Abstract
Transcriptional activation by retinoids is mediated through two families of nuclear receptors, all-trans-retinoic acid (RARs) and 9-cis retinoic acid receptors (RXRs). Conformationally restricted retinoids are used to achieve selective activation of RAR isotype α, β or γ, which reduces side effects in therapeutical applications. Synthetic retinoids mimic some of all-trans retinoic acid biological effects in vivo but interact differently with the ligand binding domain of RARα and induce distinct structural transitions of the receptor. In this report, we demonstrate that RAR-selective ligands have distinct quantitative activation properties which are reflected by their abilities to promote interaction of DNA-bound human RXRα (hRXRα)-hRARα heterodimers with the nuclear receptor coactivator (NCoA) SRC-1 in vitro. The hormone response element core motifs spacing defined the relative affinity of liganded heterodimers for two NCoAs, SRC-1 and RIP140. hRXRα activating function 2 was critical to confer hRARα full responsiveness but not differential sensitivity of hRARα to natural or synthetic retinoids. We also provide evidence showing that lysines located in helices 3 and 4, which define part of hRARα NCoA binding surface, contribute differently to (i) the transcriptional activity and (ii) the interaction of RXR-RAR heterodimers with SRC-1, when challenged by either natural or RAR-selective retinoids. Thus, ligand structure, DNA, and RXR exert allosteric regulations on hRARα conformation organized as a DNA-bound heterodimer. We suggest that the use of physically distinct NCoA binding interfaces may be important in controlling specific genes by conformationally restricted ligands.
Analysis of the mechanisms by which cognate ligands activates nuclear receptors (NRs) identified structural transitions occurring in the ligand-bound receptor (holo receptor) compared to the unliganded form (apo receptor). Considerable progress has been made upon resolution of crystal structures of the apo form of 9-cis retinoid acid receptor (retinoid X receptor) isotype α (apo-RXRα), the halo form of all-trans-retinoic acid receptor isotype γ (holo-RARγ), thyroid hormone receptor β (T3R-β), estrogen receptor (ER), and progesterone receptor bound either to natural and synthetic agonists or to antagonists (4, 5, 48, 59, 61, 62). The ligand binding domains (LBDs) of these NRs share a common structural fold defining a ligand binding pocket (LBP) and functional regions, among which the activating function 2 (AF-2) activating domain (AF2-AD), located in helix 12 of RARs, is the most conserved. Upon ligand binding, the LBD adopts a more compact structure in which the AF2-AD is folded against the LBD, creating a new interface (ligand-dependent AF-2) suitable for NR coactivator (NCoA) binding (41). Additional structural alterations lead to the disruption of the nuclear corepressor (SMRT/TRAC, N-CoR/RIP13) binding site, located at the N terminus of the LBD, allowing p/CAF binding to the DNA binding domain of RAR (3). Agonist-bound NRs bind p160 nuclear factors (SRC-1/NCoA1, p/CIP/AIB1/ACTR/TRAM1/RAC3, TIF2/GRIP1/NCoA2), other unrelated coactivators (CBP [CREB binding protein]/p300, TIF1, RIP140) and multicomponent complexes (TRAPs, DRIPs) in vitro and/or in vivo (reviewed in reference 58). Spacing of core consensus sequences LXXLL found in p160-receptor interacting domains is critical for the specific ligand-dependent interaction of these coactivators with the AF-2 of distinct NRs (10, 36, 44). p160 coactivators interact in turn with CBP/p300, generating a large complex with histone acetyltransferase activity, catalyzing a posttranslational modification of histone tails which is important for regulating RXR-RAR heterodimers to nucleosomal DNA (31). Thus, the assembly of a transcriptionally active multimeric complex sensitive to combinatorial regulation is dependent, at least in part, on the fit between the receptor AF-2 interaction surface and coactivator hydrophobic motifs.
How ligand structure specifies the use of a given nuclear corepressor and coactivator repertoire remains an open question. Comparison of crystal structures of RARy LBD bound to either all-trans retinoic acid (atRA) or 9-cis retinoic acid (9-cis RA) or a synthetic retinoid highlighted very similar structures, showing that ligands positioned in the LBP identically and induced similar structural changes in the LBD (19). This finding suggests that similar structural transitions might confer identical transcriptional properties to the holo receptor. Conversely, distinct structural arrangements would generate receptors with distinct activating, and transrepressive, activities. However, these structural data provide little information on how ligands with different structures may control the transcriptional activity of heterodimeric complexes bound to DNA, which is the transcriptionally active conformation of RARs. The dimerization partners for RAR, RXR, and DNA binding have been indeed shown to be allosteric effectors which modulate the transcriptional activity of RARs (21, 24).
Using a simple, homogenous system to compare the transactivating and transrepressive properties of synthetic and natural retinoids, we reported recently that mutating amino acids in the loop between helices 11 and 12 (L box) of human RARα (hRARα) could impinge on receptor function in a ligand structure-dependent manner (26). We showed that a single mutation in the L box not only led to distinct transactivating and transrepressive properties in the presence of atRA or CD367, a synthetic agonist with receptor binding properties similar to those of atRA, but also to different capacities to interact with NCoAs and SMRT, a nuclear corepressor (26). Ligand binding studies also suggested that retinoids were positioned differently in the LBP (27, 28), leading to the conclusion that retinoids could bind to hRARα with similar affinities but generated holo receptors with different transcriptional properties.
In this report, we were particularly interested in evaluating this latter possibility, i.e., that retinoid binding to the same receptor can elicit distinct transcriptional responses, and to elucidate the molecular basis for these differences. We first evaluated the relative efficiency and potency of several natural and synthetic retinoids to activate a simple promoter through hRXRα-hRARα heterodimers. While binding to hRARα with similar affinities, these ligands displayed different abilities to activate transcription. We compared the ability of DNA-bound hRXRα-hRARα heterodimers to recruit two transcriptional coactivators, SRC-1 and RIP140. Both proteins bind to RARs and RXRs in a ligand-dependent manner but differ in structural organization (reference 16 and references therein). SRC-1 contains four LXXLL motifs, whereas RIP140 harbors nine of these signature sequences required for coactivator binding to NRs. In addition, spacing between LXXLL motifs located in receptor interacting domains (RIDs) of these coactivators, as well as flanking sequences, are different and may therefore introduce some specificity in binding to receptors. A correlation was found with transcriptional potency and the ability of ligands to promote recruitment of SRC-1 RID, but not RIP140 RID, to hRXRα-hRARα heterodimers in vitro. Retinoic acid response elements (RAREs) acted as allosteric modulators which altered the ability of hRARα to interact with NCoAs in response to ligand binding. Mutations designed to alter hRARα AF-2 and thus its interaction with NCoAs had distinct effects depending on the activating ligand. Finally, we demonstrate that the hRXRα AF2-AD is required for maximal activation of hRARα by natural or synthetic retinoids. These results suggest that RAR transcriptional activity is controlled through ligand structure and that both DNA and its obligate dimerization partner RXR act as major allosteric effectors.
MATERIALS AND METHODS
Materials.
atRA was obtained from Sigma (Saint Quentin Fallavier, France). Other synthetic retinoids were a gift from U. Reichert, CIRD-Galderma, Valbonne, France. [35S]methionine was purchased from Amersham (Les Ulis, France). Radioinert atRA as well as antiproteases were purchased from Sigma (St. Louis, Mo.). Acrylamide-bisacrylamide mix (Protogel) was from National Diagnostics (Atlanta, Ga.). Ampicillin and kanamycin were from Appligene (Strasbourg, France). Restriction and DNA modification enzymes were from Promega (Madison, Wis.). Oligonucleotides were purchased from Eurogentec (Le Sart-Tilman, Belgium). Site-directed mutagenesis reactions were carried out with the Promega GeneEditor system. Polyethyleneimine (ExGen 500) was from EuroMedex (Souffelweyersheim, France).
Plasmids.
Constructs containing either the wild-type (wt) or mutated hRARα and wt hRXRα cDNAs subcloned into pSG5 (Stratagene) have been described elsewhere (26, 30, 47). The pGDX-hRARα vector was obtained by subcloning the hRARα cDNA as an EcoRI fragment into pGEM3Z. Mutations at K244 and K262 were generated by using the appropriate oligonucleotide containing the desired mutation and an additional silent mutation either introducing a new restriction site or inactivating a restriction site present in the wt sequence. The mutagenic primers used were K224A (5′-CATTAAGACTGTGGAATTCGCCGCGCAGCTGCCCGGC-3′ [new EcoRI site]) and K262A (5′-GATCACCCTCCTCGCGGCTGCCTGCCTGG-3′ [EcoNI site lost]) (mutations are indicated in boldface). The pGDX-hRARα vector was used as a template in these experiments. The AF-2-deleted hRXRα (dnRXR) expression vector was created by isolating the truncated hRXRα cDNA from cdmRXRαΔ19C (64) which was subcloned into the pSG5-hRXRα backbone. Glutathione S-transferase (GST)–RIP140 was a kind gift from V. Cavailles and contained a fragment of the human cDNA coding for amino acids 752 to 1158 (6). GST–SRC-1 was obtained from M. J. Tsai and B. O’Malley and encoded a fusion protein of GST with human SRC-1 spanning amino acids 382 to 842 (45). GST-CBP was obtained from T. Kouzarides and encodes a fusion protein of GST to the N-terminal domain of CBP (amino acids 1 to 1099 [2]). The reporter gene p(TREpal)3 Luc was constructed by inserting three repeats of the synthetic thyroid response element AGGTCATGACCT (TREpal motif) upstream of the adenovirus major late promoter TATA box present in the pTATA Luc vector (26). All sequences were checked by restriction analysis and automatic sequencing.
