Quantification of renal disease by glomerular injury score, percentage of glomeruli with crescents or necrosis, and interstitial disease (n = 10). Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test. IRF5, IFN regulatory factor 5.
Critical threshold level of IRF5 in B cells is necessary for the formation of spontaneous germinal center B cells, T follicular helper cells, and age-associated B cells in FcγRIIB−/−Yaa mice early in disease pathogenesis.
Our data show that the IRF5ΔB mice were protected from the development of lupus disease manifestations. To better understand mechanistically how IRF5 expression in B cells was contributing to this phenotype, we evaluated mice at 8–10 weeks of age prior to the appearance of overt signs of disease.
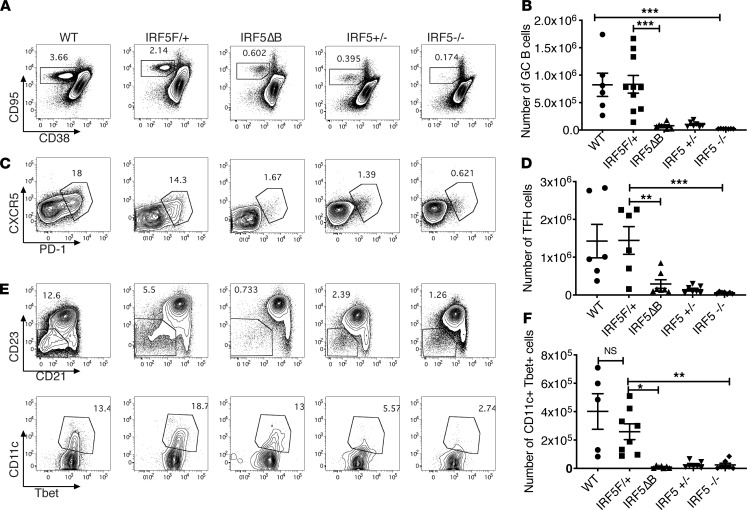
Spontaneous formation of autoimmune germinal center (GC) B cells and the presence of T follicular helper cells (Tfhs) have been shown in several mouse lupus models. These same populations are also present in human SLE (39), and it is thought that the interplay between GC B cells and Tfhs is important for the subsequent generation of pathogenic autoantibodies (40). We found that in mice as young as 8 weeks of age, there was an appreciable population of CD19+CD95+CD38– cells (GC B cells) in the spleens of FcγRIIB−/−Yaa and IRF5F/+ mice (Figure 6, A and B). Monoallelic deletion of IRF5 in B cells dramatically reduced the number of GC B cells, comparable to the reduction observed in mice that have global homozygous and heterozygous IRF5 deficiency (Figure 6, A and B). Concordantly, we observed a similar marked reduction in splenic Tfhs (CD3+CD4+CXCR5+PD-1+) in the IRF5ΔB mice compared with the FcγRIIB−/−Yaa mice and the littermate FcγRIIB−/−Yaa mice with the IRF5-floxed allele alone (Figure 6, C and D).
Figure 6. GC B cells, Tfhs, and T-bet+ B cells are reduced in the spleens of IRF5ΔB mice.
Spleen cells from 8- to 10-week-old FcγRIIB−/−Yaa WT (n = 6), IRF5F/+ (n = 10), IRF5ΔB (n = 8), IRF5+/– (global heterozygous deletion, n = 7), and IRF5–/– (global homozygous deletion, n = 7) mice were analyzed. (A and B) Representative flow cytometry plots and total numbers of CD95+CD38– GC B cells (gated on CD19+B220+ cells). (C and D) Representative flow cytometry plots and total numbers of CXCR5+PD-1+ Tfhs (gated on CD3+CD4+). (E) Upper panel indicates CD23–CD21– B cells (gated on B220+CD19+CD43–CD93 cells); lower panel indicates T-bet+CD11c+ ABCs gated on the CD23–CD21– B cells shown in the upper panel. A representative example is shown. (F) Total number of T-bet+CD11c+ ABCs. Data are shown as mean ± SEM and were analyzed using 1-way ANOVA with Tukey’s post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001. GC, germinal center; Tfhs, T follicular helper cells; IRF5, IFN regulatory factor 5; ABCs, age-associated B cells.
Age-associated B cells (ABCs) are a population of GC-derived memory B cells that are CD11c+T-bet+ and are thought to be pathogenic in autoimmune disease (41). It was recently shown that IRF5 expression in B cells is required for the in vitro differentiation of naive B cells to ABCs and that global heterozygous IRF5 deficiency results in a reduction in ABCs in the SWEF-deficient mouse model in vivo (29). We identified ABCs as CD19+B220+CD93–CD43–CD21–CD23–CD11c+T-bet+ (Figure 6E; refs. 41, 42) and found a large reduction in the number of splenic ABCs in the IRF5ΔB mice (Figure 6F). The reduction in the number of ABCs in the IRF5ΔB mice was similar to the reduction observed in mice with 1 or 2 copies of IRF5 globally deleted (Figure 6F). Taken together, these data indicate that there was a critical threshold level of IRF5, specifically in B cells, that was necessary for the formation of spontaneous GC B cells, Tfhs, and ABCs in the FcγRIIB−/−Yaa lupus model early in disease pathogenesis.
Monoallelic deletion of IRF5 reduced IL-6 and TNF-α production from B cells and reduced serum IL-6 and TNF-α levels in vivo.
