Abstract
Idiopathic pulmonary fibrosis (IPF) is an incurable genetic disease that affects 5 million people worldwide. The gain-of-function MUC5B promoter variant rs35705950 is the dominant genetic risk factor for IPF, yet has a low penetrance. This raises the possibility that other genes and transcripts affect the penetrance of MUC5B. Previously, we have shown that the concentration of Muc5b in bronchoalveolar epithelia is directly associated with the extent and persistence of bleomycin-induced lung fibrosis in mice. In this study, we investigated whether bleomycin-induced lung injury is Muc5b dependent in genetically divergent strains of mice. Specifically, mice from the eight Diversity Outbred (DO) founders were phenotyped for Muc5b expression and lung fibrosis 3 wk after intratracheal bleomycin administration. Although we identified strains with low Muc5b expression and minimal lung fibrosis (CAST/EiJ and PWK/PhJ) and strains with high Muc5b expression and extensive lung fibrosis (NZO/H1LtJ and WSB/EiJ), there also were strains that did not demonstrate a clear relationship between Muc5b expression and lung fibrosis (129S1/SvlmJ, NOD/ShiLtJ, and C57BL/6J, A/J). Hierarchical clustering suggests that other factors may work in concert with or potentially independent of Muc5b to promote bleomycin-induced lung injury and fibrosis. This study suggests that these strains and their recombinant inbred crosses may prove helpful in identifying the genes and transcripts that interact with Muc5b and cause lung fibrosis.
Keywords: diversity outbred mice, IPF, lung fibrosis, Muc5b
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic lung disease, which affects 5 million people worldwide with a median survival of 3–5 yr (1, 2) making it the most prevalent and severe form of interstitial lung disease. A common gain-of-function MUC5B promoter variant rs35705950 is the dominant risk factor, genetic or otherwise, for the development of IPF (3). The MUC5B promoter variant is associated with enhanced expression of the MUC5B transcript in lung tissue from unaffected subjects and patients with IPF (3, 4). In patients with IPF, excess MUC5B protein is especially observed in regions of lung involved in tissue fibrosis, including respiratory bronchiolar airway epithelia, type 2 alveolar epithelial cells, and epithelia composing the honeycomb cyst (5–7). Recently, we demonstrated that the concentration of Muc5b in bronchoalveolar epithelia is directly related to the extent and persistence of bleomycin-induced lung fibrosis and mortality in mice (7). However, there is no obvious explanation for the low penetrance of the MUC5B promoter variant. In humans, the minor allele of MUC5B is present in the heterozygous or homozygous state in ≈19% of the general population (3), yet IPF occurs in far less than 1% of the population (8, 9).
Here, we sought to further extend our understanding of the genetics of pulmonary fibrosis by investigating whether bleomycin-induced lung injury is Muc5b dependent in genetically divergent strains of mice. A commonly used tool to study genetic variation is fairly recently introduced the Collaborative Cross and Diversity Outbred (DO) mice, which represent a large collection of genetic deviations from eight founder inbred stains (10, 11). To explore the feasibility of the DO mice to pursue this concept, we phenotyped the response to a single dose of intratracheal bleomycin-induced injury in eight genetically divergent mouse strains that were used to create the DO population (Fig. 1A) (12, 13).
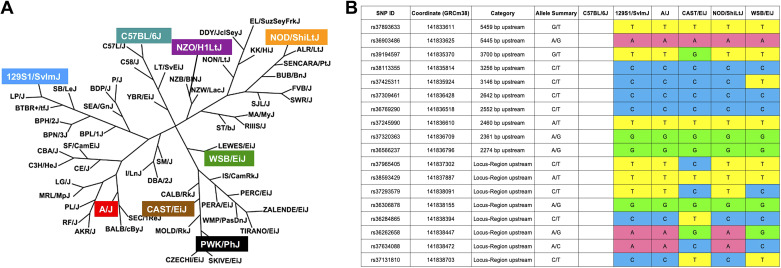
Figure 1.
Mouse family tree and single nucleotide polymorphisms (SNPs) in the Muc5b promoter region. A: the eight strains tested are distantly related. [Adapted from Petkov et al. (12) with permission from Cold Spring Harbor Laboratory Press under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license; https://creativecommons.org/licenses/by-nc/4.0/.] B: SNPs map 4 kb upstream of Muc5b gene region for six inbred mouse lines.
MATERIALS AND METHODS
Animals
Male mice from the eight Diversity Outbred founders [129S1/SvlmJ, A/J, CAST/EiJ, NZO/H1LtJ, NOD/ShiLtJ, PWK/PhJ, WSB/EiJ, and C57BL/6J inbred strains purchased from Jackson Laboratory (Bar Harbor, ME)] were phenotyped for their response to bleomycin. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Colorado. Mice were housed and their health monitored in accordance with IACUC guidelines. Mice were maintained on a 12-h light/dark cycle and fed ad libitum a normal diet of water and irradiated chow (Harlan Teklad). Moribund mice were identified by observing changes in body weight and behaviors. In accordance with veterinary care procedures at the University of Colorado, a loss of 15% body weight without recovery, hunching, fur ruffling, and lethargy were used as criteria for determining moribundity. At 9 wk of age, mice underwent intratracheal instillation of bleomycin 2.5 U/kg bleomycin (APP Pharmaceuticals, Schamburg, IL), and were euthanized by intraperitoneal injection of sodium pentobarbital followed by exsanguination.
Bleomycin Exposure
Mice were anesthetized with inhalational isoflurane (MWI Veterinary Supply Company, Boise, ID) and tracheas were directly visualized with a rodent laryngoscope (Penn Century, Wyndmoor, PA). A 22-g gavage needle was used to instill 50 µL of saline or 2.5 U/kg bleomycin solution (APP Pharmaceuticals, Schamburg, IL) directly into the trachea on day 0. Mice were harvested on day 21. Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared to n = 5–7 saline treated-control mice from each strain.
Whole Lung Lavage
Immediately following euthanasia, mouse tracheas were cannulated. Left lungs were clamped while right lungs were lavaged three times with 0.5 mL of 0.6 mM EDTA/PBS. Lavaged fluid was used for immune cells counting. Remaining lavaged fluid was divided into two aliquots and stored at −80°C.
Dot Blot Analysis
Muc5b concentration levels from mouse lung lavage were measured via dot blot analysis performed using S&S Minifold I dot blot filtration manifold (Schleicher and Schuell). Samples were diluted 1:200 and run in triplicate along with a standard sample for semiquantitative analysis, also run in triplicate. The protein samples were allowed to bind to polyvinylidene difluoride membrane, the membrane was blocked at room temperature in 5% milk in phosphate-buffered saline with 0.2% Tween for 1 h and then incubated overnight at 4°C with the Muc5b primary antibody (Santa Cruz Biotechnology) diluted 1:5,000 with Odyssey blocking buffer (LI-COR, Lincoln, NE). Secondary goat anti-rabbit IgG IRDye680RD conjugate (1:15,000; LI-COR) in Odyssey blocking buffer (LI-COR) was applied for 1 h at room temperature. Membranes were imaged with LI-COR Odyssey CLx(LI-COR), and quantitated based on fluorescence intensity using ImageStudio Lyte ver 5.2 (LI-COR, Lincoln, NE). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain. Samples were run in triplicates.
Gene Expression
The right middle lobe was homogenized in a solution of 500 µL RNA and later with b-mercaptoethanol, RNA was extracted with the RNeasy Minikit (Qiagen, Germantown, MD) and reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham MA). Muc5b expression was determined using a Muc5b (Mm00466391_m1) Taqman gene expression assay (Applied Biosystems, Foster City, CA) and normalized to PPIA (Mm02342430_g1). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 control mice from each strain. Samples were run in triplicates.
Hydroxyproline
The upper, lower, and accessory lobes of the right lung were homogenized in 550 µL PBS. Samples were then frozen at −80°C. Thawed lung homogenates were hydrolyzed overnight with a 1:1 volume of 12 N HCl at 100°C. Afterward, 5-µL samples of hydrolyzed lung and hydroxyproline standards were plated in duplicate and analyzed as described previously (7). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain. Samples were run in triplicates.