Cell culture and transfections.
HeLa cells were cultured as monolayers in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (both from BioWhittaker, Verviers, Belgium). Cells were treated with retinoids at the indicated concentrations for 16 h. Retinoids were solubilized in dimethyl sulfoxide (DMSO); vehicle final concentration was 0.1%. Transfections were carried out by the polyethyleneimine coprecipitation method as described elsewhere (26). The luciferase assay was performed with the LucLite system (Packard Instruments, Rungis, France) according to the manufacturer’s guidelines, and activity (as relative luciferase units was measured with a LumiCount plate reader (Packard).
GST pull-down experiments.
The protocol used has been published elsewhere (26). DNA-dependent protein-protein interactions were tested as follows. hRARα and 35S-labeled hRXRα were synthesized by using a Quick T7 TnT kit (Promega). The amount of receptors was quantified by trichloroacetic acid precipitation of labeled samples, and volumes were adjusted to reach identical concentrations of both receptors in the translation mix. The integrity and the ability of receptors to form heterodimers were assessed by size exclusion chromatography on a Superdex 200 HR 10/30 column (Amersham Pharmacia BioTech, Les Ulis, France). In a typical binding reaction, 5 to 10 pmol (5 μl) of hRARα and 35S-labeled hRXRα were incubated with vehicle (DMSO) or 1 μM atRA for 60 min at 4°C in a 150-μl final volume. The binding buffer consisted of 20 mM HEPES (pH 7.4), 150 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, 0.1% Nonidet P-40, and 0.1% gelatin (Bio-Rad). A 20-mer double-stranded oligonucleotide containing the TREpal response element AGGTCATGACCT was added to the binding mix to a final concentration of 0.1 μM or was substituted by other response elements when indicated in the text. Direct repeat response element sequences were as follows: DR1, cggtagGGTTCAaAGGTCActcg; DR2, cggtagGGTTCAgaAGGTCActcg; DR3, cggtagGGTTCAcgaAGGTCActcg; DR4, cggtagGGTTCAcgaaAGGTCActcg; and DR5, cggtagGGTTCAccgaaAGGTCActcg (half site sequences are indicated in uppercase). The TREpal sequence is cggtagAGGTCATGACCTctcg. In these conditions, more than 98% of receptors formed heterodimers, irrespective of the presence of the ligand (28). After a 2-h incubation on ice, 40 μl of a Sepharose–glutathione-GST–SRC-1 slurry was added to the mix and agitated slowly on a rotating wheel for 2 h at 4°C. Unbound material was removed by three successive washes of Sepharose beads by 10 volumes of ice-cold 1× phosphate-buffered saline–2 mM dithiothreitol. Resin-bound receptors were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% gel and detected by autoradiography or quantified with a PhosphorImager (Molecular Dynamics). Values were averaged from at least four independent experiments carried out with two different bacterial extracts.
RESULTS
Retinoids exhibit distinct hRARα-activating properties.
The retinoids used in this study belong to distinct structural classes, differ in the ability to activate selectively RAR isotypes (α, β, and γ [Table 1]), but share a common feature, which is the ability to bind hRARα with dissociation constants (Kd) in the nanomolar range. The natural derivatives of vitamin A (atRA and 9-cis RA) and synthetic retinoids CD367 and TTNPB bind to and activate RARα, -β and -γ. 9-cis RA is a panagonist which activates RARs and RXRs. Replacing the propenyl linker in the TTNPB structure by a carboxamide yields the RARα-selective ligand Am580. These synthetic compounds are characterized, compared to natural retinoids, by the aromatization of the polyenic chain, restricting their flexibility. Chemical structures and biological activities of these retinoids have been described in detail elsewhere (11, 38; reviewed in reference 55). CD3106 (AGN 193109) is a C-1-substituted acetylenic RAR antagonist (20), and CD2425 (AGN 190701) is an RXR-selective ligand with high affinity for α, β, and γ isoforms. A luciferase reporter construct carrying the TREpal response element was cotransfected in HeLa cells together with hRARα and hRXRα expression vectors. In these conditions, the activity of endogenous receptors was not detected (29), whereas all other reporter genes containing a direct repeat RARE were found to be sensitive to endogenous receptor activity. We note that the TREpal response element is activable by both RXR homodimers and RAR-RXR heterodimers (65). CD2425, an RXR-specific retinoid, induced reporter gene activity only weakly, and simultaneous treatment of target cells with CD2425 and Am580 led to a reporter gene activity equivalent to that observed in the presence of 9-cis RA (29). The residual activity detected in the presence of CD2425 may be attributed to RXR homodimerization, since RXR-RAR heterodimers are not responsive to RXR ligands. We conclude that our system reflects hRXRα-hRARα dimer transcriptional activity and that the TREpal response element provides allosteric regulations similar to those observed with DR2 and DR5 response elements. Dose-response curves carried out with selected retinoids from a concentration of 10−10 to 10−6 M demonstrated that no correlation could be established between the affinity of the ligand for hRARα and its ability to activate transcription from this minimal promoter, containing the TREpal response element and a TATA box. As shown in Table 1, ligand concentrations required for half-maximal activation of the reporter gene by the wt hRXRα-hRARα pair (50% effective concentration [EC50]) varied for each retinoid compared to atRA, which was chosen throughout this study as the reference ligand. Remarkably, synthetic agonists binding with a high affinity for hRARα (TTNPB, CD367, and Am580) were the most efficient at reaching 50% maximal activation, whereas atRA and 9-cis RA displayed EC50s one order of magnitude higher than those of synthetic retinoids. In contrast, these two natural stereoisomers induced higher maximal levels of reporter gene activity at 1 μM. The highest rate of activation in the presence of synthetic agonists varied from 44 to 90% compared to atRA, suggesting that maximal occupancy of hRARα by these ligands still reveals different abilities of retinoids to promote transcriptional activation. Increasing further retinoid concentrations in some cases led to a slightly higher level of reporter gene activity (10 to 15% for CD367) but in other cases inhibited cell proliferation and triggered cell death in variable ratio, resulting in an apparently decreased reporter gene activity (atRA and other synthetic retinoids). In conclusion, no clear correlation between Kd and EC50 or between EC50 and relative potency can be established. These observations showing that a given retinoid exhibits distinct quantitative activating properties (compared to other RAR-selective ligands binding to the receptor with a similar affinity) suggest that each ligand could modulate differently one or several receptor functions. Ligand binding can potentially modify multiple protein-protein interaction interfaces. First, the dimerization interface can be altered, and thus ligands might display distinct abilities to promote hRARα binding to hRXRα. Although we observed, using quantitative in vitro (GST pull-down) assays and in vivo protein-protein interaction assays (two-hybrid assay in mammalian cells), that ligand binding triggers dimerization of both partners in a DNA-independent manner, these assays did not provide evidence for a regulation at this level (11a). Second, either corepressor displacement from or coactivator recruitment to hRXRα-hRARα heterodimers bound to DNA could be altered, and we thus compared the ligands for the ability to modulate these interactions.
TABLE 1.
Transcriptional activities of hRARα challenged with natural or synthetic retinoidsa
| Ligand | Biological activity | Kd (RAR; nM) | EC50 (RAR/RXR; nM) | Relative potency (RAR/RXR; %) |
|---|---|---|---|---|
| atRA | RARα, -β, and -γ agonist | 3.3 | 10.80 | 100 |
| 9-cis RA | Panagonist | 7.1 | 18.80 | 187.8 |
| TTNPB | RARα, -β, and -γ agonist | 2.2 | 1.05 | 90.2 |
| CD367 | RARα, -β, and -γ agonist | 4.0 | 0.91 | 55.0 |
| Am580 | RARα agonist | 8.1 | 0.74 | 72.5 |
| CD3106 | RARα, -β, and -γ antagonist | 20.0 | ND | 0 |
| CD2425 | RXRα, -β, and -γ agonist | >10,000 | 10 | 15 |
The main features of each ligand (biological activity in transient transfection assays and selectivity; Kd for hRARα) are indicated. In all cases but one, Kd values were obtained as described in reference 27; data for CD3106 were taken from reference 20. HeLa cells were transfected with hRARα and hRXRα expression vectors and the TREpal-TATA Luc reporter gene. The TREpal sequence is cggtagAGGTCATGACCTctcg (26). EC50 and relative potency values were deduced from dose-response curves in which the y axis represented the average luciferase activity (expressed as the percentage of the response observed in the presence of 1 μM atRA; n = 6 from eight independent experiments), and the x axis represented concentrations of ligand. Relative potency values represent the maximal luciferase activity observed with the tested ligand relative to that observed in the presence of 1 μM atRA. EC50s represent the ligand concentration yielding half-maximal luciferase activity. Standard errors never exceeded 8%. Chemical names of retinoids: CD367, 4-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-anthracenyl)-benzoic acid; TTNPB, (E)-4[2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthyl)propen-1-y1]benzoic acid; Am580, 4-[(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)carboxamido]benzoic acid; CD2425 (AGN 190701), ((E)-2-[2-(5,6,7,8-tetrahydro-3,5,5,8,8-pentamethyl-2-naphthyl)propen-1-yl]-4-thiophenecarboxylic acid. ND, not determined.