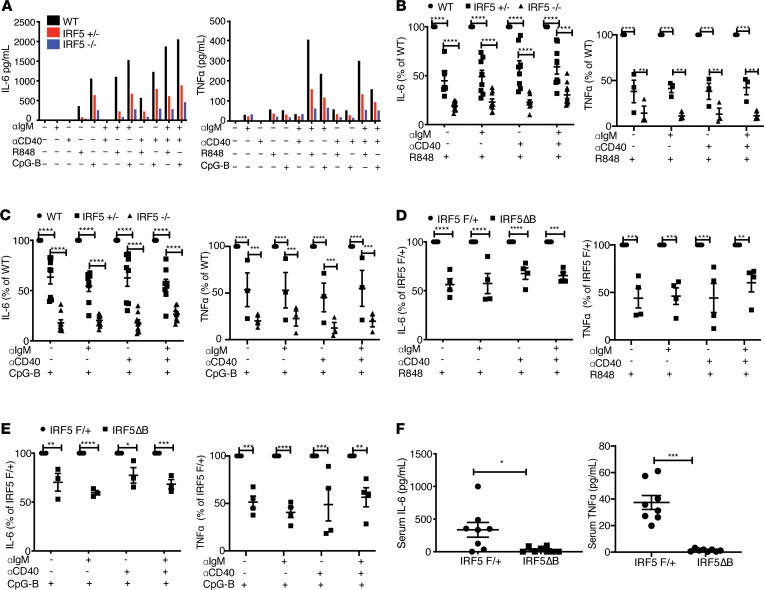
We next wanted to investigate how IRF5 expression in B cells drives the development of spontaneous GC B cells. B cell–derived IL-6 has been shown to be necessary for the development of spontaneous GCs in a mouse model of lupus and has also been suggested to be important in the development of experimental autoimmune encephalomyelitis (43, 44). B cell–derived soluble TNF is required for the development of GCs in response to T-dependent antigens and for an efficient humoral immune response (45, 46). B cell–derived soluble TNF has also been shown to mediate disease severity in a mouse model of collagen-induced arthritis via control of pathogenic autoantibody production (47). To evaluate how IRF5 monoallelic deficiency might impact B cell–intrinsic IL-6 and TNF-α production induced by the different pathways critical for GC B cell formation, we isolated B cells from 8-week-old FcγRIIB−/−Yaa, FcγRIIB−/− Yaa IRF5+/−, or FcγRIIB−/− Yaa IRF5−/− mice and stimulated the B cells with various combinations of anti-IgM antibody to model B cell receptor (BCR) activation, anti-CD40 antibody to model T cell costimulation, and the TLR7 ligand R848 or TLR9 ligand CpG-B. We found that a modest degree of IL-6 and TNF-α was induced by R848 or CpG-B alone. A substantial increase in IL-6 and TNF-α production was observed with R848 or CpG-B in combination with anti-IgM (Figure 7A). There was a gene dosage–dependent impact of IRF5 on IL-6 and TNF-α in that heterozygous deficiency of IRF5 reduced IL-6 and TNF-α production by 40%–60% in all conditions tested and homozygous deletion reduced IL-6 and TNF-α by 70%–80% (Figure 7, B and C). Notably, B cells from mice with B cell–specific heterozygous deletion of IRF5 by Mb1cre showed a similar reduction in IL-6 and TNF-α as observed in B cells from mice with germ line heterozygous deficiency of IRF5 (Figure 7, D and E). IL-12p40 production was also reduced in IRF5 heterozygous B cells in response to most stimuli combinations, but effects on RANTES, MIP-1α, and MIP-1β were more variable (Supplemental Figure 5).
Figure 7. Reduced IRF5 expression in B cells decreases IL-6 and TNF-α production in vitro, and serum IL-6 and TNF-α is reduced in IRF5ΔB mice.
(A–C) B cells were isolated from the spleens of FcγRIIB−/−Yaa mice at 8–10 weeks of age and stimulated for 24 hours with anti-IgM, anti-CD40, R848, and CpG-B alone or in combination. (A) Representative experiments showing mean IL-6 and TNF-α production by B cells from WT, IRF5+/–, and IRF5–/– mice (n = 2 for each genotype). (B and C) IL-6 (n = 8) and TNF-α (n = 3) production after R848 stimulation (B) and CpG-B stimulation (C) by B cells from WT, IRF5+/–, and IRF5–/– mice normalized to the WT control in each experiment. (D and E) IL-6 and TNF-α production by B cells from IRF5ΔB mice normalized to the littermate IRF5F/+ control in each experiment (n = 4 for each genotype). Data are shown as mean ± SEM and were analyzed using 2-way ANOVA with Tukey’s post hoc test; **P < 0.01, ***P < 0.001, ****P < 0.0001. (F) Mean serum IL-6 and TNF-α levels from 5-month-old IRF5ΔB (n = 8) and littermate IRF5F/+ (n = 8) mice. Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test; *P < 0.05, ***P < 0.001. IRF5, IFN regulatory factor 5.
We next evaluated if B cell–specific deletion of IRF5 impacted the level of serum IL-6 and/or TNF-α in the FcγRIIB−/−Yaa model. B cell–specific deletion of IL-6 has been shown to reduce serum IL-6 in another lupus model (43). We found that serum levels of both IL-6 and TNF-α were indeed decreased in 5-month-old IRF5ΔB mice compared with the FcγRIIB−/−Yaa controls (Figure 7F). These data suggest that there was a synergistic effect of both TLR7 and TLR9 signaling with BCR signaling on the production of IL-6 and TNF-α and that B cell–intrinsic IRF5 played a critical role in mediating the induction of IL-6 and TNF-α in all combinations of stimuli.
TLR7 and BCR stimulation synergized in the induction of IRF5 phosphorylation.
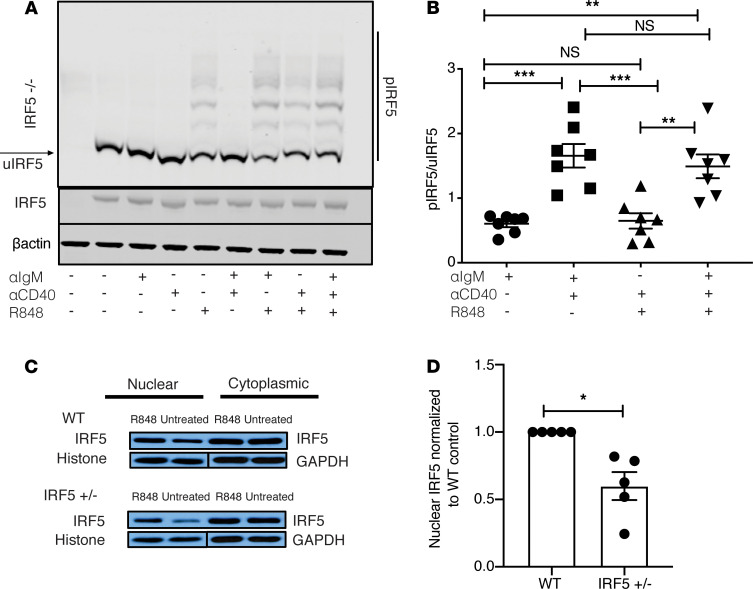
Phosphorylation of IRF5 is necessary for IRF5 dimerization and nuclear translocation (48). To determine whether the increased IL-6 and TNF-α production observed with TLR7 and BCR costimulation might be mirrored by changes in IRF5 phosphorylation, we stimulated isolated B cells from FcγRIIB−/−Yaa mice with the different combinations of anti-IgM, R848, and anti-CD40. We chose to use R848 because TLR7 gene dosage is essential for the accelerated disease in the FcγRIIB−/−Yaa mice and TLR7 is important for pathogenesis in other mouse models of lupus (16, 49). We did not detect any phosphorylation of IRF5 after stimulation with anti-IgM alone or anti-CD40 alone, consistent with the lack of IL-6 and TNF-α production observed in Figure 7A. However, we readily detected phosphorylation after stimulation with R848 alone for 2 hours (Figure 8A). The ratio of phosphorylated to unphosphorylated IRF5 was significantly increased 3-fold with the addition of anti-IgM to R848 (Figure 8B). Interestingly, the addition of anti-CD40 stimulation did not increase IRF5 phosphorylation in any of the stimulus combinations. Moreover, we demonstrated that B cells from C57BL/6 mice harboring the kinase dead mutation of IL-1 receptor–associated kinase 4 (IRAK4) completely lacked IRF5 phosphorylation in all stimulation conditions (Supplemental Figure 6A). These data indicate that TLR7 stimulation alone was sufficient to induce IRF5 phosphorylation in B cells and that the BCR and TLR7 synergized to increase IRF5 phosphorylation.
