Abstract
Both p53 and ATM are checkpoint regulators with roles in genetic stabilization and cancer susceptibility. ATM appears to function in the same DNA damage checkpoint pathway as p53. However, ATM’s role in p53-dependent apoptosis and tumor suppression in response to cell cycle dysregulation is unknown. In this study, we tested the role of murine ataxia telangiectasia protein (Atm) in a transgenic mouse brain tumor model in which p53-mediated apoptosis results in tumor suppression. These p53-mediated activities are induced by tissue-specific inactivation of pRb family proteins by a truncated simian virus 40 large T antigen in brain epithelium. We show that p53-dependent apoptosis, transactivation, and tumor suppression are unaffected by Atm deficiency, suggesting that signaling in the DNA damage pathway is distinct from that in the oncogene-induced pathway. In addition, we show that Atm deficiency has no overall effect on tumor growth and progression in this model.
The p53 tumor suppressor and ATM (mutated in human ataxia telangiectasia [AT] disease; Atm in mice) are both cancer susceptibility genes with roles in checkpoint regulation (9, 22, 23). Each is associated with distinct human genetic disorders in which patients are prone to cancer. Patients with Li-Fraumeni syndrome carry a mutant p53 allele and develop a variety of cancers, including mammary adenocarcinomas, sarcomas, brain tumors, and leukemia (21, 33). The p53 gene is also mutated in about 50% of sporadic human cancers (6, 11). AT is an autosomal recessive disease characterized by cerebellar degeneration, oculocutaneous telangiectasia, retarded growth, infertility, sensitivity to ionizing radiation (IR), and a high incidence of cancers, most commonly lymphoid malignancies (17, 30). The early deaths of most homozygous AT patients preclude an accurate assessment of the full tumor spectrum and the frequency of ATM deficiency in humans. Thus, human disease progression alone cannot predict whether p53 and ATM share tumor suppressor pathways.
p53 is involved in the cellular responses to a variety of stress signals, the best characterized of which is DNA damage (7, 32). In response to a given signal, p53 can induce cell cycle arrest or apoptosis, and these functions appear to be involved in its ability to suppress tumorigenesis. p53 deficiency can promote tumor growth by a reduction in the level of apoptosis, an event for which there would be substantial selection (12, 25, 34). Alternatively (or in addition), since p53-deficient cells are prone to genomic instability (20, 44), the loss of p53 responses may promote tumor progression through the genetic alteration of other cancer genes (8, 15, 16).
ATM is also involved in checkpoint regulation. It belongs to the phosphatidylinositol-3′ kinase superfamily, a family of signal transduction proteins with homology in their carboxyl kinase domains (29). In response to DNA damage, this 350-kDa protein kinase appears to be required for checkpoints in G1, S, and G2 phases (22, 23). Cultured cells derived from AT patients or from Atm-deficient mice are highly abnormal. These cells grow slowly and exhibit senescence prematurely (2, 17, 41). They demonstrate high rates of spontaneous apoptosis and a hypersensitivity to IR (22). Genome instability characterized by frequent chromosomal translocations and telomere defects is also commonly observed in AT cells (31, 35). Atm-deficient mice display many of the human AT phenotypes, such as retarded growth, infertility, sensitivity to IR, neurological dysfunction (although mild), and tumor proneness (2, 5, 40).
Evidence that ATM and p53 could act in the same pathway comes from studies of cell lines derived from AT patients and of knockout mice. Induction of p53 and G1 arrest in response to DNA damage is impaired in AT cell lines (14) and in mouse Atm−/− embryonic stem cells (41), embryo fibroblasts, and thymus cells (1, 2), indicating that ATM acts upstream of p53 in response to DNA damage. Both p53−/− (4, 13) and Atm−/− (2, 5, 40) mice develop thymic lymphoma, also suggesting the possibility of a common thymocyte tumor suppression pathway.
In addition to DNA damage, p53 is activated by many other signals, such as hypoxia, low ribonucleoside triphosphate levels, and oncogene-induced aberrant proliferation (7, 18). Dysregulated cell cycle activity has been shown to signal p53-dependent apoptosis and tumor suppression in vivo (12, 24, 25, 34). It is not clear whether these different stress signals are transduced to p53 via common or distinct upstream molecular pathways.