Histochemistry and Immunohistochemistry
The left lung was inflated with 4% formaldehyde at a pressure of 20 cmH2O for 5 min. The lung was then removed and placed in fresh 4% formaldehyde overnight for fixation. Left lungs were cut into uniform slices, embedded in paraffin, cut into 5-µm sections, and collected on positively charged glass slides. Hematoxylin and eosin (H&E) staining and Masson’s trichrome staining were completed using standard reagents at the University of Colorado Cancer Center Pathology Core. Slides were scanned with an Aperio CS2 before scoring with ScanScope (Leica Biosystems, CA). H&E slides were semiquantitatively scored by two separate investigators (AE, ED). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Slides were stained with 1:500 Goat-anti-Muc5b (Everest, UK), 1:500 Chicken-anti-Keratin 5 (BioLegend Inc., San Diego, CA), 1:500 Rabbit-anti-proSurfactant Protein C (Abcam, UK), and 1:20,000 DAPI (BioLegend Inc.) for histological analysis. Random fields of view (FOVs) (7–30) of both large and small airways were analyzed using ImageJ (NIH). Mice (n = 4–5) were analyzed after bleomycin-induced injury and compared with n = 4–5 saline treated-control mice from each strain.
Second Harmonic Generation Imaging and Analysis
Autofluorescence and second harmonic generation (SHG) signals were acquired using Zeiss 780 laser-scanning confocal/multiphoton-excitation fluorescence microscope with a 34-channel GaAsP QUASAR detection unit and nondescanned detectors for two-photon fluorescence (Zeiss, Thornwood, NY). Fifteen images were obtained for each lung using standardized uniform random sampling (14, 15). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Honeycomb-Like Cyst Quantification
Scanned mouse lungs stained for Muc5b overlaid with grid 0.25 mm2 in ImageJ were used to calculate area percent taken by cysts-like structures. High-power FOVs taken randomly from the regions with cysts and overlaid with grid 400 μm (2) were used to calculate cysts number and size (16). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Hierarchical Clustering
Hierarchical clustering of mouse strains based on percent change in Muc5b/Muc5b, lung injury, and fibrosis (all mean centered) was performed using the centroid clustering method and Euclidean distance metric (Partek). Mice (n = 8–13) were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Statistical Analysis
Hydroxyproline, inflammation, protein, SHG, and gene expression data were analyzed by Mann–Whitney U test and nonparametric ANOVA for multiple comparisons (GraphPad, La Jolla, CA). Slopes from correlation analysis were compared through Spearman coefficients (GraphPad).
RESULTS
Inbred Mouse Strains Have Variable Responsiveness to Bleomycin-Induced Lung Injury
Inbred founder mouse strains [129S1/SvlmJ, NZO/H1LtJ, A/J, PWK/PhJ, WSB/EiJ, NOD/ShiLtJ, CAST/EiJ, and C57BL/6J; Fig. 1A (12)] from the Diversity Outbred cross were compared from publicly available genomes (http://www.informatics.jax.org/snp) at the orthologous site for rs35705950 and found to be homozygous for the T allele (the IPF risk allele). This was confirmed by genotyping. Mice were exposed to 2.5 U/kg of bleomycin intratracheally and euthanized 3 wk post challenge. All mice survived the bleomycin challenge. All strains expressed similar levels of bleomycin hydrolase (Blmh) (Supplemental Fig. S1; all Supplemental material is available at https://doi.org/10.6084/m9.figshare.13412555), thus the differences in lung injury and fibrosis cannot be explained by differences in bleomycin metabolism. In addition, we analyzed known single nucleotide polymorphisms (SNPs) at the publicly available (http://www.informaScs.jax.org/snp) dataset of the DNA sequences for six out of eight inbred mouse stains (Fig. 1B). We found that even though there are variable SNPs present in the 4 kb region upstream of Muc5b gene, there is no common pattern which can help to explain differences in response to bleomycin-induced injury (Fig. 1B).
Left lungs were fixed and stained with H&E and were semiquantitatively scored with four measures of lung injury. Among mice treated with bleomycin, patterns of injury diverge extensively (Fig. 2A). A/J, 129S1/SvlmJ, and NZO/H1LtJ mice appeared to have the most extensive lung injury, whereas PWK/PhJ mice appeared almost unaffected by bleomycin. Lungs from 129S/SvlmJ, NZO/H1LtJ, C57BL/6J, and A/J mice presented the most amount of diffuse and peripheral injury. 129S/SvlmJ, A/J, CAST/EiJ, and NZO/H1LtJ mice demonstrated lung injury that was most associated with blood vessels and airways. Interestingly, the 129S1/SvlmJ injury began near airways and extended toward the ventral tip of the lung, distinct from all other strains. Results of the semiquantitative histological assessment of the inbred mice injury is presented in Fig. 2B. Based on the histological evaluation, strains could be classified into three groups with extensive and diffuse injury (A/J, 129S1/SvlmJ, and NZO/H1LtJ), modest extent of injury (WSB/EiJ, NOD/ShiLtJ, C57BL/6J, and CAST/EiJ), and minimal or almost no injury (PWK/PhJ). The concentration of macrophages in the saline controls was variable at baseline, but this variability was not statistically significant among the strains (Fig. 2C). The concentration of lavage macrophages was increased compared with saline controls, but varied substantially depending on strain [Fig. 2D and Supplemental Fig. S2 (top)]. Bleomycin-treated A/J, C57BL/6J, 129S1/SvlmJ, and NZO/H1LtJ lavage macrophages were higher compared with the other strains of mice (Fig. 2D). Analyzed saline-treated mice had nonstatistically significant variable total collagen levels as measured by hydroxyproline assay (Fig. 2E). Hydroxyproline assay was also used to quantify total collagen accumulation in response to bleomycin-induced injury in the right lung. We observed that lungs from CAST/EiJ, PWK/PhJ, 129S1/SvlmJ, C57BL/6J, and WSB/EiJ mouse strains had an increase in total level of collagen compared with the saline controls, with the highest increase in the WSB/EiJ mice (Fig. 2F). Lungs from NZO/H1LtJ mice had almost no change compared with their saline controls, whereas total levels of collagen in lungs from A/J and NOD/ShiLtJ mice were decreased following bleomycin administration (Fig. 2F). Interestingly, total concentrations of collagen from whole lung detected by hydroxyproline assay did not correlate with the extent of lung injury (Fig. 2 and Supplemental Fig. S3, Spearman coefficients are presented in Table 1—we found variable correlations between hydroxyproline and amount of injury, however none of the comparisons were significant by strain).
Figure 2.
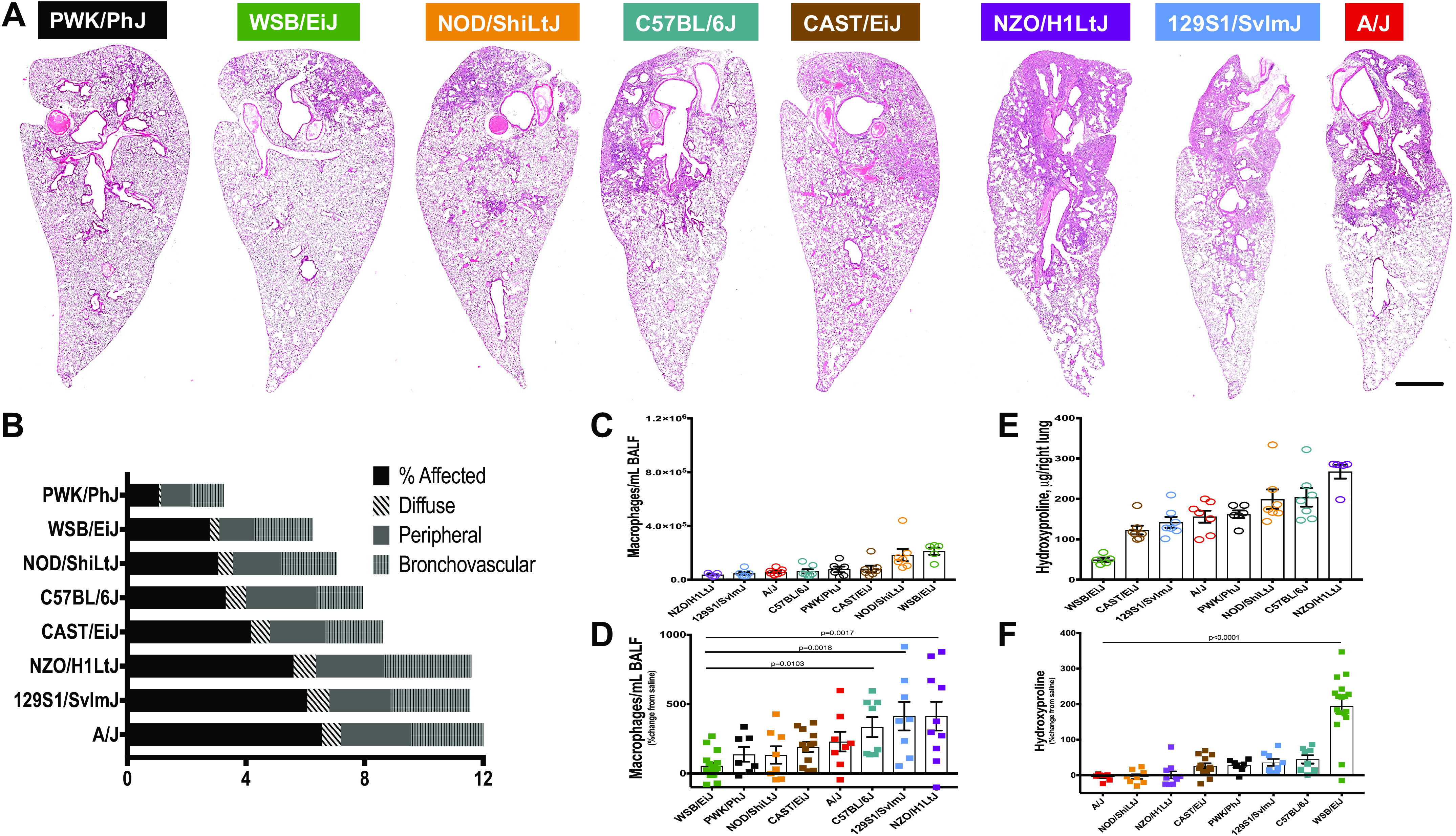
Comparisons of phenotypes after bleomycin-induced lung injury. Three weeks after a single challenge with intratracheal bleomycin (2.5 U/kg), mice showed considerable variation at baseline and injury. A: representative images of hematoxylin and eosin (H&E) stain across the eight strains of mice show differences in severity and localization of injury. The progressive amount of injured tissue (condensed, darker, with infiltrating immune cells) from PWK/PhJ (almost no injury) to A/J (about 50% of the injured tissue) is shown. Bar is 1 mm. Injury scoring (B); macrophages counts in bronchoalveolar lavage fluid (BALF) (C and D) and hydroxyproline (HP) measurements for total lung collagen (E and F). n = 8–13 mice were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Table 1.