RAR-selective agonists are equally efficient with respect to triggering SMRT release from monomeric or DNA-bound heterodimeric hRARα.
We used a quantitative protein-protein interaction assay to test retinoids for the ability to promote either SRC-1 or RIP140 binding to, or SMRT release from, RXR-RAR heterodimers synthesized by coupled in vitro transcription-translation. For this purpose, we used conditions in which the hRXRα-to-hRARα and DNA-to-receptor ratios yielded 98 to 100% heterodimeric complexes bound to DNA, as demonstrated by size exclusion chromatography and electrophoretic mobility shift assay (29). Importantly, no receptor monomer could be detected in these conditions. Thus, we generated affinity matrices with SMRT, RIP140 or SRC-1 RIDs fused to GST, which were incubated with either monomeric hRARα when indicated or performed hRXRα-hRARα dimers bound to the TREpal RARE. In preliminary experiments, the formation of heterodimers was monitored by using labeled hRARα and hRXRα (29). Note that in experiments presented below, receptor complexes in which only hRXRα was labeled were incubated with 10 μM retinoid for 2 h before adsorption to GST fusion proteins.
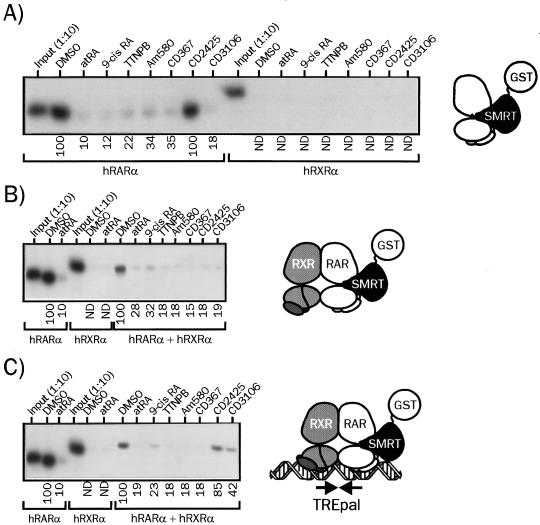
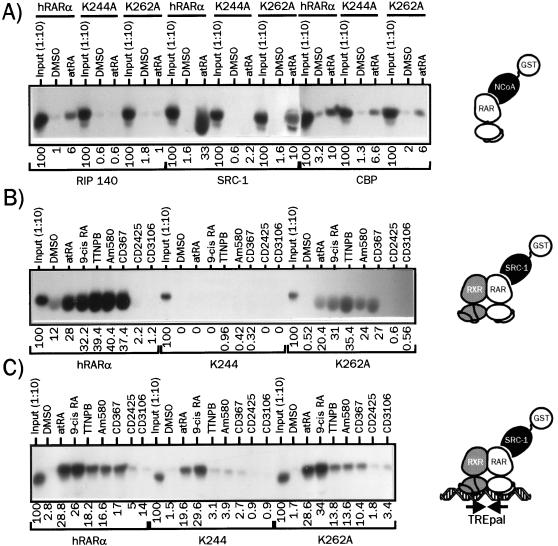
We first compared natural and synthetic retinoids for the ability to release SMRT RID fused to GST from monomeric hRARα (Fig. 1A), from hRXRα-hRARα dimers (Fig. 1B), or from heterodimers bound to the TREpal response element (Fig. 1C). As previously reported (7, 26), the monomeric, wt hRARα bound strongly to SMRT in the absence of ligand, and all retinoids induced SMRT release with similar efficiencies except CD2425, an RXR-selective retinoid. Interestingly, we note that CD3106, an RAR antagonist, was able to displace SMRT from hRARα as efficiently as agonist compounds. On the opposite, hRXRα interaction with this corepressor was not detectable. When SMRT was complexed to hRXRα-hRARα heterodimeric complexes, the ability of these ligands, except for CD2425, to mediate SMRT release was unaffected (Fig. 1B). The RXR-specific ligand was in this configuration able to promote SMRT release, suggesting that liganded RXR exerts allosteric regulation on hRARα-SMRT binding interface. The interaction of TREpal-bound heterodimers with SMRT was modulated in a manner similar to that observed with heterodimeric receptors when synthetic RAR agonists are involved (Fig. 1C). Remarkably, the RXR-selective ligand and to a lesser extent the RAR antagonist displayed decreased abilities to induce SMRT release, showing that DNA binding clearly exerts allosteric regulation on RAR nuclear corepressor binding surface. Since similar results were obtained for DR1, DR2, and DR5 RAREs (29), we conclude that whatever their structure, retinoids have similar SMRT binding interface remodelling properties and that no significant allosteric regulation can be detected in the presence of DNA and hRXRα.
FIG. 1.
Retinoids display similar efficiencies with respect to displacing SMRT from monomeric hRARα and DNA-bound hRXRα-hRARα heterodimers. (A) 35S-labeled hRARα or hRXRα was incubated in the presence of the GST-SMRT (982–1495) fusion protein for 2 h and then challenged with the indicated retinoids. SMRT-receptor complexes were isolated by adsorption of the SMRT moiety on agarose-coupled glutathione. After the beads were washed, complexes were analyzed by SDS-PAGE (8% gel), and receptor content was assayed by autoradiography. (B) 35S-labeled hRXRα and unlabeled hRARα were incubated for 2 h with GST-SMRT and then for an additional 2 h in the presence of the indicated ligand. Complexes were treated and analyzed as described above. (C) TREpal-bound heterodimers were bound to GST-SMRT and further activated by the indicated retinoids. Complexes bound to SMRT were isolated and analyzed by adsorption to agarose-linked glutathione SDS-PAGE (8% gel), and autoradiography as described above. The TREpal sequence is cggtagAGGTCATGACCTctcg. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to SMRT in the presence of the indicated ligand relative to that measured in the presence of vehicle (DMSO), defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. ND, not determined. Standard errors never exceeded 8.7%. Representative autoradiograms are shown.
DNA binding modulates the ability of structurally distinct retinoids to promote SRC-1 or RIP140 recruitment by hRXRα-hRARα heterodimers.
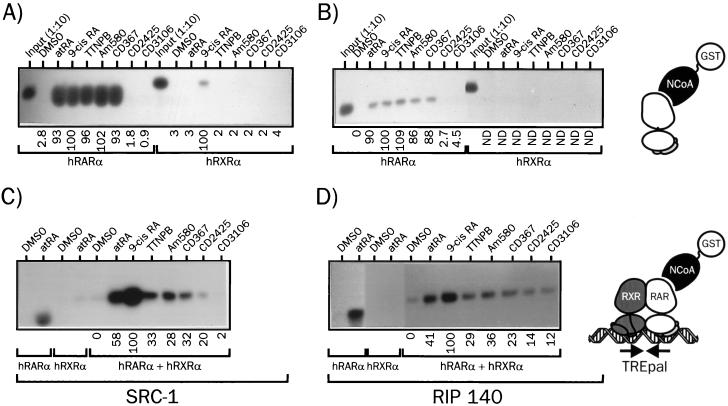
In view of the lack of difference in the abilities of natural and synthetic retinoids to promote SMRT release from hRARα, we analyzed their effects on the ligand-induced recruitment of several NCoA, in the context both of the monomeric receptor and of the DNA-bound hRXRα-hRARα heterodimer (Fig. 2). As expected, monomeric hRARα complexed to atRA recruited SRC-1 very efficiently. Similar activities were observed for its natural stereoisomer, 9-cis RA, and all RAR-selective ligands (TTNPB, CD367, and Am580). On the contrary, neither the RAR antagonist CD3106 (AGN 193109) nor the RXR-selective ligand CD2425 (AGN 190701) was able to induce SRC-1 binding to hRARα (Fig. 2A). When RIP140 RID was used as a bait, identical results were obtained, although it appeared clearly that this putative coactivator bound to hRARα with a lower affinity than SRC-1 (Fig. 2B). When similar assays were carried out with preassembled hRXRα-hRARα heterodimers on the TREpal RARE, several observations could be made. First, heterodimer binding to DNA induced a strong modulation of the capacity of ligands to bind to SRC-1 and RIP140 (Fig. 2C and D). atRA was twofold less efficient than 9-cis RA at promoting NCoA binding to DNA-bound heterodimers. 9-cis RA turned out to be the most efficient ligand, whereas synthetic RAR agonists had a clearly lower efficiency. CD3106 was unable to promote SRC-1 recruitment, in keeping with its antagonistic properties, whereas the RXR-specific ligand CD2425 enhanced only weakly SRC-1 binding to the complex. When RIP140 was used in this assay, similar results were obtained (Fig. 2D). However, we consistently observed that the RAR antagonist was in this context able to promote detectable binding of the heterodimer to RIP140, thereby excluding this protein as a potential coactivator in our system. Thus, DNA binding introduced dramatic changes in ligand capacity to promote SRC-1 recruitment, establishing a semiquantitative correlation between DNA-dependent protein-protein interaction assays and transcriptional activities in which natural retinoids were the most efficient ligands. Indeed, the 2-fold higher activity of 9-cis RA in transactivation assay is clearly correlated to its ∼2-fold higher ability to promote SRC-1 recruitment. This may obviously relate to its ability to activate both components of the dimer and thus allow binding of SRC-1 to both receptor AF-2 domains.