Based on the established link between Atm and p53 in response to DNA damage, we addressed the role of Atm in p53-dependent apoptosis and tumor suppression when induced by aberrant cell cycle activities by using the transgenic TgT121 brain tumor model. T121 is a truncated simian virus 40 large T antigen that binds and inactivates pRb and the related proteins p107 and p130 (27). Tissue-specific expression of this oncoprotein in brain choroid plexus epithelium (CP) induces the aberrant proliferation of these normally quiescent cells, and a measurable p53-dependent apoptosis pathway that suppresses tumor growth is activated (Fig. 1A). Inactivation of p53 causes an 85% reduction in CP apoptosis and accelerates tumor growth sevenfold (34). This model provides a quantitative test for p53 tumor suppression activities in vivo. In this study we assessed the impact of Atm deficiency on p53-mediated apoptosis and tumor suppression in these mice.
FIG. 1.
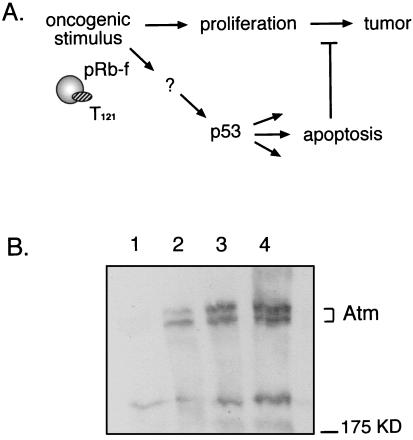
Is Atm required for p53-dependent apoptosis and tumor suppression in the TgT121 brain tumor model? (A) Transgenic expression of T121 in CP inactivates pRb family proteins (pRb-f) (including pRb, p107, and p130), resulting in abnormal proliferation and tumorigenesis. As a result, p53 is activated, causing apoptosis and attenuation of tumor growth (34). (B) Atm is expressed in T121 CP. Western immunoblotting was performed on tissue extracts by using a monoclonal antibody specific for ATM, 2C6. Lane 1, Atm−/− spleen (negative control); lanes 2 and 3, thymus and spleen tissues, respectively, of a wild-type mouse (positive control); lane 4, TgT121 CP. Two resolvable Atm-specific bands the size of mouse Atm are detectable in lanes 2 through 4 and are absent from the negative control. The position of a 175-kDa molecular mass marker is shown on the right.
MATERIALS AND METHODS
Mice.
TgT121 mice carry a transgene that expresses T121 specifically in CP under the control of lymphotropic papovavirus transcriptional signals (28). T121 consists of the simian virus 40 T-antigen N-terminal 121 amino acids with 11 C-terminal missense residues (27). The Atm−/− mice carry a truncation mutation at nucleotide 5790 caused by a PGKneo gene insertion (2). TgT121 (C57BL/6 × DBA2) mice were bred with Atm+/− mice (C57BL/6 × 129sv) to generate (TgT121 × Atm+/−) F1 mice. F1 mice were further intercrossed to generate TgT121 Atm−/− mice (C57BL/6 × DBA2 × 129sv). TgT121 and Atm−/− genotypes were determined by PCR analysis of tail DNA. TgT121 screening has been described previously (28). PCR primers were designed for genotyping Atm−/− mice. Primer pairs Atm-F (5′-GAC TTC TGT CAG ATG TTG CTG CC-3′) and Atm-B (5′-CGA ATT TGC AGG AGT TGC TGA G-3′) were used to identify the wild-type Atm allele, and Atm-F and Atm-Neo (5′-GGG TGG GAT TAG ATA AAT GCC TG-3′) were used to identify the knockout allele by performing 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The Atm-F–Atm-B pair generates a 162-bp PCR product, and the Atm-F–Atm-Neo pair generates a 441-bp PCR product.
Western blotting.
Western blotting analysis was carried out as previously described (39). Two hundred micrograms of protein from total cell lysates of fresh tissues was resolved by sodium dodecyl sulfate–5% polyacrylamide gel electrophoresis (cross-linking ratio, 29:1). Two independent anti-human ATM antibodies, 2C6 (3) and 473 (kindly provided by Eva Lee and Michael Kastan, respectively), were used separately to detect Atm protein expression in TgT121 CP. The results were the same with both reagents. The enhanced chemiluminescence system (Amersham) was used according to the manufacturer’s instructions.
Histology, S-phase, and apoptosis assays.
Brain tissues were fixed in 10% formalin, embedded in paraffin, and sectioned as previously described (34). To examine tumor size, 6-μm sections were taken from 10 successive layers at 100-μm intervals. For histology assays, sections were stained with hematoxylin and eosin as previously described (34). Bromodeoxyuridine (BrdU) labeling and immunostaining as well as the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) apoptosis assay were completed as previously described (34, 43). Quantification was made by averaging the percentage of positive cells in 10 random fields at a magnification of ×400.
In situ RNA hybridization.