Summary of correlations analyses for hydroxyproline and fibrillar collagens (SHG) to percent affected lung tissue, and Muc5b protein and Muc5b gene expression to fibrillar collagens (SHG)
| 129S1/SvlmJ | A/J | CAST/EiJ | NZO/H1LtJ | NOD/ShiLtJ | PWK/PhJ | WSB/EiJ | C57BL/6J | |
|---|---|---|---|---|---|---|---|---|
| HP vs. %Affected | ||||||||
| Slope | 8.705 ± 5.095 | 3.556 ± 1.452 | 2.196 ± 4.671 | −14.83 ± 15.81 | 3.631 ± 5.155 | 11.44 ± 12.41 | 7.311 ± 3.781 | 7.453 ± 15.61 |
| Equation | Y = 8.705 * X + 139.8 | Y = 3.556 * X + 126.8 | Y = 2.196 * X + 145.2 | Y = −14.83 * X + 353.3 | Y = 3.631 * X + 180.4 | Y = 11.44 * X + 190.1 | Y = 7.311 * X + 126.8 | Y = 7.453 * X + 270.7 |
| R square | 0.3273 | 0.5452 | 0.02162 | 0.09903 | 0.0764 | 0.1241 | 0.2108 | 0.0366 |
| Spearman r | 0.4671 | 0.4286 | 0.03873 | -0.3161 | 0.3314 | 0.3703 | 0.2706 | 0.1807 |
| P (two-tailed) | 0.2471 | 0.3536 | 0.9073 | 0.3714 | 0.419 | 0.3643 | 0.3083 | 0.6722 |
| P value summary | ns | ns | ns | ns | ns | ns | ns | ns |
| SHG vs. %Affected | ||||||||
| Slope | 0.2684 ± 0.1157 | 0.265 ± 0.08014 | 0.0863 ± 0.09017 | 0.3868 ± 0.107 | 0.07755 ± 0.03909 | 0.01541 ± 0.1753 | 0.219 ± 0.04708 | 0.02983 ± 0.1201 |
| Equation | Y = 0.2684 * X + 1.27 | Y = 0.265 * X + 0.1228 | Y = 0.0863 * X + 1.31 | Y = 0.3868 * X + 0.05676 | Y = 0.07755 * X + 0.9012 | Y = 0.01541 * X + 1.224 | Y = 0.219 * X + 0.7457 | Y = 0.02983 * X + 1.699 |
| R square | 0.4729 | 0.6456 | 0.08392 | 0.6203 | 0.3961 | 0.001286 | 0.6073 | 0.01017 |
| Spearman r | 0.5868 | 0.8623 | 0.3134 | 0.8268 | 0.4542 | 0.08939 | 0.5576 | −0.03615 |
| P (two-tailed) | 0.1353 | 0.0089 | 0.3185 | 0.0047 | 0.2619 | 0.8286 | 0.0268 | 0.9429 |
| P value summary | ns | ** | ns | ** | ns | ns | * | ns |
| Muc5b pr vs. SHG | ||||||||
| Slope | 624.1 ± 1031 | −994.1 ± 2,480 | 433.1 ± 229 | −182.9 ± 1,783 | 9,647 ± 3,018 | −1,266 ± 2,035 | 3,413 ± 1,310 | 6,497 ± 4,199 |
| Equation | Y = 6,24.1 * X + 1,446 | Y = −994.1 * X + 9,848 | Y = 433.1 * X + 142.6 | Y = −182.9 * X + 9,341 | Y = 9,647 * X − 4,752 | Y = −1,266 * X + 4,235 | Y = 3,413 * X + 3,527 | Y = 6,497 * X − 1,661 |
| R square | 0.05758 | 0.02607 | 0.2845 | 0.001314 | 0.6301 | 0.07191 | 0.3264 | 0.3238 |
| Spearman r | 0.4524 | −0.3333 | 0.5545 | 0.006061 | 0.6667 | −0.1429 | 0.6353 | 0.4643 |
| P (two-tailed) | 0.2675 | 0.4279 | 0.0818 | >0.9999 | 0.0831 | 0.7825 | 0.0097 | 0.3024 |
| P value summary | ns | ns | ns | ns | ns | ns | ** | ns |
| Muc5b RNA vs. SHG | ||||||||
| Slope | 0.3599 ± 0.1655 | −0.1165 ± 0.1389 | −0.1749 ± 0.511 | −0.5903 ± 0.2049 | −1.412 ± 0.9965 | 2.156 ± 1.757 | 0.2059 ± 0.2898 | −0.1548 ± 0.481 |
| Equation | Y = 0.3599 * X + 3.016 | Y = −0.1165 * X + 6.093 | Y = −0.1749 * X + 1.287 | Y = −0.5903 * X + 6.463 | Y = −1.412 * X + 6.781 | Y = 2.156 * X + 2.016 | Y = 0.2059 * X + 4.131 | Y = −0.1548 * X + 6.383 |
| R square | 0.4409 | 0.105 | 0.01285 | 0.5092 | 0.2506 | 0.2007 | 0.03258 | 0.01696 |
| Spearman r | 0.619 | −0.5952 | 0.01818 | −0.5273 | −0.7619 | −0.04762 | 0.1887 | −0.3095 |
| P (two-tailed) | 0.115 | 0.1323 | 0.9673 | 0.1231 | 0.0368 | 0.9349 | 0.4668 | 0.4618 |
| P value summary | ns | ns | ns | ns | * | ns | ns | ns |
HP, hydroxyproline; ns, not significant; SHG, second-harmonic generation. *P < 0.05; **P < 0.005.
The Amount and Localization of Collagen Deposition Varies across Inbred Mouse Strains in Response to Bleomycin
We sought to assess the collagen content of the lung parenchyma to more effectively compare inbred strains. Masson’s trichrome histological staining was not definitive and demonstrated inconsistent findings with hydroxyproline (Supplemental Fig. S4). Therefore, we decided to use quantitative label-free two-photon microscopy with second harmonic generation (SHG) imaging to quantitatively assess fibrillar collagen in the lung parenchyma (Fig. 3A). Fibrillar collagen accumulation was increased to variable degrees in the lung parenchyma of all examined mouse strains, except PWK/PhJ (Fig. 3B). Remarkably, fibrillar collagen accumulation in the lung parenchyma was not consistent with hydroxyproline measurements but correlated better with visual differences in H&E across the mouse strains [Fig. 3C, Spearman coefficients are presented in Table 1—we found that correlation between fibrillar collagen accumulation and amount of injury was significant for A/J (P = 0.0089), NZO/H1LtJ (P = 0.0047), and WSB/EiJ (P = 0.0268) mice].