Figure 8. TLR7 signaling is required for IRF5 phosphorylation, and IRF5 nuclear translocation is reduced in B cells from FcγRIIB−/−Yaa IRF5+/– mice.
(A and B) B cells were isolated from the spleens of FcγRIIB−/−Yaa mice at 8–10 weeks of age. (A) B cells were stimulated with anti-IgM, anti-CD40, and R848 alone or in combination for 2 hours and the protein lysate analyzed using phospho-Tag gel (upper panel) or standard gel (lower panels). B cells isolated from an IRF5-deficient (IRF5–/–) mouse are shown in the first lane. p-IRF5 denotes phosphorylated IRF5. A representative example of 7 individual experiments is shown. (B) Ratio of p-IRF5 to unphosphorylated IRF5 (u-IRF5). Intensity of p-IRF5 was normalized to the intensity of unphosphorylated IRF5 (lowest band of IRF5 on p-Tag gel as shown in A) (n = 7). (C and D) B cells from FcγRIIB−/−Yaa (WT) and FcγRIIB−/−Yaa IRF5+/– mice were stimulated for 2 hours with R848, or not stimulated (untreated), and IRF5 was probed in the nuclear and cytoplasmic fractions. (C) A representative experiment of 4 individual experiments is shown. (D) Ratio of IRF5 expression in nucleus relative to the WT after R848 stimulation; nuclear IRF5 intensity in each sample was first normalized to its own loading control (histone; n = 5). Data are shown as mean ± SEM and were analyzed using 1-way ANOVA with Tukey’s post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001. IRF5, IFN regulatory factor 5.
TLR7-induced IRF5 phosphorylation and nuclear translocation were reduced in B cells with monoallelic deletion of IRF5.
Because TLR7-induced IL-6 and TNF-α production were reduced in B cells with monoallelic deletion of IRF5 (Figure 7, A, B, and D), we hypothesized that this would be associated with a reduction in IRF5 nuclear translocation. We first examined the extent of IRF5 phosphorylation in splenic B cells from FcγRIIB−/−Yaa WT and IRF5+/− mice 2 hours after stimulation with anti-IgM, anti-CD40, and R848 alone or in combination. We found that the general pattern of IRF5 phosphorylation, as assessed by phospho-Tag gel, and the ratio of phosphorylated to unphosphorylated IRF5 did not differ between the groups (Supplemental Figure 6, B and C). However, because the total amount of IRF5 in B cells from the FcγRIIB−/− Yaa IRF5+/− mice was about half of that in the WT mice, we infer that the total amount of phosphorylated IRF5 was less in the IRF5+/− mice under these stimulation conditions. More importantly, we directly compared the ratio of nuclear IRF5 expression in B cells from the WT and IRF5+/− mice after R848 stimulation and found that nuclear IRF5 expression was reduced by approximately half in the IRF5+/− mice (Figure 8, C and D).
IRF5 expression was increased in activated B cells in vitro and in GC B cells, splenic plasmablasts, and bone marrow plasma cells in vivo.
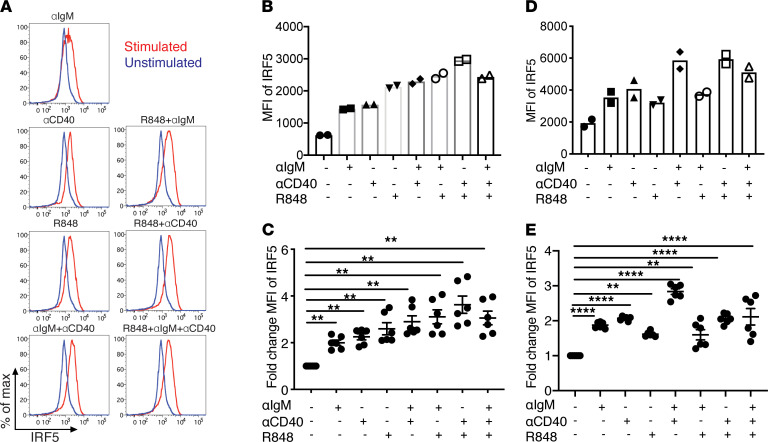
Our data in Figure 3 and Figure 6 suggest that there was a critical threshold level of IRF5 required for the generation of autoimmune GC B cells and plasma cells. We therefore hypothesized that IRF5 expression levels might be dynamically regulated after B cell activation. To determine if the expression level of IRF5 changes after B cell activation, we stimulated naive B cells isolated from FcγRIIB−/−Yaa mice using various combinations of anti-IgM, anti-CD40, and R848 and measured IRF5 expression levels by intracellular flow cytometry after 24 hours. In contrast to their lack of effect on IRF5 phosphorylation, anti-IgM and anti-CD40 alone increased IRF5 expression about 2-fold, similar to the increase induced by R848 alone (Figure 9, A–C). The various stimulus combinations induced a greater increase in IRF5 expression than the individual stimuli alone. To determine whether these findings were specific to B cells from FcγRIIB−/−Yaa mice, we repeated these studies using naive B cells isolated from C57BL/6 mice and found a similar increase in IRF5 expression in C57BL/6 B cells (Figure 9, D and E). This indicates that the increase in IRF5 expression observed in B cells under these stimulation conditions was not a unique feature of FcγRIIB−/−Yaa B cells only.
Figure 9. IRF5 expression is increased in activated B cells in vitro.
(A–E) Splenic B cells were isolated from 8- to 10-week-old FcγRIIB−/−Yaa or C57BL/6 mice and were either not stimulated or stimulated with anti-IgM, anti-CD40, and R848 alone or in combination for 24 hours. (A) Intracellular IRF5 levels were measured using flow cytometry. A representative experiment of 6 individual experiments using B cells from FcγRIIB−/−Yaa mice is shown. (B and D) MFI of IRF5 with and without stimulation in B cells from FcγRIIB−/−Yaa mice (B) and C57BL/6 mice (D) (n = 2 per strain). (C and E) Fold change of IRF5 expression normalized to unstimulated control in B cells from FcγRIIB−/−Yaa mice (C) and C57BL/6 mice (E) (n = 6 per strain). Data are shown as mean ± SEM and were analyzed using 1-way ANOVA with Tukey’s post hoc test; **P < 0.01, ****P < 0.0001. IRF5, IFN regulatory factor 5.
