Abstract
The base order-dependent component of folding energy has revealed a highly conserved region in HIV-1 genomes that associates with RNA structure. This corresponds to a packaging signal that is recognized by the nucleocapsid domain of the Gag polyprotein. Long viewed as a potential HIV-1 "Achilles heel," the signal can be targeted by a new antiviral compound. Although SARS-CoV-2 differs in many respects from HIV-1, the same technology displays regions with a high base order-dependent folding energy component, which are also highly conserved. This indicates structural invariance (SI) sustained by natural selection. While the regions are often also protein-encoding (e. g. NSP3, ORF3a), we suggest that their nucleic acid level functions can be considered potential "Achilles heels" for SARS-CoV-2, perhaps susceptible to therapies like those envisaged for AIDS. The ribosomal frameshifting element scored well, but higher SI scores were obtained in other regions, including those encoding NSP13 and the nucleocapsid (N) protein.
Keywords: Coronavirus, FORS-D analysis, RNA packaging signal, RNA structure, Sequence conservation, Stem-loop potential
Graphical Abstract
1. Introduction
After four decades of research, infection with HIV-1 can routinely be controlled but, because the virus adopts DNA-form latency, not cured. Since the newly emergent SARS-CoV-2 displays neither HIV-1-like latency nor its extreme variability and chronicity, curative treatments would seem more feasible (Rausch et al., 2020). Although both are RNA viruses, their many differences indicate that different therapeutic strategies will be required. Yet, as with HIV-1, strategies for SARS-CoV-2 have focused, with limited success, on the functions of encoded proteins rather than those of its RNA genome or of transcripts from that genome. Intriguingly, a HIV-1 vulnerability related to genome function that was long ago suggested by a nucleic acid level bioinformatics technology (Forsdyke, 1995a, Forsdyke, 2014), has recently gained support (Ingemarsdotter et al., 2018, Rein, 2020, Ding et al., 2020), and the term "Achilles heel" is increasingly employed (Ding et al., 2020, Forsdyke, 2016). The same technology now suggests similar vulnerabilities for SARS-CoV-2 (Rausch et al., 2020).
Our analytical approach requires elimination of the contribution of base composition to the energetics of folding into a higher order (stem-loop) structure of a single-stranded nucleic acid. Just as accent or dialect affects a spoken text in its entirety, so base composition tends to reflect genome-wide evolutionary pressures. Just as a local arrangement of words best conveys specific meaning to a text, so base order best reflects local evolutionary pressures. Base order is most likely to be conserved when encoding a function critical for survival. Because they do not make this fundamental distinction between base composition and order, more sophisticated methods of SARS-CoV-2 structure determination that incorporate all sequence information (Huston et al., 2020, Manfredonia et al., 2020, Tavares et al., 2021), have tended to confuse rather than clarify.
Our relatively simple assays of the base order-dependent component of the folding energy, which are devoid of redundant base compositional information (see Materials and Methods), have shown that a highly conserved region, in otherwise rapidly mutating HIV-1 genomes, associates with an RNA structure corresponding, not to a protein-encoding function, but to an RNA packaging signal. The latter is specifically recognized by the nucleocapsid domain of the Gag polyprotein (Sarni et al., 2020) and is now seen as a potential "Achilles heel" of HIV-1 that can be targeted by a recently described antiviral compound (Ingemarsdotter et al., 2018).
We here report similar highly conserved structural regions of the SARS-CoV-2 genome, one or more of which should be susceptible to targeting (Medeiros et al., 2021, Haniff et al., 2020). We identify certain open reading frames (ORFs) that, because of their conservation, have so far attracted therapeutic interest mainly related to their functions at the protein level (Robson, 2020, Issa et al., 2020), rather than at the level of the corresponding, yet highly structured, regions of the genome. The ribosomal frameshift element (FSE) that is among our results is attracting attention (Haniff et al., 2020, Kelly et al., 2020, Ziv et al., 2020, Sun et al., 2021, Li et al., 2021). Yet our analytical approach, which has been adopted by others (Andrews et al., 2018, Andrews et al., 2021, Simmonds, 2020a, Simmonds, 2020b), suggests there may be more suitable targets in other regions.
2. Materials and methods
2.1. Sequences
These were obtained from the NCBI (Bethesda) and GISAID EpiCoV (Munich) databases. The Wuhan-Hu1 sequence (GenBank NC_045512.2), deemed taxonomically prototypic (Pipes et al., 2020, Kumar et al., 2021), was compared (regarding base substitutions and folding potential) with early isolates from China (381), Italy (430), and New York, USA (932). Our "window" starting point was base 1 of the 29,903 base prototype sequence. We refer to windows by their centers. The center of the first 200 base window would be 100.
2.2. Substitution frequencies
In previous HIV-1 studies the base differences between just two individual sequences sufficed for the tabulation of a statistically significant set of base substitution frequencies (Forsdyke, 1995a). The lower mutation rates of SARS-CoV-2 strains (Robson, 2020) required the tabulation of changeable positions relative to the prototype (i.e. polymorphisms) among a set of individual sequences in a geographical region. In early September (2020), 709 Chinese sequences were filtered to remove duplicates and those with ambiguities, so yielding 381 unique sequences. These were scored for substitutable positions (whether one base could be exchanged by one of the three alternative bases) by aligning against the Wuhan prototype using Muscle software implemented in MEGA 7.0 (Edgar, 2004), with manual adjustments. This yielded a substitution value for the 200 bases in each sequence window (i.e. values ranging from zero to 200). The values were in the low range indicating less likelihood of back mutations (site saturation) in the period since divergence from the presumed prototype. Indeed, in a study of several thousand genomes 73% of sites were conserved (i.e. only 27% were polymorphic) (Simmonds, 2020b). To gain support for our results, in late September our study was extended with sets from Italy (430 from 642 downloaded) and New York, USA (932 out of 1483 downloaded). As expected, the FORS-D profiles of two early-arising members of these sets, that differed from the prototype in two (Italy) and nine (USA) bases, scarcely varied from the profile of the Chinese prototype shown here (i.e. overlapping profiles indicating within-species conservation of structure). In contrast, profiles differ greatly when different coronavirus species are compared (Simmonds, 2020b). We are concerned here with intraspecies conservation rather than interspecies conservation (Rangan et al., 2020).