Sections (6 μm) were hybridized with p21 or Bax antisense probes as previously described (26, 43). Probes were radiolabeled with α-35S-UTP and used at a concentration of 5 × 104 cpm ml−1. Autoradiography was conducted at 4°C for 3 days for the p21 probe and at 4°C for 3 weeks for the Bax probe. Signal densities were quantified with NIH Image 1.58. The threshold was set by using the sense probe signals as the background. Means were determined from 10 random areas of CP for each sample.
RESULTS
p53-dependent apoptosis does not require Atm.
A genetic approach has been taken to determine the pathway by which p53-dependent apoptosis and tumor suppression proceed in TgT121 mice. For example, by assessing the rates of apoptosis and tumor growth in a background with specific deficiencies, we previously determined that E2F1 is upstream (26) and Bax is downstream (43) of p53 in this system. The possibility that Atm could be active in TgT121 CP is demonstrated by the detection of Atm in this tissue by Western blotting analysis (Fig. 1B). A spleen extract from an Atm−/− mouse served as a negative control and showed no specific proteins the size of Atm (Fig. 1B, lane 1). In contrast, spleen and thymus tissues from a wild-type mouse (lanes 2 and 3) and CP from a TgT121 mouse (lane 4) contained a 350-kDa Atm-specific doublet detectable with two independent ATM-specific antibodies (the results presented in Fig. 1B were obtained with the 2C6 monoclonal antibody).
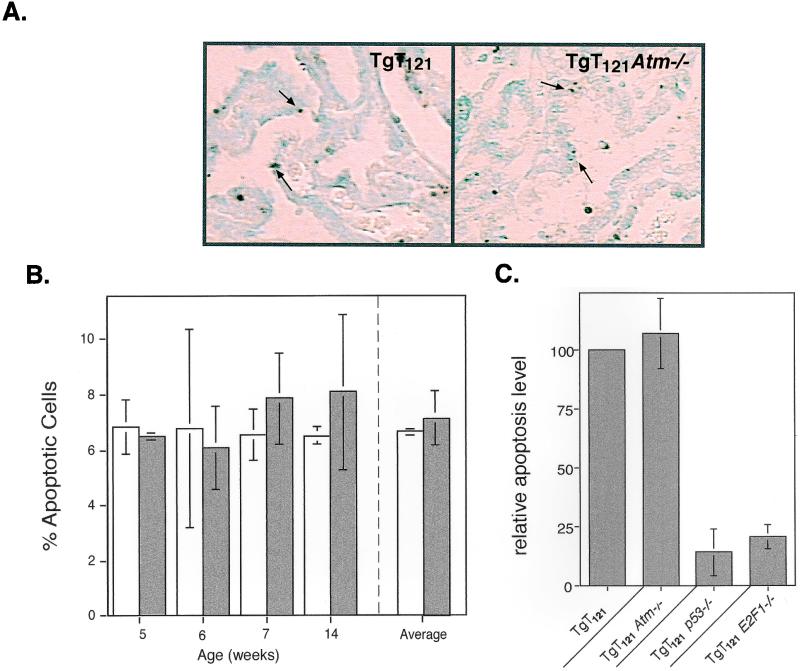
To examine whether Atm deficiency has a role in p53 tumor suppression activities, we generated TgT121 Atm−/− mice through a series of crosses by using TgT121 (28) and Atm+/− (2) mice. We first assessed the effect of Atm deficiency on T121-induced p53-dependent apoptosis by using the in situ TUNEL assay (Fig. 2A). The CP apoptotic index (AI) was determined for brain sections from TgT121 Atm−/− mice, which were compared to TgT121 Atm+/+ littermates. Previously, TgT121 CP cells were shown to have an average AI of 7.3%, 85% of which requires functional p53 (34). In the present study, the CP AI of TgT121 Atm+/+ littermate controls was similar, averaging 6.69% ± 0.17% (Fig. 2B). The CP of control Atm−/− mice appeared to be normal, with no apoptotic activity (Table 1). The AI in TgT121 Atm−/− CP was similar to that of TgT121 Atm+/+ mice (7.17% ± 0.99%) (Fig. 2; Table 1). This is in stark contrast to the dramatic decrease in AI caused by p53 deficiency (Table 1; Fig. 2C) (34). Thus, in this system, p53-dependent apoptosis in response to abnormal proliferation does not require Atm.