Figure 3.
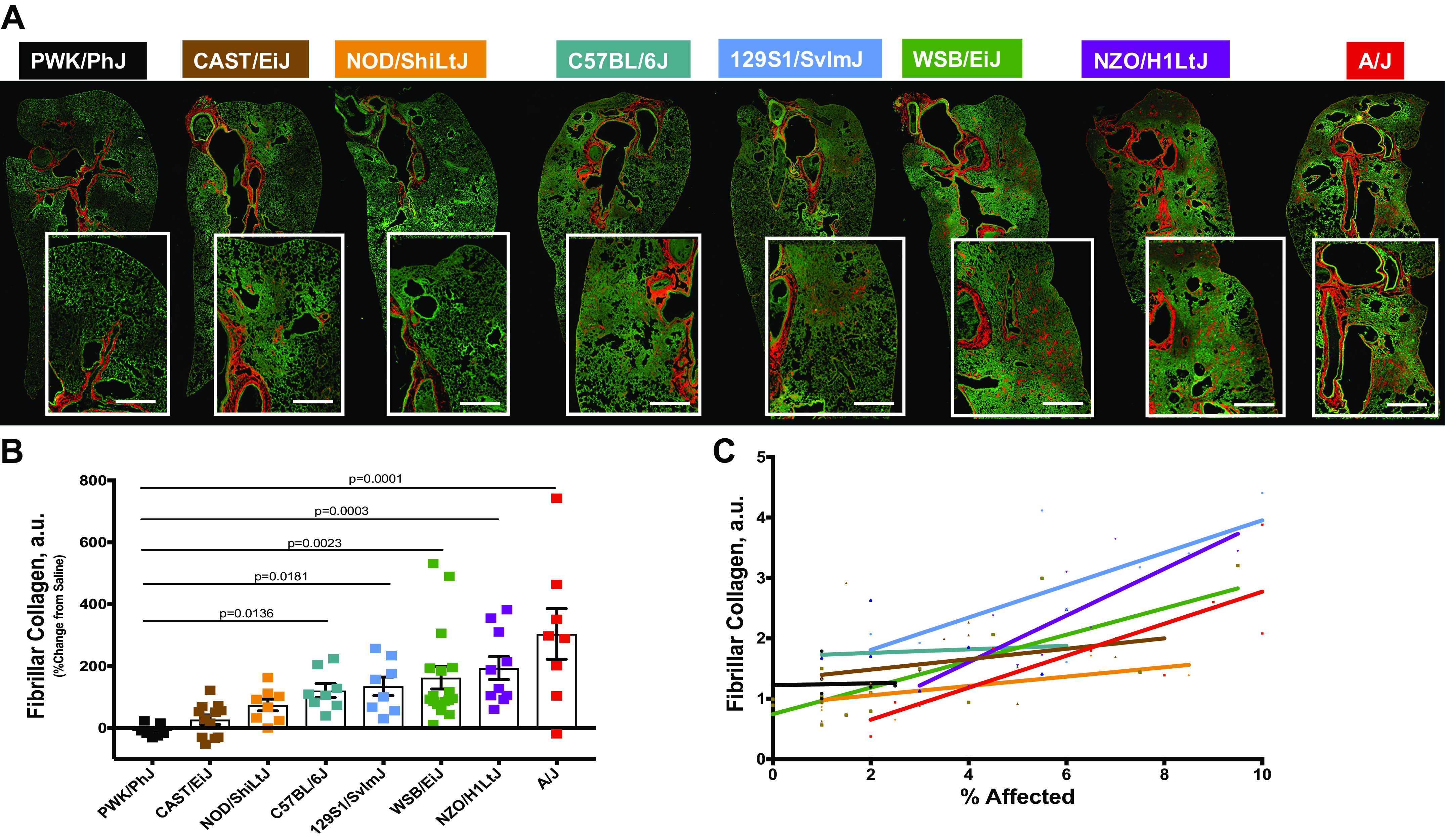
Second-harmonic generation (SHG) imaging of fibrillar collagen is a more reliable measure of parenchymal fibrosis. Fibrillar collagen was assessed in peripheral lung tissues by confocal/multi-photon fluorescence microscopy with SHG. A: representative images showing the localization of fibrillar collagen (red) around the airways and in the parenchyma following bleomycin exposure. Bar, 500 µm. Fibrillar collagen amount (B) was calculated per mouse following exposure to bleomycin or vehicle. C: fibrillar collagen accumulation correlates with the percent affected area in injured lung. n = 8–13 mice were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Inbred Mouse Strains Demonstrate Unique Relationships between Muc5b Transcript and Protein Expression and Lung Fibrosis
Secreted Muc5b protein in the lung lavage fluid was similar at the baseline (Fig. 4A) and increased in all strains following bleomycin and the mice could be separated into two groups: minimally responsive (CAST/EiJ, PWK/PhJ), and responsive (129S1/SvlmJ, A/J, NZO/H1LtJ, WSB/EiJ, NOD/ShiLtJ, and C57BL/6J) (Fig. 4B). Using qRT-PCR, we found that mice had statistically not significant variability in the Muc5b gene expression at the baseline (Fig. 4C) and that NZO/H1LtJ and WSB/EiJ mice had the most substantial increase in Muc5b expression following bleomycin administration, followed by 129S1/SvlmJ and NOD/ShiLtJ mice. Muc5b gene expression in the other strains either was unchanged or decreased up to 50% in CAST/EiJ mice (Fig. 4D). Unsupervised hierarchical clustering identified the relationship between Muc5b/Muc5b and lung fibrosis: strains with high Muc5b/Muc5b and more extensive fibrosis (WSB/EiJ, A/J, NZO/H1LtJ), low Muc5b/Muc5b and less fibrosis (CAST/EiJ and PWK/PhJ), and inconsistent relationship between Muc5b/Muc5b and the extent of lung fibrosis (NOD/ShiLtJ, C57BL/6J, and 129S1/SvlmJ) (Fig. 4E). Muc5b protein expression correlated with accumulation of fibrillar collagen and therefore the amount of lung injury (Fig. 4F), but Muc5b gene expression did not (Fig. 4G, Spearman coefficients are presented in Table 1—we found that, even though not significant, r for Muc5b protein were higher compared with Muc5b gene expression, confirming stronger correlation between Muc5b protein expression with fibrillar collagens accumulation).
Figure 4.