2.3. Base order-dependent component of folding energy
The energetics of the folding of a single-stranded nucleic acid into a stem-loop structure depend on both the composition and order of its bases. Base composition is a distinctive characteristic of a genome or large genome sector. A localized sequence (e.g. a 200 base window), which is rich in the strongly-pairing bases G and C, will tend to have a stable structure simply by virtue of its base composition, rather than of its unique base order. This high GC% value can obscure the contribution of the base order-dependent component of the folding energy, which provides a sensitive indicator of local intraspecies pressures for the conservation of function within a population (i.e. a mutated organism is eliminated by natural selection so no longer can be assayed for function in the population). In contrast, interspecies mutations tend to influence genome-wide oligonucleotide (k-mer) pressure, of which base composition (GC%) is an indicator (Aggarwala and Voight, 2016, Morozov, 2017). Assays of the latter pressure can assist the initial stages of a formal sequence alignment, but this must then be finalized by attending to local details (Edgar, 2004). Oligonucleotide (k-mer) pressure works to generate and/or sustain members of emerging species by preventing recombination with parental forms (Forsdyke, 1996, Forsdyke, 2014, Forsdyke, 2016, Forsdyke, 2019a, Forsdyke, 2019b, Forsdyke, 2021a, Forsdyke, 2021b). Its elimination facilitates focus on local folding.
Early studies of RNA virus structure by Le and Maizel (1989) were primarily concerned with the statistical significance of RNA folding, rather than with distinguishing the relative contributions of base composition and base order. However, with a pipeline between the various programs that were offered by the Wisconsin Genetics Computer Group, the base composition and base order-dependent components were separated and individually assessed (“folding of randomized sequence difference” analysis; FORS-D analysis). Departing from Le and Maizel, FORS-D values (see below) were not divided to yield Z-scores, but were simply plotted with statistical error bars (Forsdyke, 1995a, Forsdyke, 1995b, Forsdyke, 1995c, Forsdyke, 2013, Forsdyke, 2014, Forsdyke, 2016). The limits of the latter were generally close to the corresponding FORS-D values and, for clarity, are omitted here.
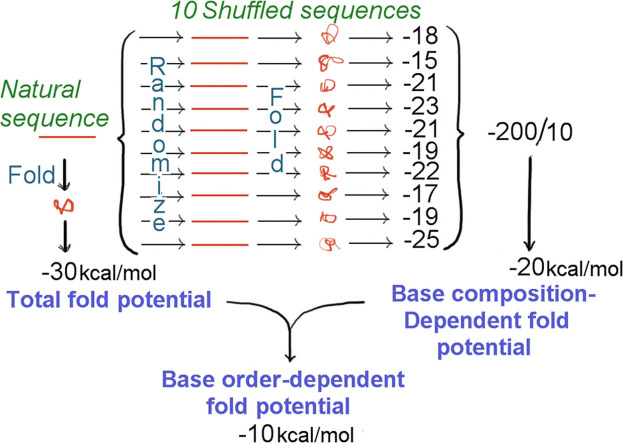
A window of 200 bases is moved along a natural sequence in 20 or 50 base steps. A folding program (RNA Structure, version 4.2; Mathews et al., 2004) is applied to the sequence in each window to obtain "folding of natural sequence" (FONS) values for each window, to which both base composition and base order will have contributed. The four bases in each sequence window are then shuffled to destroy their order while retaining base composition, and folding energy is again determined. This shuffle-and-fold "Monte Carlo" procedure is repeated ten times and the average (mean) folding value is taken as the "folding of randomized sequence mean" (FORS-M) value for that window. This reflects the contribution of base composition alone. The base order-dependent component is then derived by subtraction from the FONS value. This is the “folding of randomized sequence difference"(FORS-D) value. The sign of the difference value depends on the direction of the subtraction. (FORS-D was given positive values in the early 1990s, but the direction of subtraction was changed in subsequent work.) Fluctuations seen in FONS profiles of genomes are mostly due to changes in the FORS-D component, whereas the FORS-M component, while making a major contribution to folding energetics, is relatively constant.
Apart from the RNA Structure program, another popular folding tool, the Vienna RNA package (Lorenz et al., 2011), uses the same thermodynamic model and is known to produce very similar predictions in general.
2.4. Validation of four base shuffling
The approach was modified by others who, rather than shuffling the four bases, favored retaining some base order information. Accordingly, they shuffled groups of bases (e.g. the sixteen dinucleotides). Following disparagement of the conceptual basis of four base shuffling, which was duly clarified (Forsdyke, 2007a), the validity of single base level shuffling is now generally accepted and is being applied routinely to viral genomes (Andrews et al., 2018, Andrews et al., 2021, Simmonds, 2020a, Simmonds, 2020b). The Monte Carlo procedure can also be simplified to decrease FORS-M computational time (Chen et al., 1990) using support vector machine-based technology (Washietl et al., 2005). A later software modification ("Bodslp"), written by Professor Jian-sheng Wu (Zhang et al., 2008a, Zhang et al., 2008b), retains our Monte Carlo approach and was further developed by Professor Shungao Xu as "Random Fold-Scan" for Windows-based systems (Xu et al., 2007).
Currently, there are differences in literature terminology that may confuse. The FORS-D values derived from FORS-D analysis correspond to the minimum free energy difference ("MFED") values derived from the "GORS" analysis of Simmonds (2020a). A comment added to the latter by Forsdyke – entitled "Functional Basis for Pervasive RNA Secondary Structure" – provides further background.
In addition to assisting the study of infectious viruses and protozoa (Xue and Forsdyke, 2003), FORS-D analysis proved fruitful when applied to topics such as speciation (Forsdyke, 1996, Forsdyke, 2014, Zhang et al., 2008a), the origin of introns (Forsdyke, 1995b, Forsdyke, 1995c, Forsdyke, 2013), relating structure to recombination breakpoints and deletions (Zhang et al., 2005a, Zhang et al., 2005b), and relying on a single sequence (rather than alignments) for the determination of positive Darwinian selection (Forsdyke, 2007b). For detailed historical accounts see Forsdyke, 2016, Forsdyke, 2021a, Forsdyke, 2021b.
However, there are important caveats. For a given sequence window, output can follow only from the base order in that window. Lost are higher order structures that might occur naturally through long-range interactions (Ziv et al., 2020). Furthermore, different window sizes would lead to different predicted structures (Huston et al., 2020, Rangan et al., 2021). If the artificial demarcation of a window happens to cut between the two limbs of a natural stem-loop structure, then lost are what might have been contributed to the folding energetics had a larger window, or a different section point, been chosen. Variations in step size will generate differences in windows and hence variations in results. Such variations might be less when window margins correspond to natural section points, such as those demarcating an RNA transcript.