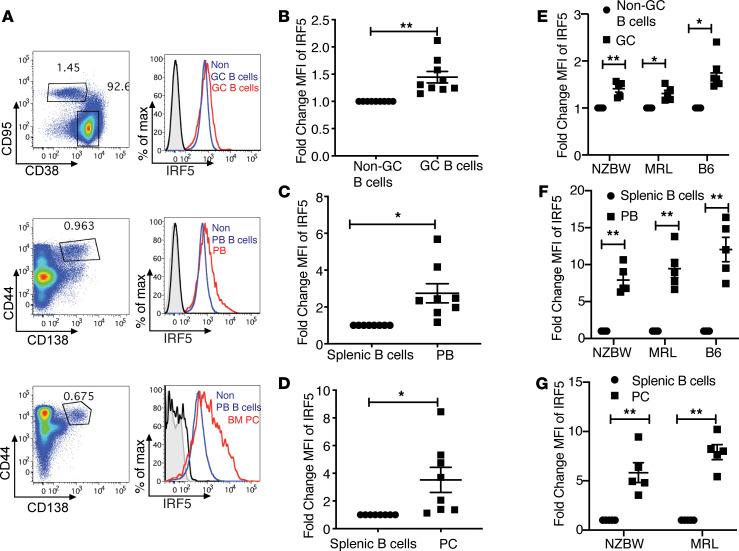
The increase in IRF5 expression after activation with anti-IgM, anti-CD40, and R848 in vitro suggested the possibility that IRF5 might be increased in GC B cells in FcγRIIB−/−Yaa mice in vivo, because TLR7 activation is thought to be important for spontaneous GC formation in other lupus models (50). To test this hypothesis, we evaluated IRF5 expression in splenic GCs of 8- to 10-week-old FcγRIIB−/−Yaa mice. We observed an approximately 1.5-fold increase in the MFI of IRF5 in GC B cells compared with non-GC B cells (Figure 10, A and B). Next, we evaluated the levels of IRF5 in plasmablasts because these cells are thought to be derived from either the GC or an extrafollicular response (1) and because it was recently reported that human plasmablasts express higher levels of IRF5 compared with naive B cells (35). We observed a roughly 2.5-fold increase in the MFI of IRF5 in plasmablasts compared with non–plasmablast B cells (CD19+; Figure 10, A and C). Furthermore, we did a similar analysis comparing the splenic CD19+ B cells with plasma cells in the bone marrow and observed an increase in IRF5 MFI in plasma cells at least as large as that observed in the splenic plasmablasts (Figure 10, A and D).
Figure 10. IRF5 expression is increased in GC B cells, splenic PBs, and BM PCs in vivo.
(A–G) Flow cytometry performed on splenocytes and bone marrow from FcγRIIB−/−Yaa mice at 8–10 weeks of age. (A) Representative examples of IRF5 expression in GC B cells (CD38–CD95+CD19+) and non-GC B cells (CD38+CD95–; upper panel); PBs (CD44+CD138+) and CD19+CD138– B cells (non-PBs; middle panel); and BM PCs (CD44+ CD138+) and non-PB B cells from spleens (lower panel). (B) Fold change in IRF5 expression in GC B cells normalized to non-GC B cells (n = 9). (C) Fold change in IRF5 expression in PBs normalized to non-PBs (n = 8). (D) Fold change in IRF5 expression in PCs normalized to splenic non-PBs (n = 8). (E–G) IRF5 expression in NZB/W mice (n = 6), MRL/lpr mice (n = 6), and C57BL/6 (n = 6) mice immunized with 4-hydroxy-3-nitrophenylacetyl coupled to chicken γ-globulin. (E) Fold change of IRF5 expression in GC B cells. (F) Fold change of IRF5 expression in PBs. (G) Fold change of IRF5 expression in plasma cells. Black histogram shows isotype control in non-GC B cells, non-PBs, or BM PCs; gray-tinted histogram shows isotype control in GC B cells, PBs, or BM PCs; blue histogram shows IRF5 expression in non-GC B cells and non-PBs; red histogram shows IRF5 expression in GC B cells, PBs, or BM PCs. Data are shown as mean ± SEM and were analyzed using 2-tailed, unpaired Welch’s t test; *P < 0.05, **P < 0.01. IRF5, IFN regulatory factor 5; GC, germinal center; PBs, plasmablasts; BM PCs, bone marrow plasma cells.
To determine whether our findings were specific to the FcγRIIB−/−Yaa model or were more generally relevant, we measured changes in IRF5 expression in GC B cells, plasmablasts, and plasma cells in 2 additional mouse lupus models, (NZBxNZW)F1 and MRL-lpr. We found that IRF5 expression was increased in these models at least as much as had been observed in the FcγRIIB−/−Yaa model (Figure 10, E–G). In addition, to determine whether our findings were restricted to lupus models or whether they might reflect GC responses more broadly, we measured changes in IRF5 expression in GC B cells and plasmablasts in C57BL/6 mice 14 days after immunization with the hapten 4-hydroxy-3-nitrophenylacetyl coupled to chicken γ-globulin. We found an increase in IRF5 expression in these cell types similar to that observed in the 3 lupus models (Figure 10, E and F).
Taken together, these data suggest that IRF5 expression levels were dynamically regulated in the GC compartment as well as during plasmablast differentiation and that this increased IRF5 expression was maintained in long-lived plasma cells.
Discussion
Global homozygous or heterozygous deficiency of IRF5 markedly reduces disease severity in mouse lupus models (5–9). However, it is not known which IRF5-expressing cell type(s) is involved in lupus pathogenesis. In this report, we demonstrate that heterozygous deficiency of IRF5 in B cells resulted in a marked reduction in lupus disease manifestations. In contrast, similar reduction of IRF5 expression in macrophages, monocytes, and neutrophils did not reduce disease severity. Although these findings do not exclude a role for IRF5 in other cell types in disease pathogenesis, they do demonstrate that there is a critical threshold level of IRF5 expression in B cells that was absolutely required.
We previously reported that heterozygous deficiency of IRF5 reduces disease in the FcγRIIB−/−Yaa mouse lupus model (5). Subsequently, it was demonstrated that global heterozygous deficiency of IRF5 also reduces disease in the Lyn-deficient mouse lupus model, the gld.apoE-deficient mouse lupus model, and in the Swap70-deficient Def6-deficient mouse lupus model (10, 28, 29). In the Swap70-deficient Def6-deficient model, the deletion of the second IRF5 allele specifically in CD21-expressing cells or in CD11c-expressing cells does not lead to further reductions in disease manifestations. In this report, we identified B cells as a critical cell type in which heterozygous deficiency of IRF5 was protective, and thus we sought to uncover possible mechanisms that might explain the profound protective effect of a 50% reduction in IRF5 expression in B cells. We found that IL-6 and TNF-α production by B cells from IRF5-heterozygous mice was reduced by about half in response to combinations of stimuli that are thought to be involved in B cell activation in the GC in lupus (51). In contrast, the expression of costimulatory molecules involved in B cell–T cell interactions and GC B cell activation, such as CD80, CD86, MHC class II, and ICOS-ligand, did not differ between B cells from IRF5-WT and IRF5-heterozygous mice (Supplemental Figure 7). B cell–intrinsic IL-6 and TNF-α production have been shown to be important in GC formation in mice immunized with T-dependent antigens and in GC formation and disease pathogenesis in lupus and other autoimmune models (43–47). It is thus plausible that the reduction in IL-6 and TNF-α production observed in our studies may explain, at least in part, the reduction in disease manifestations seen in the B cell–specific IRF5-heterozygous mice.