FIG. 2.
Assembly of hRARα into a DNA-bound heterodimer induces ligand-selective recruitment of SRC-1 and RIP140. (A) Interaction of hRARα or hRXRα with SRC-1 in the presence of various retinoids. 35S-labeled hRARα or hRXRα was incubated with the indicated activating ligand (10 μM) or vehicle (DMSO) for 2 h and then with the fusion protein GST-SRC1(382–842). Complexes were precipitated with glutathione-Sepharose beads and extensively washed. Samples were analyzed by SDS-PAGE (8% gel), and receptors were detected by autoradiography. (B) Interaction of hRARα or hRXRα with RIP140. Receptors were synthesized in vitro as described in the text and incubated in the presence of retinoids and a GST-RIP140(752–1158) fusion protein. Following washing, proteins were resolved by SDS-PAGE and revealed by autoradiography. In input lanes, 1/10 of the input from in vitro-coupled transcription-translation mix was analyzed in parallel. (C) Interaction of DNA-bound hRXRα-hRARα heterodimers with SRC-1. 35S-labeled hRXRα, hRARα, and the DNA probe TREpal were incubated as described in Materials and Methods to ensure >95% heterodimer formation. These complexes were then incubated in the presence of the activating ligand for 2 h (10 μM) and then with the GST–SRC-1 fusion protein for an additional 2 h. Complexes were precipitated and analyzed as described above. (D) Interaction of DNA-bound hRXRα-hRARα heterodimers with RIP140. The procedure was similar to that described for panel C except that SRC-1 was substituted for RIP140. The TREpal sequence is cggtagAGGTCATGACCTctcg. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to SRC-1 or RIP140 in the presence of the indicated ligand relative to that measured in the presence of 9-cis RA, defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. ND, not determined. Standard errors never exceeded 5.3%. Representative autoradiograms are shown.
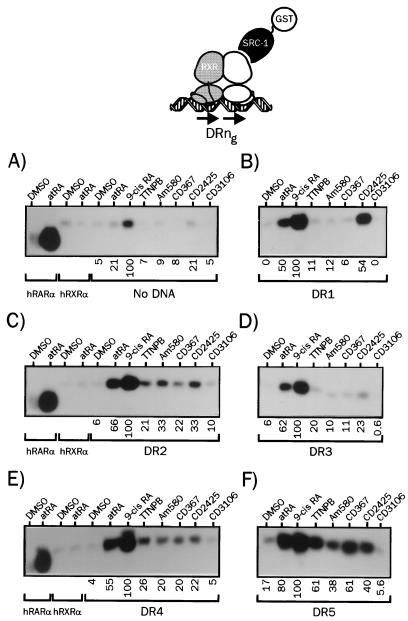
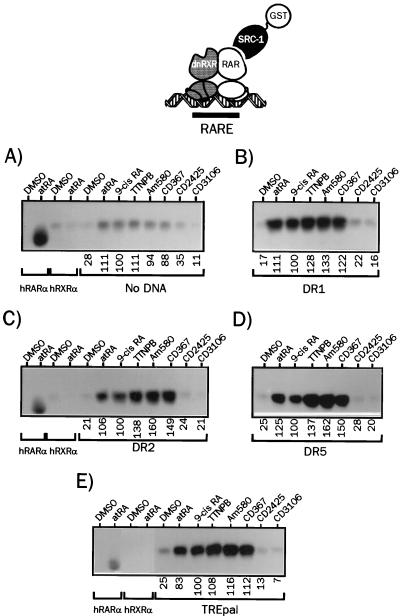
Direct repeats of the PuGG/TTCA motifs are recognition sequences for RXR-RAR heterodimers present in a number of natural promoters. They define a consensus response element to which RAR heterodimers bind strongly. Direct repeats of the PuGGTCA motifs separated by a spacer of variable length ranging from one to five nucleotides allow high-affinity binding of RXR-RAR heterodimers (37). DR1, DR2, and DR5 are the most commonly described natural RAREs, but DR3 and DR4, which mediate vitamin D3 and thyroid hormone responses, have been shown to behave as RAR-responsive elements depending on the promoter context (50). In addition, RAREs containing the AGGTCA motif as the second half site displayed significant affinities for RXR-RAR heterodimers in vitro, and cooperative bindings of RXR-RAR heterodimers to these RAREs involve distinct dimerization interfaces (37). Since DNA has been shown to be a major allosteric effector for a variety of transcription factors and NRs (reviewed in reference 32), we therefore evaluated the possible influence of various spacer lengths (and therefore of various dimerization interfaces) on the ability of retinoids to promote SRC-1 recruitment to hRXRα-hRARα heterodimers (Fig. 3). In the absence of DNA, binding of heterodimers to SRC-1 was barely detectable in the presence of RAR-specific ligands, whereas control experiments showed again a very efficient recruitment of this coactivator by monomeric hRARα (Fig. 3A). This finding confirms the specificity of our assay for receptor heterodimers. Note that only 9-cis RA was able to trigger significant SRC-1 binding, a property that could be related to its ability to bind both receptors, since coincubation of dimers with the RARα-specific ligand Am580 and the RXR-specific ligand CD2425 yielded identical results (29). hRXRα-hRARα heterodimers were then assembled on a DR1 response element on which heterodimers have a binding polarity opposite that observed with other direct repeat response elements; i.e., RAR is the 5′-bound receptor. In this configuration, SRC-1 recruitment was observed in the presence of atRA and was two- to threefold more pronounced with 9-cis RA. The RXR-specific ligand CD2425 was as efficient as atRA, whereas RAR-specific ligands displayed no activity in this system (Fig. 3B). Additional control experiments showed that heterodimers formed in these conditions, and therefore we conclude that apo-hRXRα formed, when bound to the 3′ half motif from the DR1 RARE, a high-affinity binding interface for SRC-1. We observed, however, that in experiments carried out with labeled RXR or RAR, and with labeled RAR and RXR, that the SRC-1-bound material appeared to be a mixed population consisting of about 60% RXR homodimers and 40% RXR-RAR heterodimers in the presence of CD2425 (n = 3) (29). In the presence of DR2 RARE, atRA- and 9-cis RA-complexed heterodimers bound avidly to SRC-1, and RAR-specific ligands also induced the recruitment of this coactivator, albeit with a lower efficiency (Fig. 3C). Increasing the spacer length to four nucleotides allowed stronger binding of SRC-1 to hRARα complexed to synthetic retinoids (Fig. 3E), and use of the high-affinity DR5 RARE yielded complexes that bound SRC-1 avidly (Fig. 3F). Although remaining weaker than in the presence of natural retinoids, binding of DR5-bound heterodimers to SRC-1 was the most efficient and revealed variations between ligands. Am580 turned out to be the least efficient in this configuration, whereas it was the strongest inducer in the presence of the DR2 RARE. Finally, we note that the RAR antagonist CD3106 prevented, in all configurations tested, heterodimer binding to SRC-1. This inhibition was also observed in competition experiments (29) in which heterodimers were challenged by atRA (100 nM) and CD3106 (1 μM).
FIG. 3.
The spacing between the two direct repeats of RAREs modulates ligand ability to promote SRC-1 recruitment to heterodimerized hRARα. (A) 35S-labeled hRXRα and hRARα produced by coupled in vitro transcription-translation were incubated in the presence of the indicated retinoids at 10 μM (final concentration), and complexes were bound to a GST–SRC-1 fusion protein. After a 2-h incubation, complexes were isolated by adsorption on a glutathione-linked agarose matrix and analyzed by SDS-PAGE (8% gel). (B to F) hRXRα-hRARα heterodimers associated to the indicated DRng RARE were analyzed for the ability to interact with SRC-1 in the presence of natural or synthetic retinoids as described above. The nature of the response element is indicated below each panel. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to SRC-1 in the presence of the indicated ligand relative to that measured in the presence of 9-cis RA, defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. Standard errors never exceeded 4.3%. Representative autoradiograms are shown.
In summary, ligand capacity to promote SRC-1 recruitment in vitro was found to be dramatically modulated by the response element structure to which receptor heterodimers are bound. The strongest binding always occurred with 9-cis RA and atRA, and we noted that triggering hRXRα AF-2 remodelling with CD2425 also induced SRC-1 recruitment for some response elements. This is in agreement with the weak but constant agonist activity of CD2425 detected in transient transfection assays using TREpal (Table 1) DR2 or DR5-driven tk Luc reporter genes (29). The DR5 RARE appeared to be the most efficient DNA template in that it favored a strong binding of SRC-1 to heterodimers that could relate to its higher inducibility in transcriptional activation assays.