There is also a kinetic aspect, particularly apparent with transcripts, due to the probability that the natural pattern of early 5' folding will influence later folding. Exons generally have less structure than introns (Forsdyke, 1995b, Forsdyke, 1995c) whose excision can influence the folding pattern of mature mRNAs (Saldi et al., 2021). The final result of folding may include approximation of the 5' and 3' ends of mRNA (Lai et al., 2018). While the emergence of faster computers and scalable algorithms has made feasible direct determination of entire global structures (Huang et al., 2019), the validation of their inter-loop "kissing" interactions remains problematic even for the latest technologies (Cao and Xue, 2021). Despite the limitations, determinations of local structures remain more reliable than global (Lange et al., 2012).
2.5. Validation of structural invariance index
Plots of structural invariance (SI) facilitate the identification of conserved RNA structures. Structure is reflected in the degree of negativity of FORS-D values, and invariance is reflected in limitation of recorded positive substitutions at individual base sites (polymorphisms). Thus, structure and invariance can be united numerically by addition (SI = FORS-D + Substitutions). The SI would be −20 units in the case of a FORS-D value of −20 kcal/mol in a window where there had been zero recorded substitutions (i.e. mutant forms had been eliminated by natural selection or genetic drift). If there had been 10 recorded substitutions the SI would be −10 units.
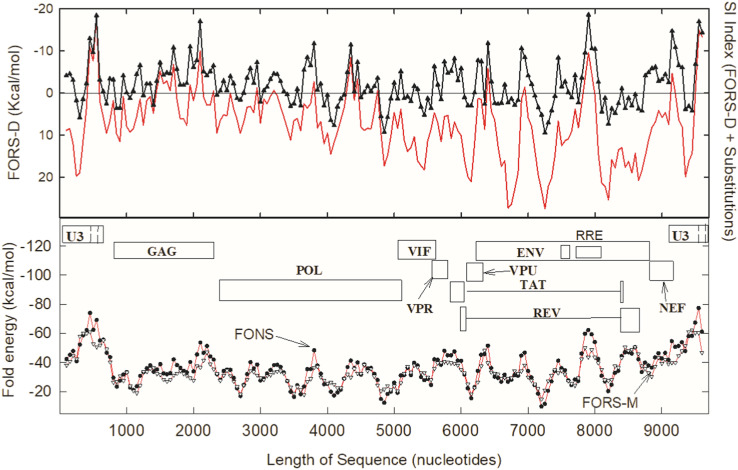
High negative SI values provide an ad hoc measure of structural conservation among a set of genomes. The validity of this approach is supported by prior work with HIV-1. Fig. 1 shows application of the index to previously reported data on HIV-1 (Forsdyke, 1995a, Forsdyke, 2014). Here a highly negative SI index value corresponds to regions recognized as likely to offer Achilles heel-like vulnerability (Forsdyke, 2016). The region around the window centered on 500 nucleotide bases is the focus of recent work (Ingemarsdotter et al., 2018, Rein, 2020, Ding et al., 2020).
Fig. 1.
Structural invariance (SI) of HIV-1 (subtype SF2) as compared with reference subtype (HXB2). The region recently recognized as a potential Achilles heel (bases 380–640), has a high negative FORS-D value (black triangles) similar to those of the RRE (Rev response element) and the 3' untranslated region (UTR). The SI index (continuous red line) indicates highest sequence conservation in the regions of the RNA packaging signal and the 3'UTR. For details see Forsdyke (1995a). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. Variation due to base order component
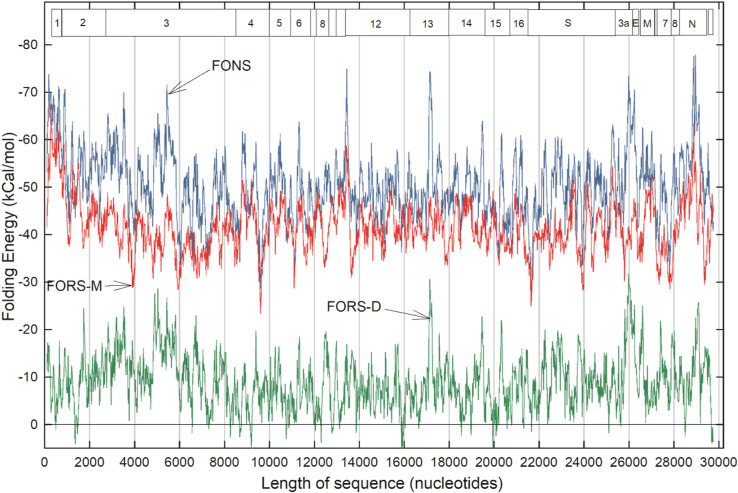
A nucleic acid folding program was applied to 200 nucleotide base "windows" that were moved along a SARS-CoV-2 sequence in 20 base steps. This provided a set of folding energy values fluctuating from −26 kCal/mol to −78 kCal/mol, with the latter negative value indicating stronger folding ( Fig. 2; blue top line). These "folding of natural sequence" (FONS) values were further dissected into their two fundamental components, that due to base composition (FORS-M) and that due to base order (FORS-D).
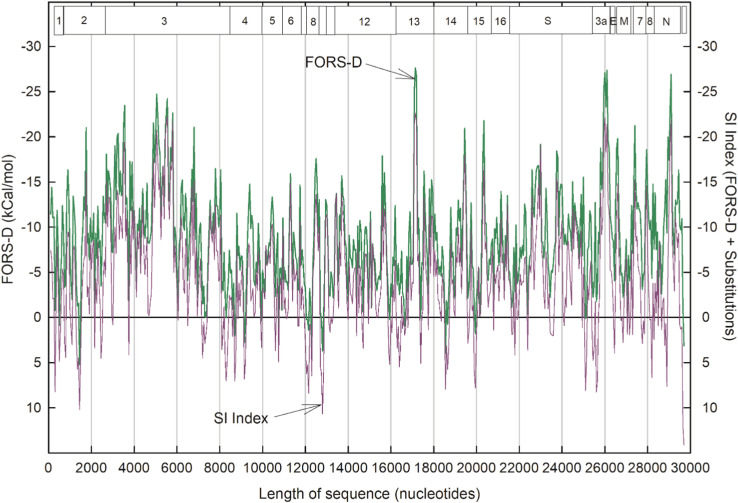
Fig. 2.