FIG. 2.
p53-mediated apoptosis does not require Atm. (A) Apoptotic cells in CP (arrows) were detected by the TUNEL assay. No qualitative changes were detected between TgT121 and TgT121 Atm−/− CP tumor cells. (B) Quantitative apoptotic indices in TgT121 and TgT121 Atm−/− CP. The percentages of apoptotic cells were determined for TgT121 (open bars) and TgT121 Atm−/− (solid bars) littermates at different ages. Each bar in the left panel represents the averaged counts from 10 fields of one sample from one mouse at the specified age. The averages of all four mice are shown at the right. The error bars represent the standard deviations among microscopic fields (left) or among mice (right). (C) Impact of Atm deficiency on apoptosis compared with impact of p53 and E2F1 deficiency. Atm deficiency has no effect on apoptosis in TgT121 CP cells, while p53 and E2F1 account for 85 and 80% of the apoptosis, respectively (26, 34).
TABLE 1.
Atm impact on tumor growth
| Genotype | % of apoptotic cellsa | % of S-phase cellsa | Type of CP neoplastic growth | No. of wks of survival (mean ± SE)b | Cause of death |
|---|---|---|---|---|---|
| Atm−/− | 0c | 0c | None | 20.2 ± 3.9 (6) | Thymoma |
| TgT121 | 100 | 100 | Slow | 31.0 ± 1.6 (16) | CP tumor |
| TgT121Atm+/− | 98c | 120c | Slow | 37.2 ± 4.21 (6) | CP tumor |
| TgT121Atm−/− | 107 ± 15 | 103 ± 33 | Slow | 22.7 ± 1.8 (15) | Thymoma |
| TgT121p53−/−d | 14 ± 10 | 109c | Rapid | 4 ± 0.2 (4) | CP tumor |
Relative to the level in TgT121 CP.
Values in parentheses are numbers of mice tested. Mean survival times of TgT121 and TgT121 Atm+/− mice are not significantly different (P = 0.1026 in an unpaired two-tailed t test). The difference may reflect background strain differences (TgT121 mice are C57BL/6 × DBA2; TgT121 Atm+/− mice are C57BL/6 × DBA2 × 129sv).
From a single sample.
Data from reference 34; TgT121 p53−/− mice are of a C57BL/6 × DBA2 × 129sv background.
Intact p53 transactivation activity in the absence of Atm.
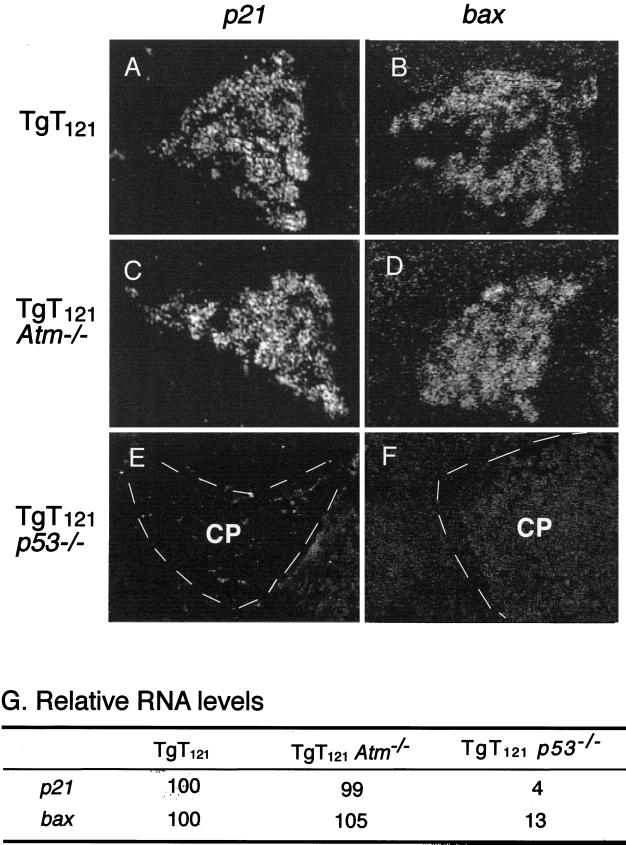
To further assess the impact of Atm deficiency on p53 function, we examined p53 transactivation activity in TgT121 Atm−/− CP. We previously showed that T121 expression in the CP induces the p53-regulated genes bax (43), p21 (26), and mdm2 (26) in a p53-dependent manner. To assess the p53 transactivation activity in the absence of Atm, bax and p21 transcript levels were measured in TgT121 Atm−/− and TgT121 Atm+/+ brain sections by in situ RNA hybridization (Fig. 3). Samples from young mice (5 to 7 weeks) were analyzed to avoid the influence of spontaneous genetic changes during tumor progression. Quantitative analysis indicated that p53-dependent p21 and bax RNA levels in the CP remain unchanged in the absence of Atm (Fig. 3A through D). The levels of p21 and bax transcripts in TgT121 Atm−/− CP were 99% ± 4.8% and 105% ± 8%, respectively, of those of TgT121 Atm+/+ littermates (Fig. 3G). All p21 expression in these cells was dependent on p53 activity (Fig. 3E), as was much of the bax expression, as previously observed (Fig. 3F). In this system, Bax is involved in p53-dependent apoptosis (43), while p21 is not (our unpublished results). Thus, Atm is dispensable for T121-induced p53 transactivation functions that are apoptosis dependent as well as independent.