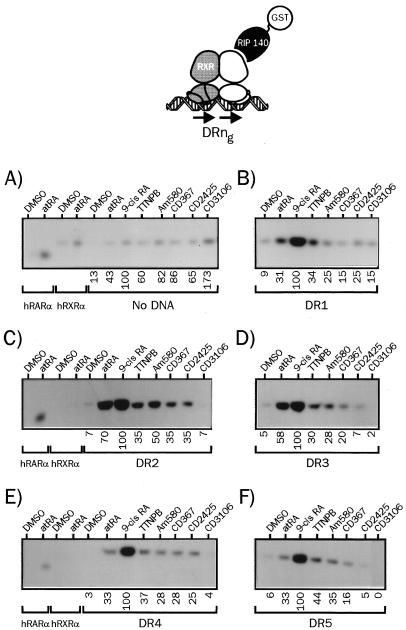
We then extended this DNA-dependent protein-protein interaction assay to RIP140 (Fig. 4), for which we have observed distinct requirement for binding to heterodimeric complexes compared to SRC-1 (see below). Such differences were also reported for the estrogen receptor (17). The lower affinity of RIP140 for hRXRα-hRARα dimers was again noted in these experiments, and receptor complexes exhibited the highest affinity for RIP140 when bound to atRA and 9-cis RA. More strikingly, the DR2 response element was the DNA template that allowed the most efficient binding of RIP140: atRA efficiently promoted SRC-1 recruitment to heterodimers bound to either the DR5, DR4, or DR1 probe (Fig. 3), whereas RIP140 bound more efficiently to atRA-complexed dimers associated to the DR2 or DR3 RARE. For other retinoids displayed, as observed for SRC-1, the ability to recruit RIP140 depended on the nature of the response element, with TTNPB being the most active synthetic ligand in most configurations. Note that CD2425 did not promote RIP140 binding to heterodimers associated with DR3 and DR5 RAREs. For other data, it is also clear that activation of both components by 9-cis RA led to a threefold potentiation of RIP140 binding on DR4 and DR5 RAREs, in opposition to SRC-1 recruitment, for which the synergy was only additive (compare Fig. 3E and F to Fig. 4E and F). Finally, we note that the antagonist CD3106 is the most efficient ligand in inducing RIP140 recruitment to heterodimers in the absence of DNA. This property may be related to the recently described repressive activity of RIP140 (25).
FIG. 4.
The nature of the RARE determines the relative affinity of liganded heterodimers for nuclear coactivators. (A) 35S-labeled hRXRα and hRARα produced by coupled in vitro transcription-translation were incubated in the presence of the indicated retinoids at 10 μM (final concentration), and complexes were bound to a GST-RIP140 fusion protein. After a 2-h incubation, complexes were isolated by adsorption on a glutathione-linked agarose matrix and analyzed by SDS-PAGE (8% gel). (B to F) hRXRα-hRARα heterodimers associated with the indicated DRng RARE were analyzed for the ability to interact with RIP140 in the presence of natural or synthetic retinoids as described above. The nature of the response element is indicated below each panel. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to RIP140 in the presence of the indicated ligand relative to that measured in the presence of 9-cis RA, defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. Standard errors never exceeded 6.2%. Representative autoradiograms are shown.
Thus, we conclude that the strength of protein-protein interaction may differ, for a given ligand and response element, from one coactivator to another, suggesting that transcriptional activation could be mediated by distinct NCoAs according to the type of RARE tethering hRXRα-hRARα dimers to retinoid-regulated promoters.
Transcriptional activation of hRARα by synthetic retinoids is compromised by mutations in helix 3 or helix 4.
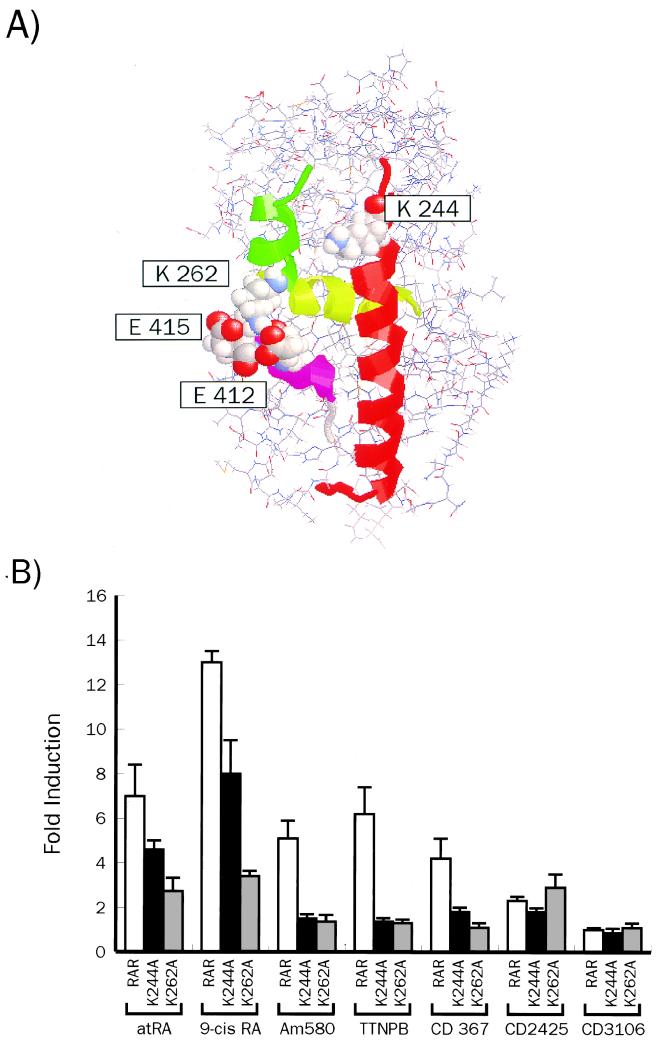
Previous work has established that the LBD of NRs undergoes structural transitions upon ligand binding. A lysine residue (K264) from helix 3 has been shown to establish salt bridges with glutamic acid side chains in the AF2-AD region of the monomeric holo-RARy LBD (48) which are essential for transcriptional activation by this receptor (Fig. 5A). The proposed stabilization of the active structure may, however, not be a feature common to all NRs, and could reflect the need for a hydrophilic region to which coactivators could bind. Such a role has been proposed for the highly conserved K366 of ER (17) and K288 of TR-β1 (14), equivalent to K244 of hRARα. Alternatively, it may also provide an alternative anchoring point for AF2-AD acidic residues (Fig. 5A). Whatever their actual role may be, we reasoned that mutations at K262 or K244 of hRARα could impinge differently on hRARα function according to the ligand used and therefore reflect a different architecture of the holo-HBD. We thus mutated K244 or K262 in hRARα and evaluated first the impact of these mutations on the transcriptional activity of hRXRα-hRARα heterodimers stimulated by natural and synthetic retinoids. When stimulated with atRA, both mutants displayed a diminished response to this ligand (60% of the wt hRARα activity). In the presence of 9-cis RA, the reporter gene activity was decreased to 50% of wt hRARα activity. More surprisingly, responsiveness to RAR-selective retinoids was abolished for the K244A receptor, whereas a lower but significant responsiveness was still observed for the K262A derivative (Fig. 5B). Thus, these amino acids, which define part of the AF-2 region of hRARα, are necessary to confer full responsiveness of hRARα to retinoids but are differently required for responsiveness to synthetic RAR-selective ligands, which displayed a strict requirement for K244.
FIG. 5.
Alteration of the nuclear coactivator binding interface inactivates selectively hRARα responsiveness to synthetic retinoids. (A) Three-dimensional modelling of the hRARα LBD showing the backbone of the polypeptide, helices 3, 4, and 12, and lysyl residues mentioned in the text (space-filling mode). The computer graphic was prepared with RasMol software. (B) Transcriptional activity of hRARα mutants K244A and K262A in the presence of natural and synthetic retinoids. HeLa cells were transfected with appropriate hRARα and hRXRα expression vectors and the TREpal-TATA Luc reporter gene (23) and stimulated with 1 μM retinoid. Luciferase activities represent induction values over the observed basal level (∼5,000 relative luciferase units in our conditions). Results represent the average values of six measurements ± standard error of the mean. Each assay was carried out six times.
Mutation of K244 or K262 abrogates SRC-1 recruitment in vitro to hRARα complexed to synthetic retinoids.
Assessing the effects of these mutations on various receptor functions, we found that both hRARα mutants displayed wt affinities for retinoids, were able to dimerize with hRXRα, and could bind as heterodimers to DNA in vitro. Furthermore, structural transitions upon ligand binding probed by limited proteolysis were identical to those observed with wt hRARα, and abilities of mutants to release SMRT were unaffected (29). Since our prediction was that these mutations could prevent NCoA recruitment, we tested first the capacity of monomeric RAR mutants to interact with RIP140, SRC-1, and CBP. Both mutations abolished RIP140 recruitment upon binding to atRA, whereas the K244A mutation selectively impaired SRC-1 binding. Interaction with the N-terminal portion of CBP was decreased by 50% for both mutants (Fig. 6A). As shown in Fig. 6B, hRXRα-hRARα dimers strongly interacted with SRC-1 in presence of natural or synthetic retinoids, whereas the hRXRα-K244A heterodimer did not recruit this coactivator regardless of the ligand used. hRXRα-K262A heterodimers recruited SRC-1 with a moderate affinity in the presence of RAR agonists, with no significant variation among ligands (Fig. 6B). Since we noted that the monomeric configuration did not reflect the transcriptional activity of receptors, we tested these mutants in the DNA-dependent protein-protein interaction assay described above, using SRC-1 as a bait (Fig. 6C). DNA-bound hRXRα-K244A as well as the hRXRα-K262A dimers interacted with SRC-1 when complexed to either atRA or 9-cis RA, as expected from transient transfections experiments (Fig. 5B). Note that the K262A derivative displayed a higher affinity for SRC-1 than did K244A. In the presence of synthetic ligands, SRC-1 did not bind to the K244A-containing heterodimer, whereas a fair level of activity was still detected with the K262A-containing heterodimers. Thus, in vitro protein-protein interaction assays established a correlation between transcriptional responsiveness and ability to recruit SRC-1. We conclude that K244 is an amino acid critical for synthetic retinoid-induced SRC-1 recruitment to hRARα.