Variations in folding energetics throughout the single-stranded RNA genome of a clade B SARS-CoV-2 sequence, deemed "prototypic" since it appeared in China early in the COVID-19 epidemic (Wuhan-Hu1; NC_045512.2). Data points for 1,484 200-base windows (beginning at base 1 and moved in 20 base steps) were connected rectilinearly. FONS values (top, blue); FORS-M values (middle, red); FORS-D values (bottom, green). Boxes indicate the locations of the nonstructural proteins (NSP 1–16) and structural proteins (S, etc.). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The composition component (Fig. 2; red middle line) is generally less negative than the FONS value (blue line), the difference being due to the contribution made by the base order-dependent component (green bottom line). Composition (FORS-M) usually makes a larger, but less variable contribution, whereas the order component (FORS-D) usually makes a smaller, but highly variable contribution. Thus, much of the fluctuation that distinguishes different parts of a genome in the FONS profile is due to the base order component. As with our previous studies of a wide variety of genomes, FORS-D values provide sensitive indicators of regional differences in the strength of folding. A notable exception is a peak (−75 kCal/mol) located in a window centered on base 13,440 at the boundary between ORFs NSP10 and NSP12, in the region of the ribosomal FSE. Here, the base composition component makes a major contribution.
In a few regions FORS-M is more negative than FONS and here FORS-D values are positive. Thus, here "nature" appears to have arranged the order of the nucleotide bases to restrain the impact of base composition on the strength of folding (i.e. encourage formation of loops that might be targeted by antisense RNAs; Forsdyke, 2007a). A need for this appears particularly evident in windows centered on the following bases: 1360 (NSP2), 9200 (NSP4), 12,300 (NSP8), 15,880 and 15,920 (NSP12), 19,940 (NSP15), 29,680 and 29,700 (3' untranslated region).
3.2. Inverse relationship between folding and substitutions
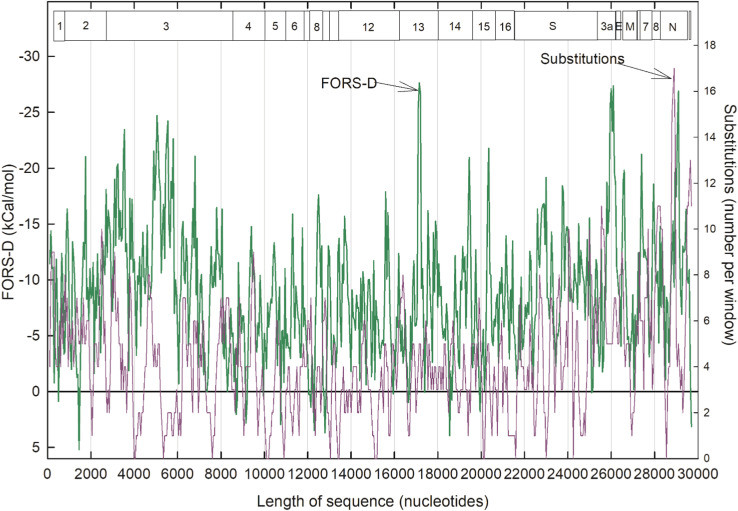
The degree of conservation in sequential windows of members of a set of sequences from China was evaluated as the number of substitutable base positions (polymorphism), relative to the prototype sequence ( Fig. 3). This was compared with corresponding FORS-D values, the profile of which differed a little from that of Fig. 2 due to different step values (see legend to figure and Materials and Methods).
Fig. 3.
Comparison of base order-dependent stem-loop potential values (FORS-D; green) for the Wuhan-Hu1 SARS-CoV-2 prototype, and base substitutions values (purple). The latter were found in the same sequence windows among a set of Chinese SARS-CoV-2 isolates. Data points for 594 200-base sequence windows (beginning at base 1 and moved in 50 base steps) were connected rectilinearly. (There were some ambiguities in substitutions in the sequences included in the first two windows, making the first two data points less reliable). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As with previous studies (Forsdyke, 1995a, Forsdyke, 1995b, Forsdyke, 1995c), there tended to be a reciprocal relationship between the base order contribution to folding energy (FORS-D) and substitutions. When one was high the other was low, and vice-versa. Negative FORS-D values were generally greatest in the region of ORF NSP3, and in narrow regions corresponding to the NSP13, ORF3a and the nucleocapsid (N) proteins. Zero substitutions were observed for windows centered on the following base nucleotide values, some isolated and some grouped (in brackets): [4000,4050], 5350, 7600, [10,100, 10,150, 10,200], 10,900, 13,050, [13,400, 13,450; close to ribosomal FSE], [15,100, 15,150, 15,200], [20,100, 20,150], 21,600, 23,000, 24,250, 28,550. On the other hand, substitutions were higher in the last part of the sequence, being particularly evident in the nucleocapsid (N) protein ORF. While the zero-substitution windows centered on bases 5350, 7600 and 21,600 appeared isolated, each had many neighboring windows with few substitutions. At some positions, low substitution values were accompanied by high negative RNA folding energy values (i.e. reciprocal relationships), indicating conservation of structural function that could be at the RNA genome level and/or in RNA transcripts from that genome.
3.3. Structural invariance index identifies potential targets
For a more focused view of the reciprocal separation of high negative folding values and corresponding numbers of substitutions (ranging from zero to high positive values) the two were added to provide the "structural invariance" (SI) index ( Fig. 4).
Fig. 4.
Data of Fig. 3 plotted with FORS-D values (green) accompanied by the structural invariance (SI) index (purple). (For plots of analogous HIV-1 data see Fig. 1). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Here, despite having some substitutions, ORFs NSP13 and NSP3 were preeminent (scoring −22.6 and −22.2 SI units, respectively). The next two best negative scores were within ORF3a (scoring −22.1 SI units) and within the N protein ORF (scoring −21.9 units). Whereas the scores for the NSP13, ORF3a and N locations were narrowly based, the entire NSP3 location tended towards high negative scores. Close downstream to the high-negative-scoring N region (window centered on base 29,100) is a region with the highest number of substitutions (see Fig. 3) centered on base 28,900 (scoring +7.6 SI units).
Windows 13,400 and 13,450 in the FSE region scored −12.5 and −13.7 units, respectively. However, there were many more windows with higher negative scores. Indeed, SI values more negative than −15.0 units were observed for windows centered on the following base nucleotide values, some isolated and some grouped (in brackets): 1750, 2700, [3500, 3550], 4900, [5000, 5050, 5100], 5200, [5450, 5500, 5550], 5800, 6800, [17,100, 17,150, 17,200], 19,450, 20,350, 23,000, 23,750, [25,950, 26,000, 26,050, 26,100], 27,400, [29,050, 29,100].