FIG. 3.
Atm deficiency does not affect p53 transactivation function in CP. Representative in situ RNA hybridization signals are shown for p21 (A, C, and E) and bax (B, D, and F) expression. p21 RNA is readily detectable in TgT121 (A) as well as in TgT121 Atm−/− (C) CP. bax RNA levels are also equally expressed in TgT121 (B) and TgT121 Atm−/− (D) cells. bax and p21 expression in TgT121 CP are both p53 dependent, as seen in TgT121 p53−/− cells (E and F). (G) Quantitative comparisons of RNA levels; signals in TgT121 are designated as 100%. The signal detected in CP by using a bax or p21 sense probe was at about the background level, ensuring the specificity of the assay. Ten fields of each sample were analyzed. Standard errors in each field are less than 10% of the relative mean value.
Effects of Atm deficiency on tumor cell proliferation and overall tumor growth.
Although the above results show that Atm is not required for p53-dependent apoptosis or transactivation, Atm could affect CP tumor growth by other mechanisms. For example, Atm deficiency could diminish tumor growth since Atm−/− fibroblasts exhibit poor growth and a reduced life span in culture (2, 41). Alternatively, Atm inactivation could accelerate tumor growth through genetic instability, as proposed for Atm deficiency-induced lymphoma and leukemia (35).
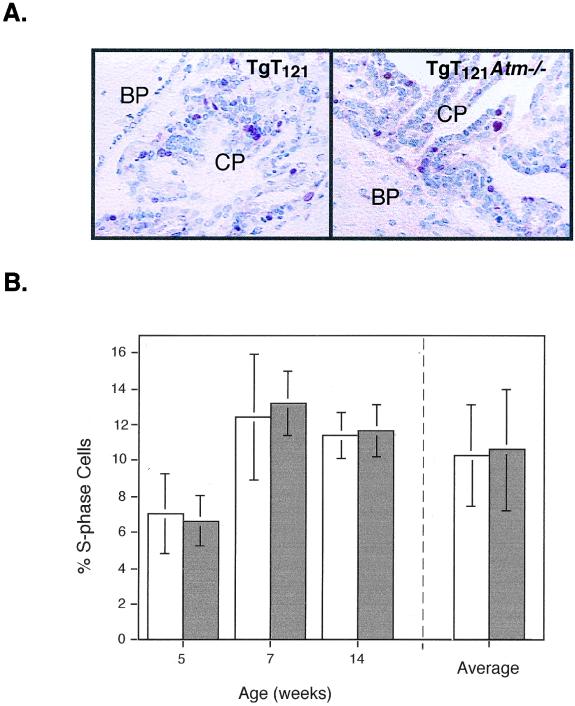
To determine whether Atm deficiency has an impact on CP tumor cell proliferation, we measured S-phase indices by in vivo BrdU incorporation (Fig. 4A). Mice were analyzed at various times during early tumor growth (5 to 14 weeks) to circumvent selective changes during tumor progression. No difference was detected in the percentage of S-phase CP in TgT121 Atm−/− and TgT121 Atm+/+ littermates (Table 1). TgT121 Atm−/− CP averaged 10.6% ± 3.4% of cells in S phase, while TgT121 Atm+/+ CP averaged 10.3% ± 2.8% of cells in S phase (Fig. 4B). Therefore, unlike fibroblast growth in culture, Atm deficiency had no measurable impact on CP tumor cell proliferation in vivo.
FIG. 4.
Effects of Atm deficiency on tumor cell proliferation. TgT121 and TgT121 Atm−/− CP were stained for BrdU incorporation to measure proliferation rates. (A) Representative views of CP from TgT121 and TgT121 Atm−/− 7-week-old littermates stained for BrdU (red), with nuclei counterstained by hematoxylin (blue). No BrdU-staining cells are present in the surrounding brain parenchyma (BP). (B) Percentage of S-phase cells in TgT121 (open bars) and TgT121 Atm−/− (solid bars) CP. Left, percentage of proliferating cells in paired littermates (each bar represents brain sections from one mouse; error bars represent the standard deviations among sets of 10 microscopic fields); right, averaged percentage of S-phase cells in these age-matched groups shown at the left (error bars represent standard deviations among individual mice).