FIG. 6.
Lysine 244 and lysine 262 are differentially involved in NCoA binding by liganded hRARα. (A) Interaction of monomeric hRARα and of K244A and K262A derivatives with RIP140, SRC-1, and CBP(1-1099). GST fusion proteins corresponding to the indicated coactivator were incubated in the presence of 35S-labeled, atRA-bound receptors. Complexes were isolated by adsorption on a glutathione-linked agarose beads and analyzed by SDS-PAGE (8% gel) as described for Fig. 1. (B) Interaction of monomeric wt and receptor mutants with SRC-1 in the presence of natural and synthetic retinoids. 35S-labeled hRARα derivatives were incubated with or without 10 μM ligand, and SRC-1-associated receptors were isolated and analyzed as described above. (C) Mutation of K244 disrupts synthetic retinoid-induced SRC-1 recruitment by DNA-bound heterodimers. The ability of each hRARα derivative to bind to SRC-1 when incorporated into a TREpal-bound heterodimer was assessed as described for Fig. 1 in the presence of the indicated ligand. In the control lanes, 1/10 of the coupled transcription-translation mix used to produce radioinert hRARα was used to assess the efficiency of synthesis by label incorporation. Products were analyzed in parallel with other samples containing labeled hRXRα. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to the indicated coactivator RID in the presence of the indicated ligand relative to total labeled receptor input, defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. Standard errors never exceeded 7.2%. Representative autoradiograms are shown.
The RXR AF2-AD is required for full responsiveness of hRARα to RAR-selective ligands.
Although initially described as a silent partner whose sole function is to increase the DNA binding affinity of RARs (21, 22), RXRs appeared to act synergistically with RARs (8, 12, 40). This synergy is dependent on the RXR AF-2 (Tc) region (8), but its contribution varies according to the structure of the response element (12, 13, 24). We were interested in testing whether the hRXRα AF-2 region participates differentially to hRARα activation by RAR-specific ligands. We therefore examined the ability of wt hRARα to activate the TREpal reporter gene in the presence of dnRXR. This hRXRα mutant, with a deletion of the 19 C-terminal amino acids (448 to 462), has been shown to bind DNA as either a homodimer or a heterodimer in vitro (65) but to display reduced transcriptional activity. The 9-cis RA-induced transcriptional activity of the wt hRARα-dnRXR was indeed much lower than that of the wt hRARα-wt hRXRα, reflecting the contribution of the liganded hRXRα to the transcriptional activity of the promoter. More surprisingly, hRXRα AF-2 deletion affected differentially the transcriptional activity induced by atRA or RAR-selective synthetic retinoids (Table 2). Indeed, atRA- and TTNPB-induced activities were the most severely affected by this mutation (−40% and −56%, respectively), whereas CD367 and Am580 displayed an 20 to 25% lower activity. However, we noted that EC50s were not affected, suggesting that this domain of RXR potentiates RAR AF2 activity without altering its sensitivity to retinoids.
TABLE 2.
The RXR AF-2 is necessary for full responsiveness of hRARα to natural and synthetic retinoidsa
| Ligand | Biological activity | Kd (RAR; nM) | EC50 (RAR/dnRXR; nM) | Relative potency (RAR/dnRXR; %) |
|---|---|---|---|---|
| atRA | RARα, -β, and -γ agonist | 3.3 | 8.70 | 46.0 |
| 9-cis RA | Panagonist | 7.1 | 12.60 | 55.4 |
| TTNPB | RARα, -β, and -γ agonist | 2.2 | 0.74 | 34.0 |
| CD367 | RARα, -β, and -γ agonist | 4.0 | 0.75 | 35.0 |
| Am580 | RARα agonist | 8.1 | 0.50 | 46.0 |
| CD3106 | RARα, -β, and -γ antagonist | 20.0 | NA | NA |
| CD2425 | RXRα, -β, and -γ agonist | >10,000 | ND | ND |
Features of retinoids are indicated as in Table 1. HeLa cells were transfected with hRARα and dnRXR expression vectors and the TREpal-TATA Luc reporter gene (26). EC50s and potency values were calculated as for Table 1 and deduced from dose-response curves as described above. NA, not detectable; ND, not determined.
Deletion of the hRXRα AF2-AD equalizes the capacity of retinoids to recruit SRC-1 by DNA-bound heterodimers.
The ability of dnRXR-hRARα heterodimers to recruit SRC-1 was characterized in different configurations. In the absence of DNA, heterodimer–SRC-1 interaction was weak and demonstrated no evidence of differences between retinoids (Fig. 7A), as shown for the wt hRXRα (Fig. 3). 9-cis RA lost its higher efficiency in this assay, as expected from the deletion of the hRXRα AF2-AD region. Upon assembly on a DR1 RARE, in contrast to its wt counterpart (Fig. 3), the dnRXR-hRARα dimer became able to recruit SRC-1 in the presence of RAR-specific retinoids. Again, no difference in the affinity for SRC-1 was noted with different retinoids, and as was also observed in the presence of DR2 (Fig. 7C), DR5 (Fig. 7D), TREpal (Fig. 7E), and DR3 and DR4 (29). This establishes a correlation between transcriptional responsiveness of dnRXR-hRARα heterodimers and their abilities to recruit SRC-1 in vitro, and so we conclude that the hRXRα AF-2 region is required for maximal activation of hRARα.
FIG. 7.
Deletion of RXR AF-2 abolishes selective retinoid-induced SRC-1 binding to hRXRα-hRARα heterodimers. (A) 35S-labeled dnRXRα and radioinert hRARα produced by coupled in vitro transcription-translation were incubated in the presence of the indicated retinoids at 10 μM (final concentration), and complexes were bound to a GST–SRC-1 fusion protein. After a 2-h incubation, complexes were isolated by adsorption on a glutathione-linked agarose matrix and analyzed by SDS-PAGE (8% gel). (B to E) dnRXRα-hRARα heterodimers associated to the indicated RARE were analyzed for the ability to interact with SRC-1 in the presence of natural or synthetic retinoids as described above. The nature of the response element is indicated below each panel. Numbers below gel lanes indicate the amount of radiolabeled receptor bound to SRC-1 in the presence of the indicated ligand relative to that measured in the presence of 9-cis RA, defined as 100%. Values represent mean data from four independent experiments carried out with two different bacterial extracts. Standard errors never exceeded 4.1%. Representative autoradiograms are shown.
DISCUSSION
Due to the highly pleiotropic effects of natural retinoids, which govern many important biological processes such as morphogenesis and cell growth, proliferation, differentiation, and death, much efforts have been devoted to the development of synthetic retinoids with the aim of improving the therapeutic properties of such compounds. The identification of RAR isotypes allowed the design of isotype-specific retinoids with improved clinical potential, and indeed targeting RAR isotypes hold great promises in the treatment of skin diseases (53, 54), and cancer chemoprevention and chemotherapy (35). Importantly, these conformationally restricted retinoids greatly facilitated the study of molecular mechanisms by which these ligands activate transcription through their cognate receptors (23, 46, 51). Transcriptional regulation by retinoid receptors is thought to be largely dependent on the ligand-dependent (AF-2), which undergoes major structural alterations upon ligand binding (reviewed in reference 41). Several lines of evidence demonstrated that these transitions are required to permit the interaction of the liganded receptor with nuclear coactivators (58). Yet, despite intense investigations, it remains unclear how ligand binding triggers these structural transitions and, importantly, whether these changes are affected by the shape of the ligand.
In this study, we demonstrated that natural and synthetic retinoids differ in the ability to activate a simple promoter, despite similar receptor binding properties. These results showed that ligands belonging to structurally distinct classes contribute differently to RAR-mediated transcription. These observations extend our previous conclusions drawn from the use of mutants of hRARα and synthetic retinoids (26), for which mutating amino acids in the helix 11-helix 12 loop had a different impact on the structure and transcriptional activity of hRARα according to the ligand used. Moreover, we (27, 28) and others (23, 56, 57) showed that structural requirements for agonist and antagonist binding to RARα were different and thus that LBPs for each ligand overlap but are not totally coincident. This body of data led us to speculate that different agonists could induce significantly different structural transitions that in turn would affect protein-protein interaction surfaces. While we observed no significant effect of the ligand structure on either heterodimerization properties or corepressor binding to hRARα, dramatic alterations of the ability of the receptor to bind to coactivators were noted. Data from transcriptional assays and DNA-dependent protein-protein interaction assays led us to several important conclusions.