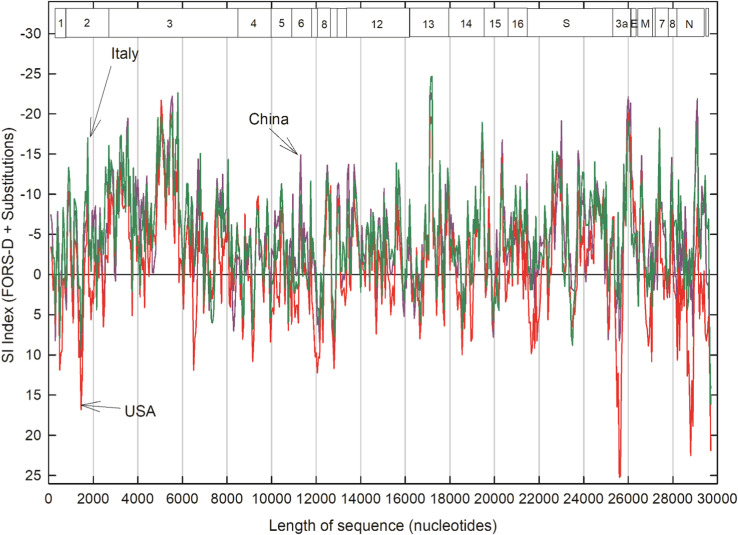
3.4. SI indices for Italy and New York (USA)
The SI profile for China (Fig. 4) was, in broad outline, confirmed with corresponding data later downloaded from Italy and New York, USA ( Fig. 5). The high negative SI indices found in the NSP3, NSP13, ORF3a and N regions, were evident with sequences for all three locations. Other regions, notably the S region, were also corroborated. The high positive SI values, indicating regions likely to have poorly conserved structures, are also corroborated at some locations. A high intraspecies mutation rate for the N protein ORF (Fig. 3) is also seen when interspecies comparisons are made with other coronavirus species, with implications for early speciation mechanisms (Dilucca et al., 2020).
Fig. 5.
Comparison of SI indices for China (381 isolates) as in Fig. 4 (purple), with SI indices for Italy (430 isolates; green) and New York, USA (932 isolates; red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Three therapeutic challenges
Mutation rates of microbial pathogens are generally higher than those of their hosts. While a microbe spreading from host-to-host can "anticipate" that it will face a succession of broadly similar challenges, in the short-term those hosts cannot likewise "anticipate" that new microbial invaders will remain as they were in previous hosts. Thus, host immune defenses may be overwhelmed. (In the long-term there is a different scenario related to innate immunity; see below). Therapeutic challenges are, first, to locate a conserved, less-variable, part of a pathogen's genome that it will have inherited sequentially from a multiplicity of past generations and so is likely to carry through to a multiplicity of future generations. Second, is to identify the corresponding primary function, be it at the genome, RNA transcript, or protein level. Third, from this knowledge (that may be incomplete; i.e. function not fully clarified), devise effective pathogen inhibition without imposing deleterious side-effects on the host.
4.2. Identification of potential RNA-level Achilles heels in HIV-1 and SARS-CoV-2
Viral vulnerability is often assumed to associate with protein-level functions (Haniff et al., 2020, Robson, 2020). However, studies of the AIDS virus have identified genome structure itself as both functional and conserved, so signifying vulnerability (Forsdyke, 1995a, Forsdyke, 2014; see Fig. 1). A genomic packaging signal for HIV-1, which is specifically recognized by the nucleocapsid domain of its Gag polyprotein, has long been recognized as a potential "Achilles heel" (Forsdyke, 1995a, Forsdyke, 2014, Forsdyke, 2016), so inviting therapeutic exploration (Ingemarsdotter et al., 2018, Rein, 2020, Ding et al., 2020). Through targeting of specific RNA conformations, Gag not only influences the assembly of HIV-1 genomic RNA into virus particles, but also regulates HIV-1 mRNA translation (Anderson and Lever, 2006). RNA conformational flexibility, facilitated by fine-tuned intermediate-strength binding, should permit easier switching between regulatory options. Indeed, small changes in target RNA structure can impede this (Ding et al., 2020). Thus, unlike most other regions of the HIV-1 genome, mutations here would likely lead to negative Darwinian selection at an early stage – hence the high conservation. Application of the same bioinformatic technology to the SARS-CoV-2 virus genome has now revealed similar potential "Achilles heels" (Fig. 4).
Lacking the chronicity of HIV-1 infection, the genome of SARS-CoV-2 should have been shaped less by adaptations to counter long-term host immune defenses. It cannot hide within its host genome in latent DNA form. Yet, the larger SARS-CoV-2 genome contains many more genes than HIV, which require differential expression according to the stage of infection. Even more complex regulatory controls can be envisaged, likely requiring conserved genome conformations at appropriate locations. Be they synonymous or non-synonymous, mutations in these structured regions could result in negative selection of the viruses in which they occurred – hence high conservation. The ribosome FSE located close to base 13,468 (Fig. 2) would seem to exemplify this (Kelly et al., 2020), and a potential targeting agent is now available (Haniff et al., 2020). However, we have here identified other structurally important regions with more base order-dependence and higher degrees of conservation (Fig. 3, Fig. 4, Fig. 5), that might, either singly or collectively, be better candidates for targeting. These broad regions, demarcated by window boundaries, should serve to focus attention on the local structural details required, not only for therapeutic purposes, but also to guide the choice of primers in diagnostic PCR assays. Thus, present goals have been achieved in that, according to our criteria, potential vulnerabilities have been identified together with their approximate locations. These should guide the application of more advanced technologies (e.g. Cao and Xue, 2021).
4.3. Base composition relates to species evolution
When determining folding energy, our approach depends on eliminating contributions of base composition which, as noted, plays an unusual role in the case of the FSE. More usually, base composition is a distinctive characteristic of entire genomes or large genome sectors, which reflects their underlying oligomer ("k-mer") content (Aggarwala and Voight, 2016; Forsdyke, 2021a, Forsdyke, 2021b; Morozov, 2017). The slow genome-wide accumulation of mutations in oligomer composition, easiest documented as changes in 1-mer frequency (base composition; GC%), can serve to initiate divergence into new species. By preventing that accumulation, potentially diverging organisms can stay within the confines of their species (Forsdyke, 1996, Forsdyke, 2019a, Forsdyke, 2019b, Forsdyke, 2021a, Forsdyke, 2021b). The presence or absence of synonymous mutations (Simmonds, 2020a, Simmonds, 2020b, Wang et al., 2021), which affect structure rather than amino acid composition, can have an important role in this process. The primary role of constancy in the base composition-related character is to prevent recombination with allied species (interspecies recombination) while facilitating the intraspecies recombination that can correct mutations, so retaining species individuality (Forsdyke, 2014, Forsdyke, 2016). Such recombination is thought likely to be initiated by "kissing" interactions between complementary sets of unpaired bases (k-mers) at the tips of stem-loop structures (Forsdyke, 1996).