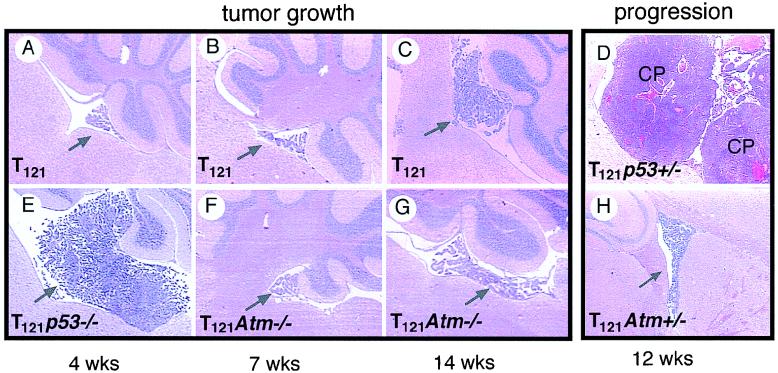
Although the apoptotic and proliferative indices of TgT121 CP were not affected by Atm inactivation, an effect on tumor morphology and progression was possible. To determine the overall impact of Atm deficiency on tumor growth, the survival times, tumor sizes, and morphologies of TgT121 Atm−/− mice were compared to those of TgT121 Atm+/+ littermates. Control TgT121 and Atm-deficient mice developed different tumor types and had different average survival times. TgT121 mice died of brain tumors with an average survival time of 31 weeks, whereas Atm-deficient mice died of thymoma with an average survival time of 20 weeks (Table 1). Like Atm−/− mice, all TgT121 Atm−/− mice developed thymic lymphoma (average survival time of 23 weeks [Table 1]). The difference between thymoma development in Atm−/− mice and that in TgT121 Atm−/− mice is not statistically significant (P = 0.5115 in an unpaired two-tailed t test) and could be due to background strain differences (C57BL/6 × 129sv versus C57BL/6 × DBA2 × 129sv) or due to T121 expression in thymocytes (28).
Although TgT121 Atm−/− mice succumbed to thymic lymphoma prior to the terminal age anticipated for mice with brain tumors, their lengthy survival indicates that Atm deficiency did not substantially accelerate the growth of T121-induced brain tumors. CP tumors present in TgT121 Atm−/− mice at the time of autopsy were comparable in size and morphology to those in TgT121 mice of similar ages (data not shown). Furthermore, assessment of tumor sizes at 4 to 20 weeks showed no significant differences between TgT121 Atm+/+ and TgT121 Atm−/− littermates (Fig. 5). The CP of Atm−/− mice was morphologically normal. These results are consistent with the observation that both apoptosis and proliferation were unaffected by Atm deficiency. In sharp contrast, TgT121 p53−/− mice develop large tumor masses within 4 weeks (Fig. 5E; Table 1) (34). Analysis of TgT121 Atm+/− mice also showed that Atm deficiency did not promote tumor progression. Tumor growth in TgT121 p53+/− mice progresses from slow growth to highly aggressive and angiogenic growth within 7 weeks, leading to the death of the mice by 12 weeks of age (Fig. 5D) (34). This occurs in all mice and correlates with a high frequency of p53 loss of heterozygocity. In contrast, no such acceleration was observed in TgT121 Atm+/− mice (Fig. 5H).
FIG. 5.
Effects of Atm deficiency on CP tumor growth and progression. Sizes of the CP masses (arrows) are shown. TgT121 mice at 7 weeks (B) and 14 weeks (C) are compared to TgT121 Atm−/− littermates (F and G, respectively). Note that p53 deficiency greatly accelerated tumor growth as indicated by the mass size at 4 weeks (E versus A). CP tumor progression to aggressive angiogenic states was observed in TgT121 p53+/− mice (D) but not in TgT121 Atm+/− mice (H). These views represent the largest mass present among step sections sampled from the full brain to ensure a fair comparison of tumor sizes.
DISCUSSION
Atm and p53.