First, the DNA-dependent protein-protein interaction assay established a clear correlation between transcriptional activity and binding of SRC-1 to DNA-bound heterodimers. Importantly, the hormone response element (HRE) was found to be critical (i) for establishing differential NCoA recruitment in response to ligand binding and (ii) according to the spacing between the two core motifs of the RARE, for modulating the relative affinity of heterodimers for NCoAs. Thus, the nature of the HRE to which hRXRα-hRARα dimers are bound, which has been shown to modulate the activity of the AF-2s of both partners (12, 24, 42), affects the capacity of RAR AF-2 to bind to coactivators and modify the relative affinity of RAR-containing heterodimers for these nuclear proteins. These observations may be of importance since we observed that putative or bona fide coactivators (RIP140, SRC-1, SRC-2, and SRC-3 [see reference 34 for nomenclature]) are coexpressed in a single cell type (29). These data also lead to the prediction that distinct direct repeat response elements would respond differently to retinoids. This hypothesis is currently under investigation but requires an experimental system in which no contribution of endogenous receptors to transcriptional activation is detectable. We indeed observed that DR1, DR2, and DR5 RARE-driven reporter genes are sensitive to endogenous receptors activity (29, 30), and all cell lines that we have tested so far coexpress several RAR and RXR isotypes (29). RAR coexpression implies that the transcriptional activity detected in response to high concentrations of retinoids (above 10−7 M) would reflect the contribution of several receptor isoforms in varying ratio, since ligands are selective only in a defined concentration range. However, we note that such differences were also described for CD367 and TTNPB in other systems using different receptor combinations and other reporter genes (1, 43, 63). Moreover, we and others also observed this differential response to natural and synthetic retinoids when studying the expression of endogenous genes driven either by a DR5 RARE (RARβ [29, 49]) or a DR2 RARE (CRABPII [49]). These observations strengthen the view that the phenomenon reported here is of general significance and can be extended to the expression of endogenous RA-responsive genes. However, it should be kept in mind that additional factors play a role in defining the ability of the receptor to elicit a transcriptional response: the actual sequence of the RARE alters the RXR-RAR DNA binding repertoire (37), and the relative ratio of corepressors, coactivators, and receptors may induce a differential response to RAR- and RXR-restricted ligands (50).
We consistently observed that SRC-1 bound more avidly to heterodimers than RIP140 and that synergistic binding triggered by the panagonist 9-cis RA was more than additive for the latter polypeptide compared to atRA-induced recruitment on DR1, DR4, and DR5 RAREs. While the molecular basis for these differences is not clear yet, we note that the SRC-1 RID used in these assays (amino acids 382 to 842) contains three LXXLL motifs separated by a 49-amino-acid spacer, while the RIP140 RID (amino acids 752 to 1158) contains only two LXXLL motifs with a spacer of 107 amino acids. In light of the recently reported three-dimensional structure of apo-peroxisome proliferator-activated receptor γ LBD complexed to a region of SRC-1 from amino acids 623 to 710 containing two LXXLL motifs (44) and of the proposed model of interaction of these two motifs with RXR-RAR heterodimers (60), one possible explanation could be that the RIP140 RAR-interacting LXXLL motif has a very low affinity for apo-RAR AF-2, whereas the RXR binding motif has a high affinity for RXR AF-2. In contrast, SRC-1 motifs would bind to both receptors with similar affinities, leading only to the observed additive effect on 9 cis-RA binding to both receptors. It is worth noting that the 9-cis RA-induced potentiation of RIP140 recruitment is not observed when heterodimers are assembled on DR2 and DR3 RAREs, arguing for an allosteric regulation through RARE half-site spacing. Finally, note that such a potentiation was observed in most cases when 9-cis RA was compared to synthetic retinoid-induced RIP140 and SRC-1 recruitment, strengthening our view that the apo-RAR structure is different in the presence of natural or synthetic agonists.
Second, the hRXRα AF2-AD appeared to be critical for maximal activation of RAR AF-2 by retinoids. The dependency of hRARα activity on the hRXRα AF2-AD was reflected by a decreased interaction of dnRXR-hRARα dimers with SRC-1, which suggests that AF2s of both partners, liganded or not, do not function autonomously. Allosteric regulation imposed by dimerization with hRXRα is thus likely to alter the structure of the RAR coactivator binding interface. Alternatively, it may suggest that liganded RAR activates RXR AF-2, in analogy with the reported allosteric regulation of the RAR AF-2 region by the RXR homodimer antagonist LG100754 (52). Such a dependency of the RAR signalling pathway on the RXR AF-2 has been documented in vivo (15), and our data therefore demonstrate a direct role for RXR AF-2 in RAR-mediated recruitment of nuclear coactivators.
Third, synthetic RAR-selective retinoids displayed differing abilities to recruit SRC-1 in vitro to DNA-bound heterodimers. The relative ability of a given ligand to recruit a coactivator could be altered by the nature of the HRE. This is indicative of the modulation of the NCoA binding interface in a ligand structure-dependent manner, a hypothesis consistent with the absolute requirement for K244 to observe synthetic retinoid-induced transcriptional activity. The K244A mutation also impaired SRC-1 recruitment, in contrast to the K262A mutant, which was found to impinge moderately and nonselectively on receptor transactivating properties. These mutations were also found to discriminate between different coactivators, showing that their interaction interfaces are not identical, as already reported for ER (17). We note that the K262A mutation in RARy yielded an inactive receptor derivative (46), while mutating the corresponding lysine in hRARα (K262 [our results]) and TR-β1 (14) showed residual activities (∼50 and 30%, respectively, of wt receptor activity). While the reason(s) for this discrepancy is not clear, the use of distinct RARs and RXRs isotypes, as well as different cellular backgrounds (simian versus human) might account for these differences.
Previous studies have demonstrated that synthetic retinoids can not only be isotype selective but also display a certain degree of selectivity toward defined receptor-RARE combinations (24). The role of the ligand structure is emphasized by our observations, which suggest that further refinement in gene selectivity could be achieved by altering NCoA interaction surfaces. Selective recruitment of p300 or CBP has indeed been shown to be required for selective activation of the gene encoding p21Cip1 and p27Kip1, respectively (18). Since transcriptional activation is the end result of multiple interactions between the receptor, its dimerization partner, DNA, and ligand, one may speculate that conformationally restricted retinoids with highly selective biological activities can be designed (24). Beside the tremendous interest for therapeutical applications, this raises the possibility that such retinoids display distinctive abilities to activate endogenous target genes, a hypothesis currently under investigation in our laboratory.
ACKNOWLEDGMENTS
We thank R. M. Evans (Salk Institute), J. D. Chen (University of Massachusetts), V. Cavailles (INSERM U148, Montpellier, France), D. D. Moore and B. W. O’Malley (Baylor College of Medicine), and B. Shroot and U. Reichert (CIRD-Galderma) for providing plasmids and ligands, and we thank E. Thoreau (CIRD-Galderma) for hRARα LBD coordinates, constant interest, and input. We are grateful to Hoffmann-La Roche for the gift of 9-cis RA. We thank C. Brand and B. Lefebvre for critical reading of the manuscript and B. Masselot for technical help.
REFERENCES
- 1.Aström A, Pettersson U, Krust A, Chambon P, Voorhees J J. Retinoic acid and synthetic analogs differentially activate retinoic acid receptor dependent transcription. Biochem Biophys Res Commun. 1990;173:339–345. doi: 10.1016/s0006-291x(05)81062-9. [DOI] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Blanco J C G, Minucci S, Lu J M, Yang X J, Walker K K, Chen H W, Evans R M, Nakatani Y, Ozato K. The histone acetylase P/CAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 6.Cavailles V, Dauvois S, Lhorset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen J Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Two distinct actions of retinoid-receptor ligands. Nature. 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 9.Chiba H, Clifford J, Metzger D, Chambon P. Specific and redundant functions of retinoid X receptor/retinoic acid receptor heterodimers in differentiation, proliferation, and apoptosis of F9 embryonal carcinoma cells. J Cell Biol. 1997;139:735–747. doi: 10.1083/jcb.139.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delescluse C, Cavey M T, Martin B, Bernard B A, Reichert U, Maignan J, Darmon M, Shroot B. Selective high affinity retinoic acid receptor alpha or beta-gamma ligands. Mol Pharmacol. 1991;40:556–562. [PubMed] [Google Scholar]
- 11a.Depoix, C. Unpublished data.