Thus, we are here concerned with localized intraspecies mutations that affect fitness, so making members of a species carrying those mutations liable to natural selection. The mutations facilitate within-species evolution rather than divergence into new species. And when that evolution has run its course, some of the polymorphic bases will have become less mutable, so will be deemed "conserved." Indeed, mutations of ORF NSP3 are high when the sequences of different coronavirus species are compared (Claverie, 2020), yet when, from intraspecies comparisons, mutations (in the form of base substitutions) are scored, they are very low (Fig. 3, Fig. 4). Our technology (see Materials and Methods) removes the base composition-dependent component of mutational changes (that relates more to interspecies evolution) and focuses on the base order-dependent component (that relates more to intraspecies evolution). It best reflects localized functions, be they encoding protein or determining the potentiality for folding into higher order structure, of linear, single-stranded, nucleic acid sequences.
4.4. Conservation as a reliable indicator
We sought regions that were both high in stem-loop potential and bereft of mutations, following the premise that conserved functions would be best targeted therapeutically, assuming the availability of pathogen-specific therapeutic agents that would not cross-react with hosts. Interference with structural nucleic acid level functions might be less likely to produce unforeseen host side-effects than with protein-level functions. But is conservation necessarily a good indicator of likely therapeutic success? Indeed, a conserved function in a pathogen could owe that conservation to the pathogen strategy of, whenever possible, mutating to resemble its host. As reviewed elsewhere (Forsdyke, 2019c), this would make it less vulnerable to host innate and acquired immune defenses. To prevent autoimmunity, the generation of immune cell repertoires involves the negative selection of self-reacting cells so creating "holes" in the repertoire that pathogens can exploit by progressive mutation towards host-self, testing mutational effectiveness a step at a time. This would make it advantageous for the host, during the process of repertoire generation, not only to negatively select immune cells of specificity towards "self," but also to positively select immune cells of specificity towards "near-self." A high level of anti-near-self immune cell clones would constitute a barrier limiting the extent of pathogen mutation towards self. The existence of such positive selection is now generally accepted, with the implication that some pathogen functions might have approached so close to host-self that targeting them therapeutically would result in cross-reactivity.
This caveat aside, we deem conservation a reliable indicator that a certain pathogen function is likely to be a suitable target for therapy. Attacking a short segment of a pathogen nucleic acid sequence is unlikely to ensnare a similar segment of its host's nucleic acid. In any case, to militate against this, the pathogen specificity of a potential therapeutic agent can be screened against the prototypic human genomic sequence (assuming it is unlikely that patient genomes will significantly depart from this).
4.5. Concluding remarks
While developments of prophylactic vaccines against infection with SARS-CoV-2 have made promising starts, mutational variants are appearing world-wide. Methods to boost post-infection host immune defenses and to directly target SARS-CoV-2 are urgently needed. These require a better understanding both of viral interactions with host innate and acquired immune systems (Forsdyke, 2019c), and of viral vulnerabilities. The latter inquiry proceeds in three steps: Find specific "Achilles heels." Design therapies to exploit them. Prove their clinical effectiveness. We have here been concerned with the first step – the identification of high SI scoring regions that may then serve to focus attention on local structural details (Wacker et al., 2020, Huston et al., 2020). Although the bioinformatic technology related to this has long been available (Forsdyke, 1995a), our claim to reveal viral "Achilles heels," as promulgated in successive textbook editions (Forsdyke, 2016), has only recently gained support (Ingemarsdotter et al., 2018, Rein, 2020, Ding et al., 2020).
This may be because the importance of removing redundant information and analyzing solely the contribution of base order to folding energy, was not fully appreciated. Even those who have employed the same technology have expressed puzzlement at the "biological purpose" of so much "pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses" (Simmonds, 2020a) and regret the "poorly understood RNA structure-mediated effects on innate and adaptive host immune responses" (Simmonds et al., 2021). Thus, we have here repeated and expanded on past clarifications (Forsdyke, 2007a, Xu et al., 2007, Zhang et al., 2008b) of the conceptual basis of a technology that has contributed to the understanding of many biological problems other than viral infections (Forsdyke, 2016). Meanwhile, it is pleasing to note that, having identified a conserved interspecies element, others are addressing the targeting and therapy steps (Lulla et al., 2021). Furthermore, with minimal evidence on targeting, much progress is being made with the therapy step (Medeiros et al., 2021, Haniff et al., 2020, Chen et al., 2020, Bush et al., 2021, Hammond et al., 2021). It is hoped that by targeting one or more of the elements within the intraspecies conserved regions in the SARS-CoV-2 genome here identified, rapid cures will be achieved.
6. Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
Authors declare no conflict of interest.
Acknowledgments
We thank Prof. Shungao Xu at Jiangsu University for software, and Ms. Le Cao and Ms. Yingying Ma at Shanghai Public Health Clinical Center, Fudan University, for their technical support. Queen's University hosts Forsdyke's webpages. The bioRxiv server hosts a preprint.
Data availability
We have uploaded all the necessary data and we have not deposited data anywhere. The RDM is not available for this study. We can provide the raw data if someone needs it.