In this study we explored the possibility that Atm functions in a p53 apoptosis and tumor suppression pathway that is induced via aberrant proliferation rather than DNA damage. The molecular signals involved in p53 induction in this case are largely unknown. One possibility is that aberrant S-phase activity generates DNA damage-like signals, in which case the pathways could be coincident. Here, we show that Atm deficiency does not impair p53 activation by inappropriate cell cycle activity in brain epithelium. The approach used in this study to study Atm has been used in previous studies to identify both upstream (26) and downstream (43) effectors in this pathway. E2F1 deficiency causes an 80% inhibition of p53-dependent apoptosis (Fig. 2C) as well as inhibition of p53 transactivation function, indicating that E2F1 lies upstream of p53 (26). Therefore, inactivation of pRb proteins by T121 leads to E2F1 activation, which then induces p53 function (26). In contrast, this report shows that Atm is not required in this pathway—p53-dependent apoptosis, transactivation, and tumor suppression are fully active in the absence of Atm. Together, these data indicate that different stress signals are transduced to p53 via distinct pathways.
Comparison of these results with those of previous reports addressing the role of ATM in p53-dependent activities largely supports the idea that the distinction observed here is based on the inducing signal rather than cell type or biological response (i.e., apoptosis or growth arrest). So far, ATM has been implicated only in DNA damage-induced p53 responses, including the induction of both growth arrest and apoptosis. Atm is required for DNA damage-induced p53 induction and G1 arrest in human (23) and mouse (2) fibroblasts and in mouse embryonic stem cells (41) and thymus tissue (1). It is also required for IR-induced p53-dependent apoptosis of neurons in the developing central nervous system, since the irradiation of newborn mice induces widespread central nervous system apoptosis, coincident with p53 induction. This apoptosis is dramatically reduced in both p53-deficient and Atm-deficient mice (10).
In the thymus, IR-induced p53-dependent apoptosis is not Atm dependent in vivo (1, 10), although the p53-dependent G1/S checkpoint does require Atm (1). Therefore, within a given tissue, distinct p53-mediated biological effects are induced by apparently distinct pathways upstream of p53 (1). Two reports show a partial reduction in the IR-induced p53-dependent apoptosis of Atm−/− thymus cells (41) or thymocytes (37). This contradiction could indicate differences in thymocyte subpopulations or could result from in vitro versus in vivo approaches. In the present study, we have examined in vivo apoptotic responses to show that Atm is dispensable for p53-mediated apoptosis and tumor suppression in brain epithelium. We cannot exclude the possibility that Atm-related factors fully compensate for the absence of Atm.
Whether Atm and p53 participate in the same tumor suppression pathways in other cell types is not known. Although both Atm- and p53-deficient mice develop thymic lymphomas with high frequency, in Atm p53 double-null mice, tumor growth is accelerated (37, 42), indicating that Atm and p53 cooperate in thymocyte tumor suppression, rather than fall within a linear pathway. In addition, V(D)J recombination-associated translocations are frequent in thymomas from Atm-deficient mice (2) but not in thymomas from p53-deficient mice (19). Therefore, in mouse thymocytes, where both Atm and p53 are proven tumor suppressors, the pathways may be distinct.
Atm and tumor growth.
In the present study Atm deficiency did not affect CP tumor cell proliferation or tumor growth despite the fact that the proliferation of Atm−/− fibroblasts was impaired and senescence occurred prematurely. This lack of Atm effect on CP tumor cell proliferation could reflect cell-type-specific differences. In Atm−/− fibroblasts, proliferative defects can be rescued by the inactivation of either p21 or p53 (36, 38, 42). CP tumor cells express p21 abundantly and possess functional p53. However, pRb (a known target of cyclin-dependent kinases inhibited by p21) is inactivated by T121 in CP tumor cells, possibly masking any growth-inhibitory effects of Atm deficiency. Finally, we considered the possibility that genetic instability induced by Atm deficiency could result in measurable effects on tumor progression. TgT121 p53+/− CP tumors progress to highly aggressive states with frequent p53 loss of heterozygocity (34) and widespread aneuploidy (our unpublished results). No such accelerated progression was observed in TgT121 Atm+/− mice.
In summary, our results suggest that distinct signal transduction pathways may induce p53, depending on the cellular insults. While Atm appears to be involved in p53 activation by DNA damage, it is not required for p53 activation by aberrant cell cycle activity in brain epithelium. Furthermore, when these tumors are initiated by the inactivation of the pRb family, Atm deficiency has no further impact on tumor growth and progression.
ACKNOWLEDGMENTS
We thank Le Zhang for excellent technical assistance, Michael Kastan and Eva Lee for providing reagents, and Eva Lee for communication of unpublished data.
This work was supported by grants from the National Institutes of Health (CA65773 and CA46283) to T.V.D.