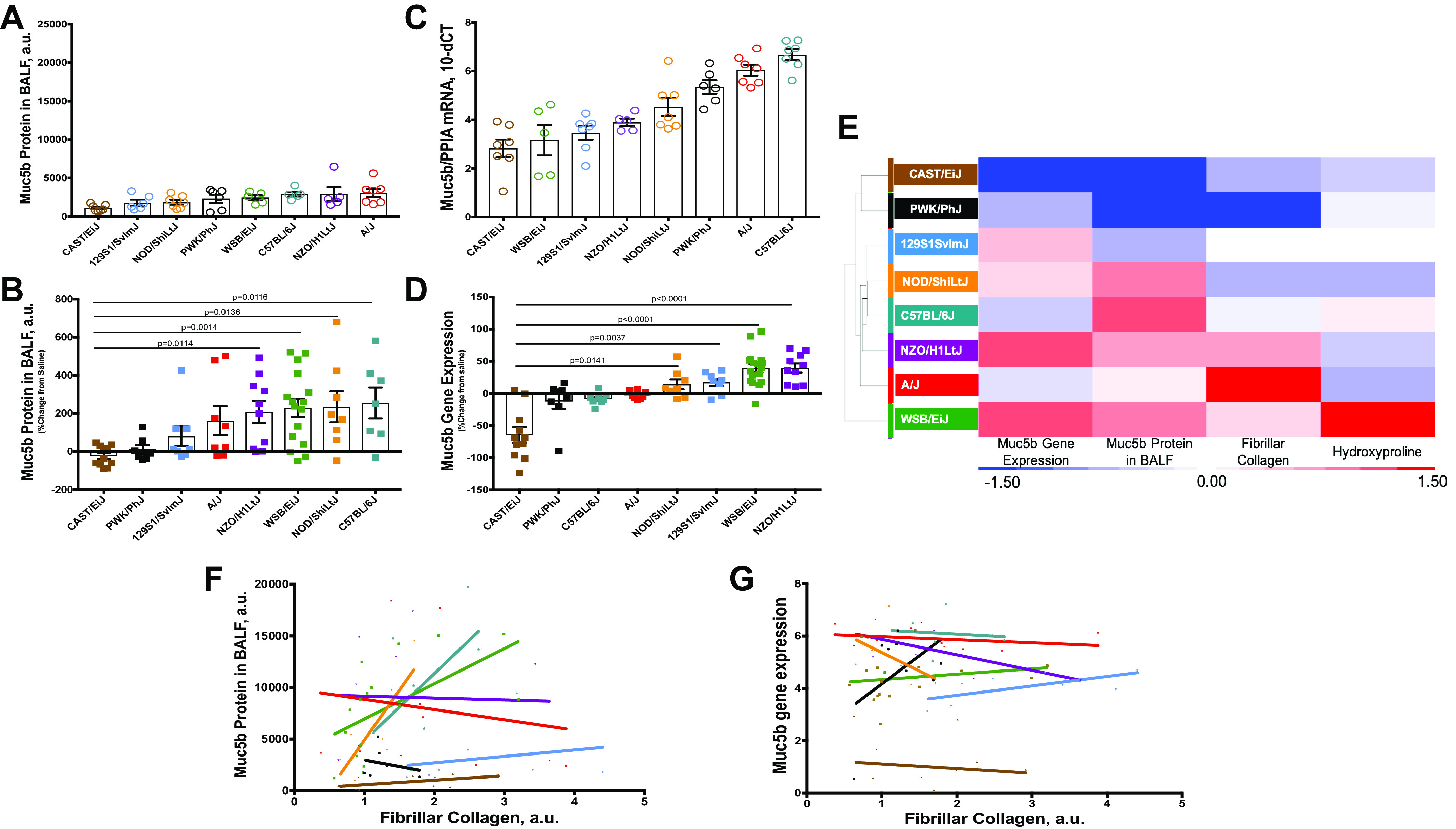
Muc5b gene expression and protein secretion do not fully correlate with other measures of injury and fibrosis. Muc5b protein in bronchoalveolar lavage fluid (BALF) (A and C, Supplemental Fig. S5, top row) and whole lung gene expression (B and D, Supplemental Fig. S5, bottom row) vary at baseline and after injury among mouse strains and their correlation with percent affected area in injured lung (F and G, correspondingly). Hierarchical clustering of mouse strains based on %changes in Muc5b gene expression, protein, and measures of lung fibrosis (E). n = 8–13 mice were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
Intracellular Muc5b Protein Expression Does Not Correlate with the Extent of Fibrosis
Since we found that concentration of secreted Muc5b protein correlated better with the extent of lung fibrosis, we performed an additional analysis for the number of Muc5b expressing cells and Muc5b protein expression distribution and how it changed at 3 wk after bleomycin-induced lung injury (Fig. 5A). We found that mice had statistically not significant variability in the Muc5b intensity and number of Muc5b expressing cells in proximal (Fig. 5, B and C) and distal airways (Fig. 5, D and E) at the baseline. However, in general, distal airways had less Muc5b expressing cells with less signal intensity compared with proximal airways (Fig. 5, B and D, correspondingly). We observed augmented Muc5b mean intensity (corresponding to the concentration of protein) in the proximal airways of C57BL/6J, CAST/EiJ, and NOD/ShiLtJ mouse strains, compared with the matching saline exposures (Fig. 5F). Number of Muc5b expressing cells increased in the proximal airways of C57BL/6J, CAST/EiJ, NOD/ShiLtJ, and WSB/EiJ mouse strains, compared with their corresponding saline values (Fig. 5G). Whereas the other strains had a decreased mean intensity and the number of Muc5b expressing cells in the proximal airways. In the distal airways, Muc5b concentration was increased only in CAST/EiJ mice (Fig. 5H), but the number of Muc5b expressing cells was elevated in the NOD/ShiLtJ, PWK/PhJ, and C57BL/6J mice (Fig. 5I). Surprisingly, we have not observed a relation between Muc5b protein expression or number of Muc5b expressing cells and lung injury in either proximal or distal airways at 3 wk following bleomycin exposure.
Figure 5.
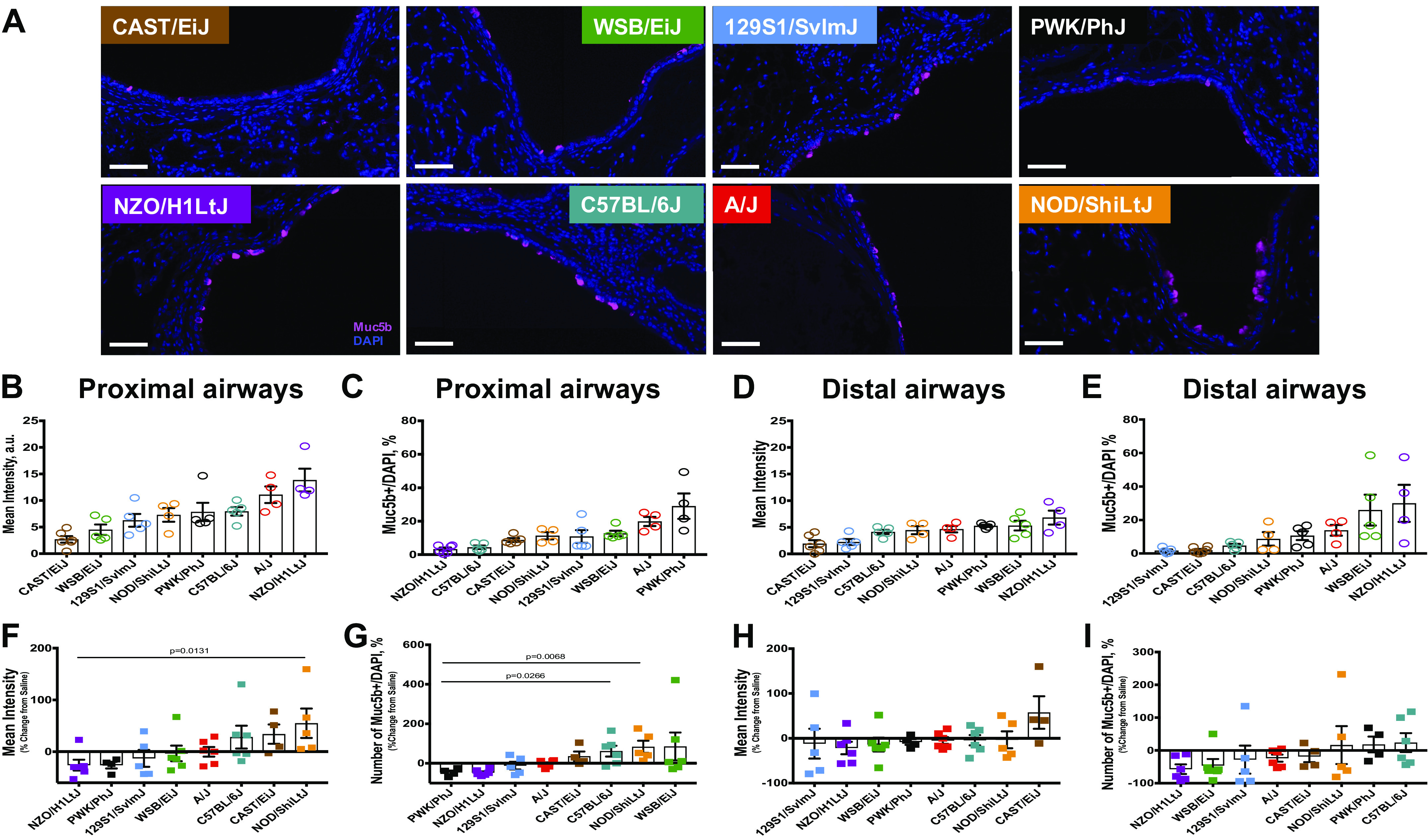
Airways Muc5b expression. A: representative images for Muc5b (purple) staining in the proximal airways for 8 strains of mice. Tissues were counterstained with nuclear staining (DAPI, blue). Bar, 100 µm. Quantification for Muc5b mean intensity in proximal (B and F, Supplemental Fig. S6, top row) and distal (C and G, Supplemental Fig. S7, top row) airways, and number of Muc5b expressing cells in proximal (D and H, Supplemental Fig. S6, bottom row) and distal (E and I, Supplemental Fig. S7, bottom row) airways. n = 4–5 mice were analyzed after bleomycin-induced injury and compared with n = 4–5 saline treated-control mice from each strain.