References
- Aggarwala V., Voight B.F. An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat. Genet. 2016;48:349–355. doi: 10.1038/ng.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.C., Lever A.M.L. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R.J., Roche J., Moss W.N. ScanFold: an approach for genome-wide discovery of local RNA structural elements – applications to Zika virus and HIV. PeerJ. 2018;6 doi: 10.7717/peerj.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R.J., O’Leary C.A., Tomkins V.S., Peterson J.M., Haniff H.S., Williams C., et al. A map of the SARS-CoV-2 RNA structurome. NAR Genom. Bioinform. 2021;3(2):1–14. doi: 10.1093/nargab/lqab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J.A., Williams C.C., Meyer S.M., Tong Y., Haniff H.S., Childs-Disney J.L., Disney M.D. Systematically studying the effect of small molecules interacting with RNA in cellular and preclinical models. ACS Chem. Biol. 2021;16:1111–1127. doi: 10.1021/acschembio.1c00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Xue Y. Characteristic chemical probing patterns of loop motifs improve prediction accuracy of RNA secondary structures. Nucleic Acids Res. 2021;49:4294–4307. doi: 10.1093/nar/gkab250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-H., Le S.-Y., Shapiro B., Currey K.M., Maizel J.V. A computational procedure for assessing the significance of RNA secondary structure. Comput. Appl. Biosci. 1990;6:7–18. doi: 10.1093/bioinformatics/6.1.7. [DOI] [PubMed] [Google Scholar]
- Chen W., Feng P., Liu K., Wu M., Lin H. Computational identification of small interfering RNA targets in SARS-CoV-2. Virol. Sin. 2020;35:359–361. doi: 10.1007/s12250-020-00221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J.-M. A putative role of de-mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 Nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12:646. doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12:1–21. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P., Kharytonchyk S., Waller A., Mbaekwe U., Basappa S., Kuo N., Frank H.M., Quasney C., Kidane A., Swanson C., Van V., Sarkar M., Cannistraci E., Chaudhary R., Flores H., Telesnitsky A., Summers M.F. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA. 2020;117:17737–17746. doi: 10.1073/pnas.2008519117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsdyke D.R. Reciprocal relationship between stem-loop potential and substitution density in retroviral quasispecies under positive Darwinian selection. J. Mol. Evol. 1995;41:1022–1037. doi: 10.1007/BF00173184. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. A stem-loop "kissing" model for the initiation of recombination and the origin of introns. Mol. Biol. Evol. 1995;12:949–958. doi: 10.1093/oxfordjournals.molbev.a040273. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. Conservation of stem-loop potential in introns of snake venom phospholipase A2 genes: an application of FORS-D analysis. Mol. Biol. Evol. 1995;12:1157–1165. [Google Scholar]
- Forsdyke D.R. Different biological species "broadcast" their DNAs at different (G+C)% "wavelengths". J. Theor. Biol. 1996;178:405–417. doi: 10.1006/jtbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. Calculation of folding energies of single-stranded nucleic acid sequences: conceptual issues. J. Theor. Biol. 2007;248:745–753. doi: 10.1016/j.jtbi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. Positive Darwinian selection. Does the comparative method rule? J. Biol. Syst. 2007;15:95–108. [Google Scholar]
- Forsdyke D.R. Introns first. Biol. Theor. 2013;7:196–203. [Google Scholar]
- Forsdyke D.R. Implications of HIV RNA structure for recombination, speciation, and the neutralism-selectionism controversy. Microbes Infect. 2014;16:96–103. doi: 10.1016/j.micinf.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. third ed. Springer; New York: 2016. Evolutionary Bioinformatics; pp. 241–244. [Google Scholar]
- Forsdyke D.R. Success of alignment-free oligonucleotide (k-mer) analysis confirms relative importance of genomes not genes in speciation. Biol. J. Linn. Soc. 2019;128:239–250. [Google Scholar]
- Forsdyke D.R. Hybrid sterility can only be primary when acting as a reproductive barrier for sympatric speciation. Biol. J. Linn. Soc. 2019;128:779–788. [Google Scholar]
- Forsdyke D.R. Two signal half century: from negative selection of self-reactivity to positive selection of near-self reactivity. Scand. J. Immunol. 2019;89 doi: 10.1111/sji.12746. [DOI] [PubMed] [Google Scholar]
- Forsdyke D.R. Neutralism versus selectionism: Chargaff's second parity rule, revisited. Genetica. 2021;149:81–88. doi: 10.1007/s10709-021-00119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsdyke D.R. Complementary oligonucleotides rendered discordant by single base mutations may drive speciation. Biol. Theor. 2021:16. doi: 10.1007/s13752-021-00380-z. [DOI] [Google Scholar]
- Hammond S.M., Aartsma-Rus A., Alves S., Borgos S.E., Buijsen R.A.M., Collin R.W.J., Covello G., Denti M.A., Desviat L.R., Echevarría L., Foged C., Gaina G., Garanto A., Goyenvalle A.T., Guzowska M., Holodnuka I., Jones D.R., Krause S., Lehto T., Montolio M., Van Roon-Mom W., Arechavala-Gomeza V. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med. 2021;13:13243. doi: 10.15252/emmm.202013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniff H.S., Tong Y., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O'Leary C.A., Benhamou R.I., Moss W.N., Disney M.D. Targeting the SARS-CoV‑2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 2020;6:1713–1721. doi: 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Zhang H., Deng D., Zhao K., Liu K., Hendrix D.A., Mathews D.H. LinearFold: linear-time approximate RNA folding by 5'-to-3' dynamic programming and beam search. Bioinformatics. 2019;35:i295–i304. doi: 10.1093/bioinformatics/btz375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston N.C., Wan H., Strine M.S., Tavares R., de C.A., Wilen C., Pyle A.M. Comprehensive in-vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2020;81:584–598. doi: 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingemarsdotter C.K., Zeng J., Long Z., Lever A.M.L., Kenyon J.C. An RNA-binding compound that stabilizes the HIV-1 gRNA packaging signal structure and specifically blocks HIV-1 RNA encapsidation. Retrovirology. 2018;15:25. doi: 10.1186/s12977-018-0407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: Nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.A., Olson A.N., Neupane K., Munshi S., San Emeterio J., Pollack L., Woodside M.T., Dinman J.D. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2) J. Biol. Chem. 2020;295:10741–10748. doi: 10.1074/jbc.AC120.013449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tao Q., Weaver S., Sanderford M., Caraballo-Ortiz M.A., Sharma S., Pond S., Miura S. An evolutionary portrait of the progenitor SARS-CoV-2 and its dominant offshoots in COVID-19 pandemic. Mol. Biol. Evol. 2021;38:3046–3059. doi: 10.1093/molbev/msab118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.-J.C., Kayedkhordeh M., Cornell E.V., Farah E., Bellaousov S., Rietmeijer R., Salsi E., Mathews D.H., Ermolenko D.N. mRNAs and lncRNAs intrinsically form secondary structures with short end-to-end distances. Nat. Commun. 2018;9:4328. doi: 10.1038/s41467-018-06792-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S.L., Maticzka D., Möhl M., Gagnon J.N., Brown C.M., Backofen R. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012;40:5215–5226. doi: 10.1093/nar/gks181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.-Y., Maizel J.V. A method for assessing the statistical significance of RNA folding. J. Theor. Biol. 1989;138:495–510. doi: 10.1016/s0022-5193(89)80047-5. [DOI] [PubMed] [Google Scholar]
- Li, Y., Garcia, G., Arumugaswami, V., Guo, F., 2021. Structure-based design of antisense oligonucleotides that inhibit SARS-CoV-2 replication, bioRxiv, August 24, doi: 10.1101/2021.08.23.457434.