In the inactivated state, IRF5 resides in the cytoplasm as a monomer. IRF5 activation requires phosphorylation of the monomer, leading to either IRF5 homodimerization or heterodimerization of IRF5 with other IRF or NF-κB family members, followed by nuclear translocation and the induction of target gene transcription (17). We found that the amount of IRF5 in the nucleus after TLR7 activation was approximately 50% lower in B cells from IRF5-heterozygous FcγRIIB−/−Yaa mice than in B cells from IRF5-sufficient FcγRIIB−/−Yaa mice. Mechanistically, this may explain the reduced production of IL-6 and TNF-α by the IRF5-heterozygous B cells.
The IRF5 polymorphisms associated with an increased risk of developing SLE and other autoimmune diseases are in the noncoding region of the IRF5 gene and show some association with increased IRF5 expression and/or functional change (3, 20). We found that BCR activation alone or CD40 activation alone increased IRF5 protein about 2-fold but did not induce IRF5 phosphorylation. Thus, increased IRF5 expression, per se, did not result in IRF5 activation. In contrast, TLR7 activation alone induced both increased IRF5 expression and IRF5 phosphorylation, and BCR activation, although not able to induce IRF5 phosphorylation independently, synergized with TLR7 activation to further increase both IRF5 expression and phosphorylation. These findings were not just restricted to B cells from an autoimmune disease–prone background but were also observed in B cells from C57BL/6 mice. Intriguingly, we also observed that IRF5 phosphorylation in all stimulation conditions was IRAK4 dependent. Consistent with these in vitro findings, we observed an increase in IRF5 expression in GC B cells in vivo early in disease development, prior to the appearance of overt signs of autoimmunity, with even larger increases evident in plasmablasts and bone marrow plasma cells. We corroborated these findings in 3 different lupus mouse models and in C57BL/6 mice immunized with a T-dependent antigen, suggesting that the increase in IRF5 in these compartments during B cell activation could have been a general feature in B cell biology.
IRF5 exerts a number of effects in B cells, including class-switching to IgG2a/IgG2c by binding to the IgG2a promoter, regulation of the transcription factor Blimp1, and regulation of T-bet–expressing ABCs (6, 23, 29). Although these B cell–intrinsic effects likely make significant contributions to disease pathogenesis, our data suggest that there were also critical roles for B cell–intrinsic IRF5 more proximally in the autoimmune response. The generation of high-affinity autoantibodies and long-lived plasma cells in lupus is dependent on the development of a robust GC response, with TLR7 and TLR9 signaling in B cells likely being required for this response (51). We found a dramatic reduction in the number of GC B cells prior to the onset of overt autoimmune disease in the mice with heterozygous deficiency of IRF5 in B cells. This was accompanied by an equally marked reduction in the number of Tfhs, a cell type that provides the signals to B cells that are required for GC formation, affinity maturation, and the development of most high-affinity autoantibodies and memory B cells (52, 53). This suggests that B cell–intrinsic IRF5 may be required for the generation of Tfhs in our lupus model. Early Tfh differentiation is regulated by IL-6, ICOS, IL-2, and T cell receptor signal strength in mouse models (52). This early Tfh cell differentiation is generally thought to be the result of naive T cell interactions with DCs. However, B cells can be the dominant antigen-presenting cells that activate naive CD4+ T cells in certain circumstances, and B cells activated by TLR9 and TLR7 ligands in the context of viral infection are sufficient to induce Tfh development in the absence of DCs (54).
In summary, we have demonstrated that there was a critical threshold level of IRF5 in B cells that was required for disease development in the FcγRIIB−/−Yaa mouse lupus model and that IRF5 nuclear translocation was substantially reduced in B cells from IRF5-heterozygous mice after TLR7 activation. We also showed in multiple lupus models and in immunization studies that the level of IRF5 expression was dynamically regulated throughout the B cell activation process, increasingly and progressively from GC B cells to mature plasma cells. Furthermore, IRF5 phosphorylation was increased in a synergistic manner by activation signals believed to be important in the generation of autoreactive GC B cells. The fact that a 50% reduction in B cell IRF5 expression was sufficient to largely abrogate disease development suggests that targeting IRF5 in B cells may have been an effective therapeutic approach in lupus.
Methods
Generation of IRF5-floxed mice.
The vector to target IRF5 was pEZ-Frt-LoxP-DT (courtesy of Klaus Rajewsky, Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany). Three DNA fragments of the C57BL/6 mice IRF5 locus were inserted into the cloning site of the vector pEZ-Frt-LoxP-DT: 1.6 kb of the region upstream of the IRF5 exon 3, 2 kb of the region between exon 3 to exon 6, and 2.8 kb of the region downstream of exon 6 (Supplemental Figure 2). The linearized vector was electroporated into embryonic stem (ES) cells derived from C57BL/6 mice in the Transgenic Mouse Core at Brigham and Women’s Hospital (Boston, Massachusetts, USA), and correctly targeted ES cell colonies were identified by PCR. Selected ES cells were injected into blastocysts. Chimeras were bred with C57BL/6 mice to obtain mice with the conditional IRF5 allele in the germ line as determined by PCRs. The mice were further crossed with ROSA26:FLPe knockin mice (Jackson Laboratory, 9086) to delete the neomycin-resistant gene between Frt sites. IRF5 LoxP positioning was confirmed by PCR and Southern blotting analysis.
Animals.
WT (IRF5+/+) FcγRIIB−/−Yaa, IRF5–/– FcγRIIB–/–Yaa, and IRF5+/– FcγRIIB–/–Yaa mice were all generated as previously described (5). FcγRIIB–/–Yaa Mb1cre/+ IRF5fl/+ (IRF5ΔB) were generated by crossing IRF5fl/fl mice on a FcγRIIB–/–Yaa background. Mice that expressed cre under the direction of the Ig-α promoter (mb1-cre mice) were provided by Michael Reth (University of Freiburg, Freiburg, Germany; ref. 32) and were also crossed on a FcγRIIB–/–Yaa background. Mb1cre/+ FcγRIIB–/–Yaa males were then crossed to FcγRIIB–/– IRF5fl/fl females. The final experimental and littermate control mice were generated by breeding Mb1cre/+ FcγRIIB−/−Yaa males with FcγRIIB−/− IRF5fl/+ females. B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice were purchased from Jackson Laboratory. Mice that expressed cre under the direction of the LysM promoter (LysMcre mice) were purchased from Jackson Laboratory (B6.129P2-Lyz2tm1(cre)Ifo/J; strain number 004781; ref. 37). The final experimental and littermate control mice were generated in a similar manner to the Mb1cre study detailed above, except that the LysMcre/+ FcγRIIB–/–Yaa males were bred with FcγRIIB–/– IRF5fl/fl females, not FcγRIIB–/– IRF5fl/+ females. Mice deficient in IRAK-4KN/KN were provided by Shizuo Akira (Osaka University, Osaka, Japan; ref. 55). (NZBxNZW)F1 mice and MRL-lpr mice were purchased from Jackson Laboratory (stock numbers 100008 and 000485, respectively). All mice used in this study, apart from (NZBxNZW)F1 and MRL-lpr, were backcrossed more than 9 generations on a C57BL/6 background.