Muc5b Expression and Accumulation in Parenchymal Cysts May Be a Consequence of Lung Injury and Fibrosis
Analyzing lung injury on H&E-stained sections, we observed cysts formation in lung parenchyma of the Diversity Outbred mouse strains. When we stained the lung sections for Muc5b, we detected that the majority of the formed cyst-like structures were lined with Muc5b expressing cells and filled with Muc5b secreted protein (Fig. 6A), similar to what we previously observed in honeycomb cysts in patients with IPF (6). Previously, we and others had shown that cysts structures can be lined with keratin 5-positive basal (both human and mice) (16, 17) and type 2 alveolar epithelial cells (human) (7). Therefore, we stained the lung sections for Muc5b, keratin 5 and prosurfactant protein C (SPC) to identify the cell types forming Muc5b-expressing cysts in the DO founder mice. We have found that NOD/ShiLtJ, C57BL/6J, 129S1/SvlmJ, NZO/H1LtJ, WSB/EiJ, and A/J mice had both types of cysts—lined with keratin 5-positive basal and type 2 alveolar epithelial cells (SPC positive), at different proportions and with different amount of Muc5b filling the lumen of the cysts-like structures. We quantified the area with the Muc5b expressing cells and filled cyst-like structures present in the lungs (Fig. 6B), their ratio to histologically unaffected lung tissues (Fig. 6C), number of cyst-like structures in per high power field of view (Fig. 6D), and average cyst size (Fig. 6E). We found that CAST/EiJ mice did not develop cyst-like structures lined with Muc5b expressing cells and only one of the PWK/PhJ mice had Muc5b filled cysts. In contrast, NOD/ShiLtJ, C57BL/6J, 129S1/SvlmJ, NZO/H1LtJ, WSB/EiJ, and A/J mice had established honeycomb-like cysts structures with increased areas of cysts in the WSB/EiJ and A/J mice. Furthermore, we have observed a progressive increase of the cysts, lined with keratin 5-positive basal cells in addition to an increased Muc5b expression and accumulation within keratin-5-positive cysts like structures. Average number of Muc5b expressing/filled cyst-like structures in the high-power field of view and their average sizes were more similar between the mouse strains. Remarkably, mice with the highest number of areas with cyst-like structures and their ratios to the histologically unaffected lungs had the highest amount of lung injury and fibrillar collagen accumulation.
Figure 6.
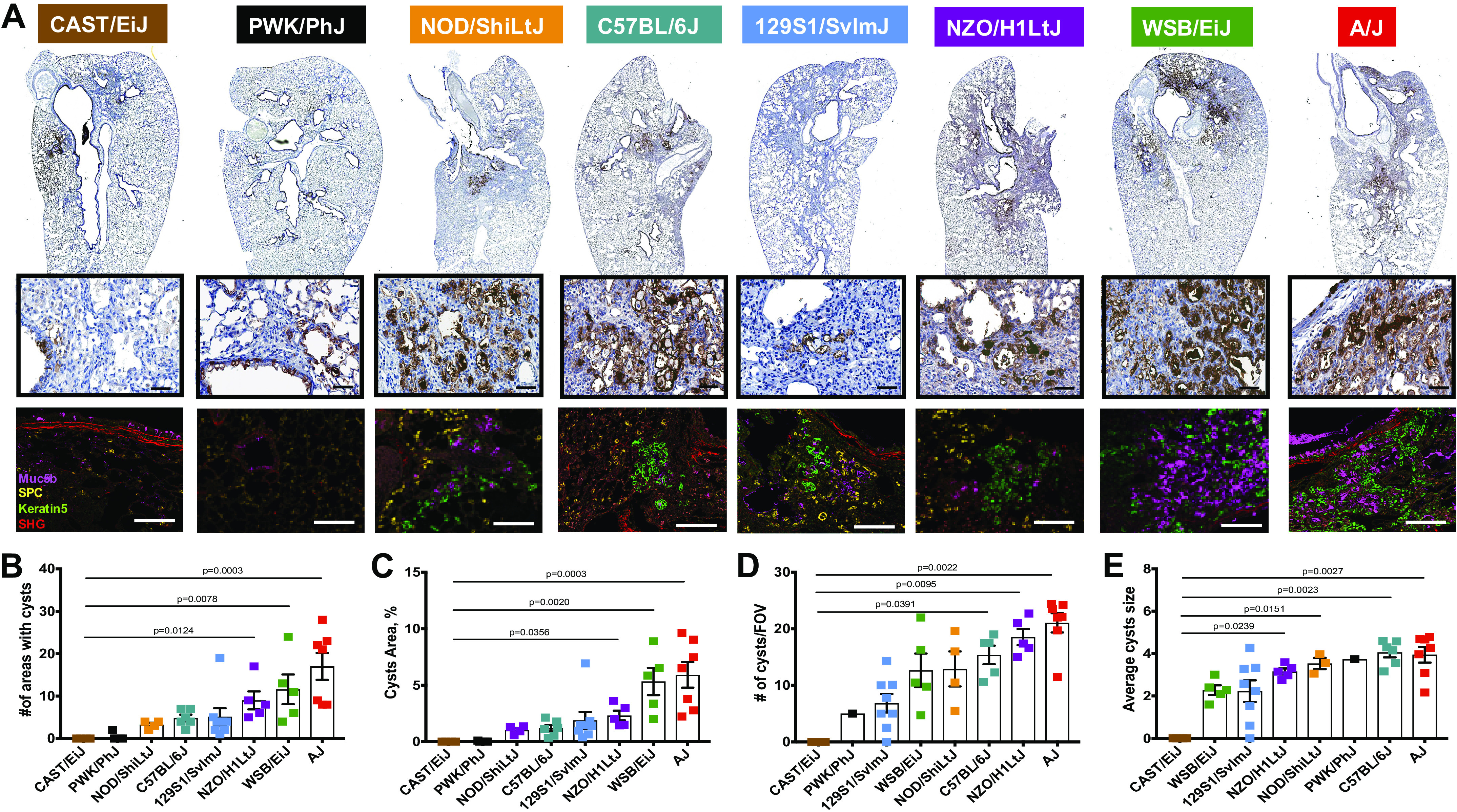
Muc5b expressing honeycomb-like cysts. A: representative images for Muc5b immunohistochemistry (IHC) staining in eight mouse strains (lower magnification; top row) and regions with cyst-like structures shown at higher magnification (middle row). Immunofluorescent images for representative cyst-like regions with Muc5b (purple), proSPC (yellow), keratin5 (green), and SGH (red) signals in all analyzed mouse strains (bottom row). Bar, 60 µm. Quantification for the number of cyst-like structures (B), area % taken by cyst-like structures (C), number of cyst-like structures in fields of view (FOV; D), and average cyst-like structures size (E). n = 8–13 mice were analyzed after bleomycin-induced injury and compared with n = 5–7 saline treated-control mice from each strain.
DISCUSSION
Our findings indicate that other factors may work in concert with or potentially independent of Muc5b to promote bleomycin-induced lung injury and fibrosis. Although we identified strains with low Muc5b/Muc5b expression and minimal lung fibrosis (CAST/EiJ and PWK/PhJ) and strains with high Muc5b/Muc5b expression and extensive lung fibrosis (NZO/H1LtJ and WSB/EiJ), there also were strains that did not demonstrate a clear relationship between Muc5b/Muc5b expression and lung fibrosis (129S1/SvlmJ, NOD/ShiLtJ, and C57BL/6J, A/J). These findings demonstrate that the diversity outbred founder mouse strains and especially the diversity outbred mouse population may provide insight into the role of Muc5b/Muc5b in lung injury and repair, and fibroproliferative responses.
The relationship between MUC5B overproduction and IPF is complex. We have previously shown that Muc5b transgenic mice are more responsive to bleomycin; however, at least in the first several months of life, Muc5b transgenic mice do not spontaneously develop lung fibrosis (7). Similarly, the human gain-of-function MUC5B promoter variant that causes overexpression of MUC5B in the distal airspace is the dominant risk factor for the development of IPF, yet does not appear on its own to be sufficient to cause pulmonary fibrosis. Despite the limitation that IPF is likely underdiagnosed (8, 18–21), the MUC5B promoter variant appears to have a low penetrance in causing IPF. Thus, it is likely that other genes, proteins, and/or environmental exposures could interact with the MUC5B promoter variant to cause IPF only in individuals with additional risk factors. Our findings suggest that the etiology of IPF will be best understood by identifying the genes, transcripts, proteins, and environmental exposures that interact with MUC5B and contribute to the development of IPF.