REFERENCES
- 1.Barlow C, Brown K, Deng C, Tagle D, Wynshaw-Boris A. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- 2.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J N, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 3.Chen G, Lee E Y H P. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J Biol Chem. 1996;271:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- 4.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A J, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 5.Elson A, Wang Y, Daugherty C J, Morton C C, Zhou F, Campos-Torres J, Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 7.Hansen R, Oren M. p53: from inductive signal to cellular effect. Curr Opin Genet Dev. 1997;7:46–51. doi: 10.1016/s0959-437x(97)80108-6. [DOI] [PubMed] [Google Scholar]
- 8.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 9.Hawley R S, Friend S H. Strange bedfellows in even stranger places: the role of ATM in meiotic cells, lymphocytes, tumors, and its functional links to p53. Genes Dev. 1996;10:2383–2388. doi: 10.1101/gad.10.19.2383. [DOI] [PubMed] [Google Scholar]
- 10.Herzog K H, Chong M J, Kapsetaki M, Morgan J I, Mckinnon P J. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 11.Hollstein M, Soussi T, Thomas G, von Brevern M-C, Bartsch H. p53 gene alterations in human tumors: perspectives for cancer control. Recent Results Cancer Res. 1997;143:369–389. doi: 10.1007/978-3-642-60393-8_26. [DOI] [PubMed] [Google Scholar]
- 12.Howes K A, Ransom N, Papermaster D S, Lasudry J G H, Albert D M, Windle J J. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 13.Jacks T, Remington L, Williams B, Scmitt E, Halachmi S, Bronson R, Weinberg R. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 14.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 15.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 17.Lavin M F, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 18.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 19.Liao M-J, Zhang X-X, Hill R, Gao J, Qumsiyeh M B, Nichols W, Van Dyke T. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol Cell Biol. 1998;18:3495–3501. doi: 10.1128/mcb.18.6.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 21.Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, Friend S H. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 22.Meyn M S. Ataxia-telangiectasia and cellular responses to DNA damage. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- 23.Morgan S E, Kastan M B. p53 and ATM: cell cycle, cell death, and cancer. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 24.Morgenbesser S, Williams B, Jacks T, DePinho R. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 25.Pan H, Griep A E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 26.Pan H, Yin C, Dyson N, Harlow E, Yamasaki L, Van Dyke T. A key role for E2F1 in p53-dependent apoptosis and cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 27.Pipas J M, Peden K W C, Nathans D. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol Cell Biol. 1983;3:203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sáenz Robles M T, Symonds H, Chen J, Van Dyke T. Induction versus progression of brain tumor development: differential functions for the pRB- and p53-targeting domains of simian virus 40 T antigen. Mol Cell Biol. 1994;14:2686–2698. doi: 10.1128/mcb.14.4.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 30.Sedgwick R P, Boder E. Ataxia-telangiectasia. In: Vinken P J, Bruyn G W, Klawans H L, editors. Handbook of clinical neurology. New York, N.Y: Elsevier; 1991. pp. 347–423. [Google Scholar]
- 31.Smilenov L B, Morgan S E, Mellado W, Sawant S G, Kastan M B, Pandita T K. Influence of ATM function on telomere metabolism. Oncogene. 1997;15:2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- 32.Smith M L, Fornace A J., Jr Genomic instability and the role of p53 mutations in cancer cells. Curr Opin Oncol. 1995;7:69–75. [PubMed] [Google Scholar]
- 33.Srivastava S, Zou Z Q, Pirollo K, Blattner W, Chang E H. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 34.Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 35.Taylor A M R, Metcalfe J A, Thick J, Mak Y-F. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 36.Wang Y A, Elson A, Leder P. Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proc Natl Acad Sci USA. 1997;94:14590–14595. doi: 10.1073/pnas.94.26.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westphal C H, Rowan S, Schmaltz C, Elson A, Fisher D E, Leder P. atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat Genet. 1997;16:396–401. doi: 10.1038/ng0897-397. [DOI] [PubMed] [Google Scholar]
- 38.Westphal C H, Schmaltz C, Rowan S, Elson A, Fisher D E, Leder P. Genetic interactions between atm and p53 influence cellular proliferation and irradiation-induced cell cycle checkpoints. Cancer Res. 1997;57:1664–1667. [PubMed] [Google Scholar]
- 39.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development, and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Ashley T, Brainerd E E, Bronson R T, Meyn M S, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Yang E M, Brugarolas J, Jacks T, Baltimore D. Involvement of p53 and p21 in cellular defects and tumorigenesis in Atm−/− mice. Mol Cell Biol. 1998;18:4385–4390. doi: 10.1128/mcb.18.7.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin C, Knudson C M, Korsmeyer J S, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 44.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]