Bone marrow chimera generation.
Bone marrow cells were isolated from FcγRIIB–/–Yaa IRF5+/+ and FcγRIIB–/–Yaa IRF5–/– donors and washed once in PBS followed by RBC lysis using RBC lysing buffer (MilliporeSigma, R7757). Donor marrow cells (106 cells per mouse) were injected into the tail veins of B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) recipient mice after cesium irradiation of recipient mice with 2 doses of 500 rad separated by 3 to 4 hours. Recipient mice were subsequently placed in irradiated cages, and water was supplemented with sulfamethoxazole/trimethoprim (Hi-Tech Pharmaceuticals) (at 2/0.4 mg/mL). Immune cell engraftment was subsequently confirmed using flow cytometry of PBMCs and antibodies specific for CD45.1 (clone A20; BioLegend) and CD45.2 (clone 104; BioLegend).
Immunization.
C56BL/6 mice (8–10 weeks old) were immunized i.p. with 50 g 4-hydroxy-3-nitrophenylacetyl coupled to chicken γ-globulin at a substitution rate of 16 (Biosearch Technologies) in alum (56). Spleens were harvested on day 14.
B cell isolation and stimulation.
Resting B cells were purified from the spleens of WT FcγRIIB–/–Yaa, IRF5–/– FcγRIIB–/–Yaa, IRF5+/– FcγRIIB–/–Yaa, IRF5F/+ FcγRIIB–/–Yaa, and IRF5ΔB mice by negative selection using MACS cell separation with anti-CD43 beads (Miltenyi Biotec). Cells were plated at 1–2 × 105 per well in duplicate or triplicate in RPMI 1640 supplemented with 10% FBS, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C and stimulated with 0.05 μM R848 (InvivoGen), 10 μg/mL anti-IgM [F(ab′)2 fragment; μ chain specific; Jackson ImmunoResearch], and 2 μg/mL anti-CD40 antibody (clone HM40-3, BD Biosciences) alone or in combination. Supernatants were collected 24 hours later and cytokines evaluated by ELISAs (R&D Systems and BioLegend) and ProcartaPlex multiplex (Invitrogen). Cells were evaluated by flow cytometry for activation markers and intracellular levels of IRF5.
Western blot and phospho-tag immunoblot analysis.
Splenic B cells were lysed in RIPA buffer (MilliporeSigma) that included phosphatase and protease inhibitors (MilliporeSigma). For standard Western blot analysis, samples were separated on a 4%–12% Bis-Tris gel (Invitrogen) using MOPS buffer (Invitrogen). For phospho-tag immunoblot analysis, SDS-PAGE was performed using SuperSep Phos-tag 7.5% precast gel (Fujifilm Wako Chemicals USA Corporation). Samples were transferred to nitrocellulose membranes using iBlot (Invitrogen) and probed with anti-IRF5 antibody (catalog ab181553, Abcam) and anti–β-actin antibody (clone 8H10D10, Cell Signaling Technology). Samples were imaged using the Odyssey CLx (LI-COR) and quantified using ImageStudio.
Preparation of cytosolic and nuclear protein fractions.
Cytosolic and nuclear protein fractions were prepared using a slight modification of the protocol described by Liou et al. (57). Splenic B cells were washed once with cold PBS and resuspended in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT) with 0.1% Triton X-100, phosphatase inhibitors (MilliporeSigma), and protease inhibitors (Calbiochem). Cells were lysed on ice for 10 minutes. After centrifugation at 6000g for 15 minutes at 4ºC, the supernatant was saved as the cytosolic fraction. The nuclear pellet was washed twice with buffer A to remove any residual cytosolic fraction proteins and then resuspended in buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 0.5 mM DTT) with protease inhibitors on ice. The nuclear pellet was sonicated briefly before incubation on ice for 20 minutes. The insoluble debris were removed by centrifugation at 15,000g for 15 minutes at 4ºC, and the supernatant was saved as the nuclear fraction. The protein concentration was determined with Pierce BCA Protein Assay Kit (Life Technologies).
Flow cytometry.
Splenocytes were labeled with antibodies against CD4 (clone GK1.5, BD Biosciences), CD8 (clone 53-6.7, BioLegend), and CD3 (clone 17A2, BD Biosciences) to identify T cell populations; against CD19 (clone 1D3, BD Biosciences) and B220 (clone RA3-6B2, BD Biosciences) to identify B cells; and against CD44 (clone IM7, BD Biosciences) and CD62L (clone MEL-14, BioLegend) to identify naive T cells, activated T cells, and memory T cells. Biotin-labeled anti-CXCR5 (clone 2G8, BD Biosciences) was used in combination with streptavidin-BV711 (BD Biosciences, catalog 563262), CD3, PD-1 (clone J43, BD Biosciences), and CD4 to identify Tfhs. ABC B cells were identified by first gating on CD19 and B220, then gating out CD43+ (clone S7, BD Biosciences) and CD93+ (clone AA4.1, BD Biosciences) cells, followed by gating on CD21– (clone 7G6, BD Biosciences) and CD23– (clone B3B4, BD Biosciences) negative cells. Finally, CD11c (clone HL3, BD Biosciences) and Tbet (clone eBio4B10, eBioscience) delineated Tbet+ ABCs. The FoxP3 fix/perm from eBioscience (catalog 00-5523-00) was used for the intracellular staining of Tbet. For staining of spleen plasmablasts, antibodies against CD3, CD19, B220, CD44, and CD138 (clone 281-2, BD Biosciences) were used. For staining of bone marrow plasma cells, antibodies against CD4, CD8, Gr-1 (clone RB6-8C5, BioLegend), and F4/80 (clone BM8, BioLegend) were used to gate out non-B cells, and plasma cells [CD138 (281-2)+B220−] were identified using antibodies against B220 and CD138. Bone marrow plasma cells were also identified using same stain as used for spleens. Antibodies against CD19, CD38 (clone 90, BioLegend), CD95 (clone Jo2, BD Biosciences), and B220 were used to define germinal center B cells. Antibodies against CD80 (clone 16-10A1, BioLegend), CD86 (clone GL-1, BioLegend), CD275 (clone HK5.3, BioLegend), and MHCII (clone M5/114.15.2, eBioscience) were used to evaluate activation. B cells were first gated on CD19+ and live cells (live/dead, catalog 65-0866-14, eBioscience). Immunofluorescence was measured with an LSRII (BD Biosciences). Sorting experiments were done on BD FACSAria III (BD Biosciences). The data were analyzed using FlowJo software (Tree Star).