Previously, variants of different genes, including desmoplakin (DSP) variants and genes regulating telomere length (TERT, TERC, OBFC1), have been identified associated with IPF (22–25). DSP expression is higher in IPF and was found in proximal and distal airways, as well as areas of honeycomb cysts (26). Desmoplakins are part of desmosomes, which are essential for epithelial cells barrier function and wound repair (27–29). Individuals with MUC5B promoter variant have increased MUC5B mucin expression in the airway cells and cells lining honeycomb cysts walls (26). In addition, mucus accumulation in the lumen of distal airways and cysts could potentially lead to impaired mucus clearance and recurrent injury/repair cycles, leading to aberrant lung regeneration and fibrosis. In fact, our findings in Muc5b transgenic mice suggest that decreased mucociliary clearance may, in part, be responsible for the development of Muc5b-induced lung fibrosis (7). Both, DSP and MUC5B/MUC5B are overexpressed in honeycomb cysts (6, 7, 26), suggesting that the interaction of these genes or proteins may provide insight into the development of IPF. Telomeres in the alveolar epithelial cells type II of patients with IPF have consistently been reported to be shortened and may play a role in lung remodeling and fibrosis (30). Recently, we identified that both, secretory and alveolar type II cells can line honeycomb cysts walls in human lungs and express increased amount of MUC5B (7). In aggregate, these findings suggest that interactions or associations of MUC5B with other genes might play a role in the IPF development and could be distinctive in different cell types.
In addition, here we have found that mice with the highest concentration of cyst-like structures had the most extensive lung injury and fibrillar collagen accumulation. Moreover, level of parenchymal injury was best correlated with fibrillar collagen accumulation, measured by advanced two-photon microscopy technique second harmonic generation (SHG) and amount of areas with cysts-like structures, expressing and filled with Muc5b. Interestingly, hydroxyproline analysis did not demonstrate a correlation with lung injury and overall had less sensitivity detecting parenchymal fibrosis development, compared with trichrome and SHG. This inconsistency might be justified by that, even though the highest amount of hydroxyproline is found in collagen, this amino acid is present on all types of collagens (both, intra- and extracellular) and some other proteins, like elastin (31, 32). Besides, mice demonstrated different types of collagen accumulation around proximal airways and blood vessels, which were analyzed by hydroxyproline (HP) assay together with parenchymal fibrosis, whereas parenchymal and bronchovascular fibrosis can be analyzed separately using histology (all intra- and extracellular collagens via trichrome staining) or just extracellular fibrillar collagens accumulation (via SHG). Moreover, SHG in addition to being quantitative was shown to be more sensitive technique, compared with regular histological analysis (33).
Remarkably, we found that cysts number and size seemed to correlate less with the amount of parenchymal fibrosis. These observations raise interesting questions about the etiology of cysts—when do the cysts form; which cell types are responsible for their origin and formation? Do cells lining cysts express Muc5b themselves or do they differentiate into Muc5b expressing cells? Do these cells have different genes/proteins expression and physiological conditions? For example, it was shown that in C57 mice both, alveolar epithelial type II cells and keratin 5 expressing basal cells contribute to the honeycomb-like cysts formation and development and some of them have shown to acquire local hypoxia-inducing ER stress (34, 35). Further studies are investigating how these at least two cell types will differ in Muc5b/Muc5b expression and function and how it will influence parenchymal fibrosis development.
Although future studies in large populations of patients with IPF may reveal the critical biological mechanisms that interact with the gain-of-function MUC5B promoter variant to cause IPF, genetically engineered strains of mice could be essential in identifying the genes, proteins, and/or environmental exposures, contributing to lung fibrosis initiation and development. Moreover, in an alternative approach such as usage of the Diversity Outbred mice, which can be more beneficial than regular crosses, chromosomal regions controlling the genetic variance of complex traits can be mapped as quantitative trait loci (QTLs). Multiple studies have used inbred mice after bleomycin-induced injury to understand the mechanism of fibrosis development and identify novel genes that might be involved in these processes. They found number of gene candidates, which might be responsible for differential fibrosis development in divergent inbred mouse stains; however, they have lacked the translational insights made possible by an understanding of the role of the MUC5B variant in human fibrotic lung disease, used high doses of bleomycin for single or serial intratracheal bleomycin instillation (36), employed intraperitoneal injections (37), or facilitated chronic exposure to bleomycin through osmotic mini-pumps (38–40), which would be expected to alter the type of injury and progression of the disease. Moreover, we examined a broader spectrum of lung injury, including fibrosis and honeycomb development with different epithelial cell types lining, as well as the effects of Muc5b/Muc5b expression. Our results suggest that diversity outbred mice may have, in addition to different Muc5b/Muc5b expression, different time-course of the injury and different genes interplaying with Muc5b/Muc5b that could be helpful in further understanding the genetics of IPF.
DATA AVAILABILITY
The data that support the findings of this study are available upon request.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S7: https://doi.org/10.6084/m9.figshare.13412555.
GRANTS
This research was supported by National Heart, Lung, and Blood Institute (NHLBI) Grants R01-HL097163, R01-HL149836, P01-HL092870, UH3-HL123442, and DoD W81XWH-17-1-0597. This study was also supported by Rocky Mountain Neurological Disorders Core Grant Number P30 NS048154 and by Diabetes Research Center Grant Number P30 DK116073 and by the Cancer Center Support Grant P30 CA046934.
DISCLAIMERS
The content is the authors’ sole responsibility and does not necessarily represent official NIH views.
DISCLOSURES
D. A. Schwartz is an employee of Eleven P15, Inc.—a company with mission of early detection and early intervention of IPF. I. V. Yang receives consulting fees from Eleven P15, outside the submitted work. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
I.V.Y. and D.A.S. conceived and designed research; E.D., A.M.E., C.E.H., and N.H. performed experiments; E.D., A.M.E., C.E.H., and J.S.K. analyzed data; E.D., M.I.S., J.S.K., I.V.Y., and D.A.S. interpreted results of experiments; E.D. prepared figures; E.D. and A.M.E. drafted manuscript; E.D., A.M.E., C.E.H., J.S.K., I.V.Y., and D.A.S. edited and revised manuscript; E.D., A.M.E., C.E.H., N.H., M.I.S., J.S.K., I.V.Y., and D.A.S. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank the Advanced Light Microscopy Core part of NeuroTechnology Center at University of Colorado Anschutz Medical Campus. The authors also appreciate the contribution to this research made by E. Erin Smith, Allison Quador, and Jessica Arnold of the University of Colorado Cancer Center Pathology Shared Resource.