- 12.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- 14.Feng W J, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 15.Feng X, Peng Z H, Di W, Li X Y, Rochette-Egly C, Chambon P, Voorhees J J, Xiao J H. Suprabasal expression of a dominant-negative RXR alpha mutant in transgenic mouse epidermis impairs regulation of gene transcription and basal keratinocyte proliferation by RAR-selective retinoids. Genes Dev. 1997;11:59–71. doi: 10.1101/gad.11.1.59. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptor. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Henttu P M A, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki H, Eckner R, Yao T-P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9 cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 19.Klaholz B P, Renaud J P, Mitschler A, Zusi C, Chambon P, Gronemeyer H, Moras D. Conformational adaptation of agonists to the human nuclear receptor RAR gamma. Nat Struct Biol. 1998;5:199–202. doi: 10.1038/nsb0398-199. [DOI] [PubMed] [Google Scholar]
- 20.Klein E S, Pino M E, Johnson A T, Davies P J, Nagpal S, Thacher S M, Krasinski G, Chandraratna R A. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J Biol Chem. 1996;271:22692–22696. doi: 10.1074/jbc.271.37.22692. [DOI] [PubMed] [Google Scholar]
- 21.Kurokawa R, Direnzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld M G, Heyman R A, Glass C K. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 23.Lamour F P, Lardelli P, Apfel C M. Analysis of the ligand-binding domain of human retinoic acid receptor alpha by site-directed mutagenesis. Mol Cell Biol. 1996;16:5386–5392. doi: 10.1128/mcb.16.10.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Vista-Picard N, Hobbs P D, Pfahl M, Dawson M I. The receptor-DNA complex determines the retinoid response: a mechanism for the diversification of the ligand signal. Mol Cell Biol. 1996;16:4137–4146. doi: 10.1128/mcb.16.8.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C H, Chinpaisal C, Wei L N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol Cell Biol. 1998;18:6745–6755. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefebvre B, Mouchon A, Formstecher P, Lefebvre P. H11-H12 loop retinoic acid receptor mutants exhibit distinct trans-activating and trans-repressing activities in the presence of natural or synthetic retinoids. Biochemistry. 1998;37:9240–9249. doi: 10.1021/bi9804840. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre B, Mouchon A, Formstecher P, Lefebvre P. Distinct modes of interaction of the retinoic acid receptor alpha with natural and synthetic retinoids. Mol Cell Endocrinol. 1998;139:161–169. doi: 10.1016/s0303-7207(98)00065-3. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre B, Rachez C, Formstecher P, Lefebvre P. Structural determinants of the ligand-binding site of the human retinoic acid receptor alpha. Biochemistry. 1995;34:5477–5485. doi: 10.1021/bi00016a019. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, P., et al. Unpublished data.
- 30.Lefebvre P, Gaub M P, Tahayato A, Rochette-Egly C, Formstecher P. Protein phosphatases 1 and 2A regulate the transcriptional and DNA binding activities of retinoic acid receptors. J Biol Chem. 1995;270:10806–10816. doi: 10.1074/jbc.270.18.10806. [DOI] [PubMed] [Google Scholar]
- 31.Lefebvre P, Mouchon A, Lefebvre B, Formstecher P. Binding of retinoic acid receptor heterodimers to DNA—a role for histones NH2 termini. J Biol Chem. 1998;273:12288–12295. doi: 10.1074/jbc.273.20.12288. [DOI] [PubMed] [Google Scholar]
- 32.Lefstin J A, Yamamoto K R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 33.Leng X, Tsai S Y, O’Malley B W, Tsai M-J. Ligand-dependent conformational changes in thyroid hormone and retinoic acid receptors are potentially enhanced by heterodimerization with retinoic X receptor. J Steroid Biochem Mol Biol. 1993;46:643–661. doi: 10.1016/0960-0760(93)90306-h. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Chen J D. The receptor-associated coactivator 3 activates transcription through CREB-binding protein recruitment and autoregulation. J Biol Chem. 1998;273:5948–5954. doi: 10.1074/jbc.273.10.5948. [DOI] [PubMed] [Google Scholar]
- 35.Love J M, Gudas L J. Vitamin A, differentiation and cancer. Curr Opin Cell Biol. 1994;6:825–831. doi: 10.1016/0955-0674(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 36.MacInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assamunt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mader S, Chen J Y, Chen Z P, White J, Chambon P, Gronemeyer H. The patterns of binding of RAR, RXR and TR homodimers and heterodimers to direct repeats are dictated by the binding specificities of the DNA binding domains. EMBO J. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin B, Bernardon J M, Cavey M T, Bernard B, Carlavan I, Charpentier B, Pilgrim W R, Shroot B, Reichert U. Selective synthetic ligands for human nuclear retinoic acid receptors. Skin Pharmacol. 1992;5:57–65. doi: 10.1159/000211018. [DOI] [PubMed] [Google Scholar]
- 39.Means A L, Gudas L J. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–233. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- 40.Minucci S, Leid M, Toyama R, Saintjeannet J P, Peterson V J, Horn V, Ishmael J E, Bhattacharyya N, Dey A, Dawid I B, Ozato K. Retinoid X receptor (RXR) within the RXR-retinoic acid receptor heterodimer binds its ligand and enhances retinoid-dependent gene expression. Mol Cell Biol. 1997;17:644–655. doi: 10.1128/mcb.17.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 42.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs—evidence for 2 autonomous transactivation functions (AF-1 and AF-2) and heterodimerization invivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagpal S, Athanikar J, Chandraratna R A S. Separation of transactivation and AP1 antagonism functions of retinoic acid receptor alpha. J Biol Chem. 1995;270:923–927. doi: 10.1074/jbc.270.2.923. [DOI] [PubMed] [Google Scholar]
- 44.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor y. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 45.Oňate S A, Tsai S Y, Tsai M J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 46.Ostrowski J, Roalsvig T, Hammer L, Marinier A, Starrett J E, Yuo K L, Reczek P R. Serine 232 and methionine 272 define the ligand binding pocket in retinoic acid receptor subtypes. J Biol Chem. 1998;273:3490–3495. doi: 10.1074/jbc.273.6.3490. [DOI] [PubMed] [Google Scholar]
- 47.Rachez C, Sautiere P, Formstecher P, Lefebvre P. Identification of amino acids critical for the DNA binding and dimerization properties of the human retinoic acid receptor alpha. Importance of lysine 360, lysine 365, and valine 361. J Biol Chem. 1996;271:17996–8006. doi: 10.1074/jbc.271.30.17996. [DOI] [PubMed] [Google Scholar]
- 48.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 49.Roy D Y, Taneja R, Chambon P. Synergistic activation of retinoic acid (RA)-responsive genes and induction of embryonal carcinoma cell differentiation by RA receptor alpha (RAR alpha)-, RAR beta-, or RAR gamma-selective ligand in combination with a retinoid X receptor-specific ligand. Mol Cell Biol. 1995;15:6481–6487. doi: 10.1128/mcb.15.12.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanguedolce M V, Leblanc B P, Betz J L, Stunnenberg H G. The promoter context is a decisive factor in establishing selective responsiveness to nuclear class II receptors. EMBO J. 1997;16:2861–2873. doi: 10.1093/emboj/16.10.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scafonas A, Wolfgang C L, Gabriel J L, Soprano K J, Soprano D R. Differential role of homologous positively charged amino acid residues for ligand binding in retinoic acid receptor alpha compared with retinoic acid receptor beta. J Biol Chem. 1997;272:11244–11249. doi: 10.1074/jbc.272.17.11244. [DOI] [PubMed] [Google Scholar]
- 52.Schulman I G, Li C, Schwabe J W, Evans R M. The phantom ligand effect: allosteric control of transcription by the retinoic X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 53.Shroot B, Michel S. Pharmacology of retinoids in the skin: In vitro and in vivo assays. Eur J Med Chem. 1995;30:S487–S503. [Google Scholar]
- 54.Shroot B, Michel S. Pharmacology and chemistry of adapalene. J Am Acad Dermatol. 1997;36:S96–S103. doi: 10.1016/s0190-9622(97)70050-1. [DOI] [PubMed] [Google Scholar]
- 55.Shudo K, Kagechika H. Structural evolution of retinoids. Adv Drug Res. 1993;24:81–119. [Google Scholar]
- 56.Tate B F, Allenby G, Janocha R, Kazmer S, Speck J, Sturzenbecker L J, Abarzua P, Levin A A, Grippo J F. Distinct binding determinants for 9-cis retinoic acid are located within AF-2 of retinoic acid receptor-alpha. Mol Cell Biol. 1994;14:2323–2330. doi: 10.1128/mcb.14.4.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate B F, Grippo J F. Mutagenesis of the ligand binding domain of the human retinoic acid receptor alpha identifies critical residues for 9-cis-retinoic acid binding. J Biol Chem. 1995;270:20258–20263. doi: 10.1074/jbc.270.35.20258. [DOI] [PubMed] [Google Scholar]
- 58.Torchia J, Glass C K, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 59.Wagner R L, Apriletti J W, Mcgrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 60.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 61.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 62.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 63.Xiao J H, Durand B, Chambon P, Voorhees J J. Endogenous retinoic acid receptor (RAR)-retinoid X receptor (RXR) heterodimers are the major functional forms regulating retinoid-responsive elements in adult human keratinocytes. Binding of ligands to RAR only is sufficient for RAR-RXR heterodimers to confer ligand- dependent activation of hRAR beta 2/RARE (DR5) J Biol Chem. 1995;270:3001–3011. doi: 10.1074/jbc.270.7.3001. [DOI] [PubMed] [Google Scholar]
- 64.Zavacki A M, Lehmann J M, Seol W, Willson T M, Kliewer S A, Moore D D. Activation of the orphan receptor RIP14 by retinoids. Proc Natl Acad Sci USA. 1997;94:7909–7914. doi: 10.1073/pnas.94.15.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X K, Salbert G, Lee M O, Pfahl M. Mutations that alter ligand-induced switches and dimerization activities in the retinoid X receptor. Mol Cell Biol. 1994;14:4311–4323. doi: 10.1128/mcb.14.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]