Intracellular flow cytometry.
B cells or splenocytes were first stained with extracellular antigens. Cells were then fixed for 15 minutes at room temperature with BioLegend fixative. Cells were then washed with BioLegend permeabilize buffer, and then cells were incubated with anti-IRF5 (clone W16007B, BioLegend) or isotype control (R&D Systems, catalog IC006P) diluted in 1× perm buffer. To identify neutrophils, monocytes, and pDCs, splenocytes were stained with antibodies against CD11b (M1/70), Ly6c (HK1.4), Ly6G (1A8), PDCA1 (eBio927), CD19, and CD3.
Histology.
At the end of the experiment at 4–5 months of age, kidneys were harvested from FcγRIIB–/–Yaa IRF5F/+ control and FcγRIIB−/−Yaa IRF5ΔB mice and fixed in formalin or snap-frozen in OCT compound. Formalin-fixed and paraffin-embedded kidneys were sectioned at 8 μm and then stained with H&E. Kidney disease was assessed by a blinded investigator. To this end, randomly selected areas of cortex were examined, and at least 50 glomeruli from each animal were scored. Glomerular and interstitial disease were evaluated as previously described (5, 58, 59).
IHC.
Mouse kidneys were snap-frozen in OCT (Tissue-Tek, Sakura Finetek) and stored at –80°C. Seven-micrometer cryosections were cut and fixed with methanol and acetone (1:1), blocked with 1% BSA, and then stained with Alexa Fluor 594–conjugated donkey anti-mouse IgG (Invitrogen, catalog A21203) and FITC-conjugated goat anti-mouse C3 (Cappel Laboratories, catalog 0855500) at 4°C overnight. After washing with Tris-buffered saline, the stained sections were scanned with a Nikon Deconvolution Wide-Field Epifluorescence System. Fluorescence intensity, representing IgG and C3 deposition, was measured using ImageJ (NIH) and analyzed with GraphPad Prism. Representative images were acquired using a Zeiss LSM 710 confocal microscope.
Serological assays.
IgG isotypes and IgM were measured by ELISA established using antibodies from BD Biosciences and Southern Biotech. ANA titer was measured by immunofluorescence using HEp2-coated slides (INOVA Diagnostics, Inc.) as described (5, 59). The antinucleosome antibody ELISA was developed using nucleosome antigen ATN02-05 (2 μg/mL; Arotec Diagnostics Limited) and HRP-conjugated anti–mouse IgG antibody (1:3000, MilliporeSigma, catalog A8924). Anti-nucleosome antibody (clone PL2-3, gift from Marc Monestier, Temple University, Philadelphia, Pennsylvania, USA) was used as the positive control for quantification. The anti-Sm/RNP ELISA was developed using Sm/RNP antigen ATR01 (2 μg/mL; Arotec Diagnostics Limited) and HRP-conjugated anti–mouse IgG antibody (1:3000, MilliporeSigma). Anti-Sm/RNP antibody (Y2) was used as the positive control for quantification.
Statistics.
P values were calculated using a 2-tailed, unpaired Welch’s t test, or a 1-way ANOVA with Tukey’s post hoc test. P values less than 0.05 were considered significant. Statistics were calculated using GraphPad Prism.
Study approval.
All animal studies were performed under protocols that were approved by the IACUC at Boston University and the University of Massachusetts Medical School.
Author contributions
AP, KY, UK, RGB, and IRR designed the study. AP, KY, ACB, VS, PS, KN, BKH, YX, YT, and RGB performed the experiments and analyzed the data. GAV optimized experimental protocols. AP and IRR wrote the manuscript. AP, KY, VS, PS, BKH, GAV, YX, UK, YT, RGB, and IRR read and commented on various drafts of the manuscript. For the 2 co–first authors, the assignment of authorship order was made after discussion with, and with the full agreement of, both co–first authors. The assignment was based on their overall contributions to the various components of the experimental work and manuscript preparation, recognizing that both co–first authors made very substantial and critical contributions to the manuscript.
Supplementary Material
Acknowledgments
We thank Arlene Sharpe and the Transgenic Mouse Core at Brigham and Women’s Hospital, Boston, for help with making the IRF5-floxed mouse line. We thank Michael Reth for providing the Mb1cre mice. We thank Tadatsugu Taniguchi for providing the IRF5-deficient mice and Tak Mak for permission to use the IRF5-deficient mice. We thank Ann Marshak-Rothstein and Jeffrey Browning for their careful reading of the manuscript and helpful comments. We thank Christopher Gould at BioLegend for providing the PE-conjugated anti-IRF5 antibody prior to commercial release for testing and validation. This work was supported by the following NIH grants: R01AI130199 (IRR), R01DK095058 (RGB), K01AR060857 (KY).
Version 1. 07/01/2021
In-Press Preview
Version 2. 08/09/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, Pellerin et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2021;6(15):e141395.https://doi.org/10.1172/jci.insight.141395.
Contributor Information
Alex Pellerin, Email: apelleri@bu.edu.
Kei Yasuda, Email: kei.yasuda.01@gmail.com.
Abraham Cohen-Bucay, Email: ACOHENBUCAY@mgh.harvard.edu.
Vanessa Sandra, Email: vs2718@cumc.columbia.edu.
Prachi Shukla, Email: pishukla7@gmail.com.
Barry K. Horne Jr, Email: bkhorne@bu.edu.
Gregory A. Viglianti, Email: gviglian@bu.edu.
Yao Xie, Email: xieyao@bu.edu.
Ulf Klein, Email: U.P.Klein@leeds.ac.uk.
Ying Tan, Email: ytan@bu.edu.
Ramon G. Bonegio, Email: bonegio@bu.edu.
Ian R. Rifkin, Email: irifkin@bu.edu.