REFERENCES
- 1.Raghu G, Chen SY, Hou Q, Yeh WS, Collard HR. Incidence and prevalence of idiopathic pulmonary fibrosis in US adults 18–64 years old. Eur Respir J 48: 179–186, 2016. doi: 10.1183/13993003.01653-2015. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson JP, McKeever TM, Fogarty AW, Navaratnam V, Hubbard RB. Increasing global mortality from idiopathic pulmonary fibrosis in the twenty-first century. Ann Am Thorac Soc 11: 1176–1185, 2014. doi: 10.1513/AnnalsATS.201404-145OC. [DOI] [PubMed] [Google Scholar]
- 3.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, Evans CM, Garantziotis S, Adler KB, Dickey BF, du Bois RM, Yang IV, Herron A, Kervitsky D, Talbert JL, Markin C, Park J, Crews AL, Slifer SH, Auerbach S, Roy MG, Lin J, Hennessy CE, Schwarz MI, Schwartz DA. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med 364: 1503–1512, 2011. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helling BA, Gerber AN, Kadiyala V, Sasse SK, Pedersen BS, Sparks L, Nakano Y, Okamoto T, Evans CM, Yang IV, Schwartz DA. Regulation of MUC5B expression in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 57: 91–99, 2017. doi: 10.1165/rcmb.2017-0046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakano Y, Yang IV, Walts AD, Watson AM, Helling BA, Fletcher AA, Lara AR, Schwarz MI, Evans CM, Schwartz DA. MUC5B promoter variant rs35705950 affects MUC5B expression in the distal airways in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 193: 464–466, 2016. doi: 10.1164/rccm.201509-1872LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, Schwarz MI, Schwartz DA, Reynolds SD. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One 8: e58658, 2013. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock LA, Hennessy CE, Solomon GM, Dobrinskikh E, Estrella A, Hara N, Hill DB, Kissner WJ, Markovetz MR, Grove Villalon DE, Voss ME, Tearney GJ, Carroll KS, Shi Y, Schwarz MI, Thelin WR, Rowe SM, Yang IV, Evans CM, Schwartz DA. Muc5b overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat Commun 9: 5363, 2018. doi: 10.1038/s41467-018-07768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson AL, Swigris JJ, Lezotte DC, Norris JM, Wilson CG, Brown KK. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med 176: 277–284, 2007. doi: 10.1164/rccm.200701-044OC. [DOI] [PubMed] [Google Scholar]
- 9.Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 174: 810–816, 2006. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 10.Bogue MA, Churchill GA, Chesler EJ. Collaborative cross and diversity outbred data resources in the mouse phenome database. Mamm Genome 26: 511–520, 2015. doi: 10.1007/s00335-015-9595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, et al. ; Complex Trait Consortium . The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137, 2004. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 12.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 14: 1806–1811, 2004. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Church GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447, 2012. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhlfeld C, Ochs M. Quantitative microscopy of the lung: a problem-based approach. Part 2: stereological parameters and study designs in various diseases of the respiratory tract. Am J Physiol Lung Cell Mol Physiol 305: L205–L221, 2013. doi: 10.1152/ajplung.00427.2012. [DOI] [PubMed] [Google Scholar]
- 15.Ochs M, Muhlfeld C. Quantitative microscopy of the lung: a problem-based approach. Part 1: basic principles of lung stereology. Am J Physiol Lung Cell Mol Physiol 305: L15–L22, 2013. doi: 10.1152/ajplung.00429.2012. [DOI] [PubMed] [Google Scholar]
- 16.Kurche JS, Dobrinskikh E, Hennessy CE, Huber J, Estrella A, Hancock LA, Schwarz MI, Okamoto T, Cool CD, Yang IV, Evans CM, Schwartz DA. Muc5b enhances murine honeycomb-like cyst formation. Am J Respir Cell Mol Biol 61: 544–546, 2019. doi: 10.1165/rcmb.2019-0138LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517: 621–625, 2015. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunninghake GM, Hatabu H, Okajima Y, Gao W, Dupuis J, Latourelle JC, Nishino M, Araki T, Zazueta OE, Kurugol S, Ross JC, San José Estépar R, Murphy E, Steele MP, Loyd JE, Schwarz MI, Fingerlin TE, Rosas IO, Washko GR, O'Connor GT, Schwartz DA. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med 368: 2192–2200, 2013. doi: 10.1056/NEJMoa1216076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araki T, Putman RK, Hatabu H, Gao W, Dupuis J, Latourelle JC, Nishino M, Zazueta OE, Kurugol S, Ross JC, San José Estépar R, Schwartz DA, Rosas IO, Washko GR, O'Connor GT, Hunninghake GM. Development and progression of interstitial lung abnormalities in the Framingham Heart Study. Am J Respir Crit Care Med 194: 1514–1522, 2016. doi: 10.1164/rccm.201512-2523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kropski JA, Pritchett JM, Zoz DF, Crossno PF, Markin C, Garnett ET, Degryse AL, Mitchell DB, Polosukhin VV, Rickman OB, Choi L, Cheng DS, McConaha ME, Jones BR, Gleaves LA, McMahon FB, Worrell JA, Solus JF, Ware LB, Lee JW, Massion PP, Zaynagetdinov R, White ES, Kurtis JD, Johnson JE, Groshong SD, Lancaster LH, Young LR, Steele MP, Phillips Iii JA, Cogan JD, Loyd JE, Lawson WE, Blackwell TS. Extensive phenotyping of individuals at risk for familial interstitial pneumonia reveals clues to the pathogenesis of interstitial lung disease. Am J Respir Crit Care Med 191: 417–426, 2015. doi: 10.1164/rccm.201406-1162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King TE Jr, Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ; ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 183: 788–824, 2011. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armanios MY, Chen JJ-L, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA 3rd, Lansdorp PM, Greider CW, Loyd JE. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med 356: 1317–1326, 2007. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 23.Kropski JA, Reiss S, Markin C, Brown KK, Schwartz DA, Schwarz MI, Loyd JE, Phillips JA 3rd, Blackwell TS, Cogan JD. Rare genetic variants in PARN are associated with pulmonary fibrosis in families. Am J Respir Crit Care Med 196: 1481–1484, 2017. doi: 10.1164/rccm.201703-0635LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, Weissler J, Fitzgerald J, Kershaw C, Klesney-Tait J, Mageto Y, Shay JW, Ji W, Bilguvar K, Mane S, Lifton RP, Garcia CK. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet 47: 512–517, 2015. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA 104: 7552–7557, 2007. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathai SK, Pedersen BS, Smith K, Russell P, Schwarz MI, Brown KK, Steele MP, Loyd JE, Crapo JD, Silverman EK, Nickerson D, Fingerlin TE, Yang IV, Schwartz DA. Desmoplakin variants are associated with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 193: 1151–1160, 2016. doi: 10.1164/rccm.201509-1863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asimaki A, Saffitz JE. Remodeling of cell-cell junctions in arrhythmogenic cardiomyopathy. Cell Commun Adhes 21: 13–23, 2014. doi: 10.3109/15419061.2013.876016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzumcu A, Norgett EE, Dindar A, Norgett EE, Dindar A, Uyguner O, Nisli K, Kayserili H, Sahin SE, Dupont E, Severs NJ, Leigh IM, Yuksel-Apak M, Kelsell DP, Wollnik B. Loss of desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J Med Genet 43: e5, 2006. doi: 10.1136/jmg.2005.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta 1778: 572–587, 2008. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Naikawadi RP, Disayabutr S, Mallavia B, Donne ML, Green G, La JL, Rock JR, Looney MR, Wolters PJ. Telomere dysfunction in alveolar epithelial cells causes lung remodeling and fibrosis. JCI Insight 1: e86704, 2016. doi: 10.1172/jci.insight.86704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietz AA, Lubrano T, Covault HP, Rubinstein HM. Correct for hydroxyproline in elastin when measuring collagen in tissues with a high elastin content. Clin Chem 28: 1709, 1982. doi: 10.1093/clinchem/28.7.1709. [DOI] [PubMed] [Google Scholar]
- 32.Bentley JP, Hanson AN. The hydroxyproline of elastin. Biochim Biophys Acta 175: 339–344, 1969. doi: 10.1016/0005-2795(69)90011-7. [DOI] [PubMed] [Google Scholar]
- 33.Ranjit S, Dobrinskikh E, Montford J, Dvornikov A, Lehman A, Orlicky DJ, Nemenoff R, Gratton E, Levi M, Furgeson S. Label-free fluorescence lifetime and second harmonic generation imaging microscopy improves quantification of experimental renal fibrosis. Kidney Int 90: 1123–1128, 2016. doi: 10.1016/j.kint.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee DK, Gotts JE, Matthay MA, Shannon JM, Chapman HA, Vaughan AE. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol 19: 904–914, 2017. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burman A, Kropski JA, Calvi CL, Serezani AP, Pascoalino BD, Han W, Sherrill T, Gleaves L, Lawson WE, Young LR, Blackwell TS, Tanjore H. Localized hypoxia links ER stress to lung fibrosis through induction of C/EBP homologous protein. JCI Insight 3: e99543, 2018. doi: 10.1172/jci.insight.99543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung MP, Monick MM, Hamzeh NY, Butler NS, Powers LS, Hunninghake GW. Role of repeated lung injury and genetic background in bleomycin-induced fibrosis. Am J Respir Cell Mol Biol 29: 375–380, 2003. doi: 10.1165/rcmb.2003-0029OC. [DOI] [PubMed] [Google Scholar]
- 37.Gelinas R, Chesler EJ, Vasconcelos D, Miller DR, Yuan Y, Wang K, Galas D. A genetic approach to the prediction of drug side effects: bleomycin induces concordant phenotypes in mice of the collaborative cross. Pharmgenomics Pers Med 4: 35–45, 2011. doi: 10.2147/PGPM.S22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paun A, Lemay AM, Tomko TG, Haston CK. Association analysis reveals genetic variation altering bleomycin-induced pulmonary fibrosis in mice. Am J Respir Cell Mol Biol 48: 330–336, 2013. doi: 10.1165/rcmb.2012-0078OC. [DOI] [PubMed] [Google Scholar]
- 39.Bergeron ME, Stefanov A, Haston CK. Fine mapping of the major bleomycin-induced pulmonary fibrosis susceptibility locus in mice. Mamm Genome 29: 670–679, 2018. doi: 10.1007/s00335-018-9774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stefanov AN, Fox J, Depault F, Haston CK. Positional cloning reveals strain-dependent expression of Trim16 to alter susceptibility to bleomycin-induced pulmonary fibrosis in mice. PLoS Genet 9: e1003203, 2013. doi: 10.1371/journal.pgen.1003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available upon request.