- Lorenz R., Bernhart S.H., Siederdissen C.H. zu, Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla V., Wandel M.P., Bandyra K.J., Ulferts R., Wu M., Dendooven T., Yang X., Doyle N., Oerum S., Beale R., O’Rourke S.M., Randow F., Maier H.J., Scott W., Ding Y., Firth A.E., Bloznelyte K., Luisi B.F. The stem loop 2 motif is a site of vulnerability for SARS-CoV-2. J. Virol. 2021;95 doi: 10.1128/JVI.00663-21. e00663-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M., Incarnato D. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020;48:12436–12452. doi: 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros I.G., Khayat A.S., Stransky B., Santos S., Assumpção P., de Souza J.E.S. A small interfering RNA (siRNA) database for SARS-CoV-2. Sci. Rep. 2021;11:8849. doi: 10.1038/s41598-021-88310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov A.A. k-mer distributions of aminoacid sequences are optimized across the proteome. bioRxiv. 2017 doi: 10.1101/190280. [DOI] [Google Scholar]
- Pipes L., Wang H., Huelsenbeck J.P., Nielsen R. Assessing uncertainty in the rooting of the SARS-CoV-2 phylogeny. Mol. Biol. Evol. 2020;38:1537–1543. doi: 10.1093/molbev/msaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R., Watkins A.M., Chacon J., Kretsch R., Kladwang W., Zheludev I.N., Townley J., Rynge M., Thain G., Das R. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res. 2021;49:3092–3108. doi: 10.1093/nar/gkab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch J.W., Capoferria A.A., Katusiime M.G., Patro S.C., Kearney M.F. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:24614–24616. doi: 10.1073/pnas.2017726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A. The heart of the HIV RNA packaging signal? Proc. Natl. Acad. Sci. USA. 2020;117:19621–19623. doi: 10.1073/pnas.2013378117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson B. The use of knowledge management tools in viroinformatics. Example study of a highly conserved sequence motif in Nsp3 of SARS-CoV-2 as a therapeutic target. Comput. Biol. Med. 2020;125 doi: 10.1016/j.compbiomed.2020.103963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T., Riemondy K., Erickson B., Bentley D.L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell. 2021;81:1789–1801. doi: 10.1016/j.molcel.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarni S., Biswas B., Liu S., Olson E.D., Kitzrow J.P., Rein A., Wysocki V.H., Musier-Forsyth K. HIV-1 Gag protein with or without p6 specifically dimerizes on the viral RNA packaging signal. J. Biol. Chem. 2020;295:14391–14401. doi: 10.1074/jbc.RA120.014835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, P., 2020a. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses – an endeavour to understand its biological purpose, bioRxiv, 17 June, doi: 10.1101/2020.06.17.155200. [DOI] [PMC free article] [PubMed]
- Simmonds P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. mBio. 2020;11 doi: 10.1128/mBio.01661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Williams S., Harvala H. Understanding the outcomes of COVID-19 – does the current model of an acute respiratory infection really fit? J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Abriola L., Niederer R.O., Pedersen S.F., Alfajaro M.M., Silva Monteiro V., Wilen C.B., Ho Y.C., Gilbert W.V., Surovtseva Y.V., Lindenbach B.D., Guo J.U. Restriction of SARS-CoV-2 replication by targeting programmed −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R., de C.A., Mahadeshwar G., Wan H., Huston N.C., Pyle A.M. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. J. Virol. 2021;95 doi: 10.1128/JVI.02190-20. e02190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker A., Weigand J.E., Akabayov S.R., Altincekic N., Bains J.K., Banijamali E., Binas O., Castillo-Martinez J., Cetiner E., Ceylan B., Chiu L.Y., Davila-Calderon J., Dhamotharan K., Duchardt-Ferner E., Ferner J., Frydman L., Fürtig B., Gallego J., Grün J.T., Hacker C., Haddad C., Hähnke M., Hengesbach M., Hiller F., Hohmann K.F., Hymon D., de Jesus V., Jonker H., Keller H., Knezic B., Landgraf T., Löhr F., Luo L., Mertinkus K.R., Muhs C., Novakovic M., Oxenfarth A., Palomino-Schätzlein M., Petzold K., Peter S.A., Pyper D.J., Qureshi N.S., Riad M., Richter C., Saxena K., Schamber T., Scherf T., Schlagnitweit J., Schlundt A., Schnieders R., Schwalbe H., Simba-Lahuasi A., Sreeramulu S., Stirnal E., Sudakov A., Tants J.N., Tolbert B.S., Vögele J., Weiß L., Wirmer-Bartoschek J., Wirtz Martin M.A., Wöhnert J., Zetzsche H. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. 2020;48:12415–12435. doi: 10.1093/nar/gkaa1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Pipes L., Nielsen R. Synonymous mutations and the molecular evolution of SARS-Cov-2 origins. Virus Evol. 2021;7 doi: 10.1093/ve/veaa098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H.Y., Forsdyke D.R. Low complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptations. Mol. Biochem. Parasitol. 2003;128:21–32. doi: 10.1016/s0166-6851(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Xu S.G., Wei J.F., Zhang C.Y. A FORS-D analysis software “Random_fold_scan” and the influence of different shuffle approaches on FORS-D analysis. J. Jiangsu Univ. 2007;17:461–466. [Google Scholar]
- Zhang C.-Y., Wei J.-F., He S.-H. Local base order influences the origin of ccr5 deletions mediated by DNA slip replication. Biochem. Genet. 2005;43:229–237. [PubMed] [Google Scholar]
- Zhang C.-Y., Wei J.-F., He S.-H. The key role for local base order in the generation of multiple forms of China HIV-1 B'/C intersubtype recombinants. BMC Evol. Biol. 2005;5:53. doi: 10.1186/1471-2148-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu S., Wei J.-F., Forsdyke D.R. Microsatellites that violate Chargaff's second parity rule have base order-dependent asymmetries in the folding energies of complementary DNA strands and may not drive speciation. J. Theor. Biol. 2008;254:168–177. doi: 10.1016/j.jtbi.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang C.-Y., Wei J.-F., Wu J.-S., Xu W.-R., Sun X., He S.-H. Evaluation of FORS-D analysis: a comparison with the statistically significant stem-loop potential. Biochem. Genet. 2008;46:29–40. doi: 10.1007/s10528-007-9126-6. [DOI] [PubMed] [Google Scholar]
- Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol. Cell. 2020;80:1067–1077. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have uploaded all the necessary data and we have not deposited data anywhere. The RDM is not available for this study. We can provide the raw data if someone needs it.