References
- 1.Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012;18(6):871–882. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsokos GC. Autoimmunity and organ damage in systemic lupus erythematosus. Nat Immunol. 2020;21(6):605–614. doi: 10.1038/s41590-020-0677-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eames HL, et al. Interferon regulatory factor 5 in human autoimmunity and murine models of autoimmune disease. Transl Res. 2016;167(1):167–182. doi: 10.1016/j.trsl.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Barnes BJ. Genetic versus non-genetic drivers of SLE: implications of IRF5 dysregulation in both roads leading to SLE. Curr Rheumatol Rep. 2019;21(1):2. doi: 10.1007/s11926-019-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richez C, et al. IFN regulatory factor 5 is required for disease development in the FcgammaRIIB-/-Yaa and FcgammaRIIB-/- mouse models of systemic lupus erythematosus. J Immunol. 2010;184(2):796–806. doi: 10.4049/jimmunol.0901748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savitsky DA, et al. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc Natl Acad Sci U S A. 2010;107(22):10154–10159. doi: 10.1073/pnas.1005599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tada Y, et al. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 2011;63(3):738–748. doi: 10.1002/art.30183. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, et al. Pleiotropic IFN-dependent and -independent effects of IRF5 on the pathogenesis of experimental lupus. J Immunol. 2012;188(8):4113–4121. doi: 10.4049/jimmunol.1103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D, et al. Irf5-deficient mice are protected from pristane-induced lupus via increased Th2 cytokines and altered IgG class switching. Eur J Immunol. 2012;42(6):1477–1487. doi: 10.1002/eji.201141642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ban T, et al. Lyn kinase suppresses the transcriptional activity of IRF5 in the TLR-MyD88 pathway to restrain the development of autoimmunity. Immunity. 2016;45(2):319–332. doi: 10.1016/j.immuni.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434(7030):243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 12.Schoenemeyer A, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280(17):17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda K, et al. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178(11):6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- 14.Paun A, et al. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J Biol Chem. 2008;283(21):14295–14308. doi: 10.1074/jbc.M800501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryzhakov G, et al. Activation and function of interferon regulatory factor 5. J Interferon Cytokine Res. 2015;35(2):71–78. doi: 10.1089/jir.2014.0023. [DOI] [PubMed] [Google Scholar]
- 16.Christensen SR, Shlomchik MJ. Regulation of lupus-related autoantibody production and clinical disease by Toll-like receptors. Semin Immunol. 2007;19(1):11–23. doi: 10.1016/j.smim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15(11):1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura T, et al. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 19.Barnes BJ, et al. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276(26):23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 20.Matta B, Barnes BJ. Coordination between innate immune cells, type I IFNs and IRF5 drives SLE pathogenesis. Cytokine. 2020;132:154731. doi: 10.1016/j.cyto.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yasuda K, et al. Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int Immunol. 2013;25(5):295–306. doi: 10.1093/intimm/dxs114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krausgruber T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12(3):231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 23.Lien C, et al. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci U S A. 2010;107(10):4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang CM, et al. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012;13(5):421–430. doi: 10.1038/gene.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorner T, Lipsky PE. Beyond pan-B-cell-directed therapy - new avenues and insights into the pathogenesis of SLE. Nat Rev Rheumatol. 2016;12(11):645–657. doi: 10.1038/nrrheum.2016.158. [DOI] [PubMed] [Google Scholar]
- 26.Barnas JL, et al. B cell targeted therapies in autoimmune disease. Curr Opin Immunol. 2019;61:92–99. doi: 10.1016/j.coi.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teichmann LL, et al. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity. 2013;38(3):528–540. doi: 10.1016/j.immuni.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watkins AA, et al. IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J Immunol. 2015;194(4):1467–1479. doi: 10.4049/jimmunol.1402807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manni M, et al. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat Immunol. 2018;19(4):407–419. doi: 10.1038/s41590-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A, et al. Follicular dendritic cell activation by TLR ligands promotes autoreactive B cell responses. Immunity. 2017;46(1):106–119. doi: 10.1016/j.immuni.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8(7):665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 32.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103(37):13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishman D, Isenberg DA. Splenic involvement in rheumatic diseases. Semin Arthritis Rheum. 1997;27(3):141–155. doi: 10.1016/S0049-0172(97)80013-3. [DOI] [PubMed] [Google Scholar]
- 34.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358(9):929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 35.De S, et al. B cell-intrinsic role for IRF5 in TLR9/BCR-induced human B cell activation, proliferation, and plasmablast differentiation. Front Immunol. 2017;8:1938. doi: 10.3389/fimmu.2017.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weening JJ, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. 2004;15(2):241–250. doi: 10.1097/01.ASN.0000108969.21691.5D. [DOI] [PubMed] [Google Scholar]
- 37.Clausen BE, et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 38.Abram CL, et al. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods. 2014;408:89–100. doi: 10.1016/j.jim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corsiero E, et al. Ectopic lymphoid structures: powerhouse of autoimmunity. Front Immunol. 2016;7:430. doi: 10.3389/fimmu.2016.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco P, et al. T follicular helper (Tfh) cells in lupus: activation and involvement in SLE pathogenesis. Eur J Immunol. 2016;46(2):281–290. doi: 10.1002/eji.201545760. [DOI] [PubMed] [Google Scholar]
- 41.Knox JJ, et al. T-bet+ memory B cells: generation, function, and fate. Immunol Rev. 2019;288(1):149–160. doi: 10.1111/imr.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, et al. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood. 2011;118(5):1294–1304. doi: 10.1182/blood-2011-01-330530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arkatkar T, et al. B cell-derived IL-6 initiates spontaneous germinal center formation during systemic autoimmunity. J Exp Med. 2017;214(11):3207–3217. doi: 10.1084/jem.20170580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barr TA, et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J Exp Med. 2012;209(5):1001–1010. doi: 10.1084/jem.20111675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endres R, et al. Mature follicular dendritic cell networks depend on expression of lymphotoxin beta receptor by radioresistant stromal cells and of lymphotoxin beta and tumor necrosis factor by B cells. J Exp Med. 1999;189(1):159–168. doi: 10.1084/jem.189.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumanov AV, et al. Cellular source and molecular form of TNF specify its distinct functions in organization of secondary lymphoid organs. Blood. 2010;116(18):3456–3464. doi: 10.1182/blood-2009-10-249177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruglov A, et al. Contrasting contributions of TNF from distinct cellular sources in arthritis. Ann Rheum Dis. 2020;79(11):1453–1459. doi: 10.1136/annrheumdis-2019-216068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Pelaez M, et al. Protein kinase IKK beta-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci U S A. 2014;111(49):17432–1747. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27(5):801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan H, et al. TLR7, a third signal for the robust generation of spontaneous germinal center B cells in systemic lupus erythematosus. Cell Mol Immunol. 2018;15(3):286–288. doi: 10.1038/cmi.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeFranco AL. Germinal centers and autoimmune disease in humans and mice. Immunol Cell Biol. 2016;94(10):918–924. doi: 10.1038/icb.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seth A, Craft J. Spatial and functional heterogeneity of follicular helper T cells in autoimmunity. Curr Opin Immunol. 2019;61:1–9. doi: 10.1016/j.coi.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong S, et al. B cells are the dominant antigen-presenting cells that activate naive CD4+ T cells upon immunization with a virus-derived nanoparticle antigen. Immunity. 2018;49(4):695–708. doi: 10.1016/j.immuni.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Kawagoe T, et al. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J Exp Med. 2007;204(5):1013–1024. doi: 10.1084/jem.20061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson SM, et al. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204(9):2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liou HC, et al. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14(8):5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bossaller L, et al. Overexpression of membrane-bound fas ligand (CD95L) exacerbates autoimmune disease and renal pathology in pristane-induced lupus. J Immunol. 2013;191(5):2104–2114. doi: 10.4049/jimmunol.1300341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins AA, et al. Evaluating the role of nucleic acid antigens in murine models of systemic lupus erythematosus. Methods Mol Biol. 2014;1169:143–158. doi: 10.1007/978-1-4939-0882-0_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.