Abstract
We established and elaborated on a method to enrich the membrane proteome of mature pollen from economically relevant crop using the example of Solanum lycopersicum (tomato). To isolate the pollen protein fraction enriched in membrane proteins, a high salt concentration (750 mM of sodium chloride) was used. The membrane protein-enriched fraction was then subjected to shotgun proteomics for identification of proteins, followed by in silico analysis to annotate and classify the detected proteins.
Keywords: Membrane proteome, Pollen, Tomato, Proteomics
Background
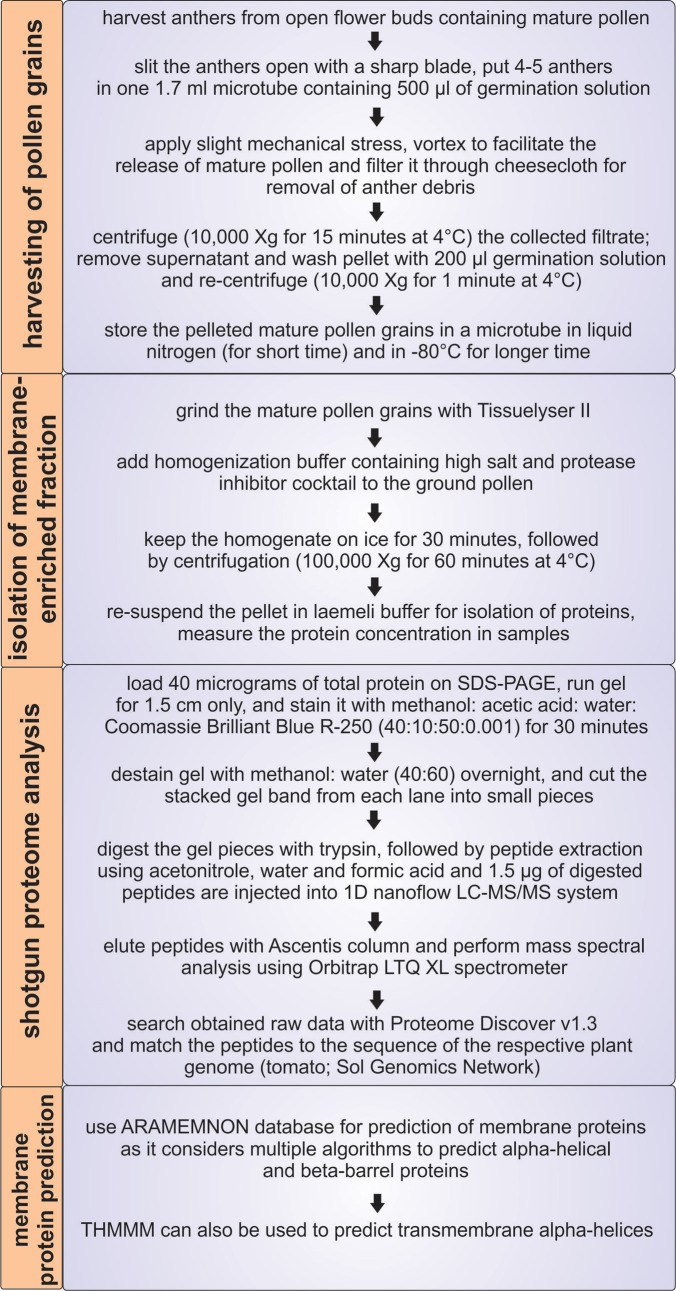
As proper distribution of proteins and solutes between different cellular compartments or the insertion of newly-synthesized proteins into membranes is largely dependent on membrane proteins, the membrane proteome is central for maintenance of cellular and organellar homeostasis ( Paul et al., 2013 ; 2014 and 2016a). Considering the importance of membrane proteins in general, these are also essential for pollen function and development ( Paul et al., 2016b ). Many global pollen proteomic studies have been performed in the past ( Chaturvedi et al., 2013 and 2016); however, information about the intracellular distribution of proteins and the composition of the membrane proteome in pollen is rarely discussed ( Pertl et al., 2009 ). One reason might be the low abundance and solubility of membrane proteins. Here we describe a protocol to isolate and analyze a protein fraction enriched in membrane proteins from mature pollen, which was established for tomato (Figure 1).
Figure 1. Overview of the protocol for isolation of the membrane proteome of mature tomato pollen.
Materials and Reagents
1.7 ml microtubes (Corning, Axygen®, catalog number: MCT-175-C)
Pipette tips
Cheesecloth Miniwipes (Fisher Scientific, catalog number: 06-665-28)
Sharp blade (Red Devil, catalog number: 3272)
Bond-Elute C-18 SPEC plate (Agilent Technologies, Santa Clara, CA, USA)
Seeds of the cultivar of choice. For the method development–seeds of Solanum lycopersicum cv. Moneymaker (Accesion: LA2706) and cv. Red setter were used
Liquid nitrogen
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-500)
Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P9599)
6x Laemmli buffer (Alfa Aesar, catalog number: J61337)
Urea (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 29700)
Acrylamide/bis-acrylamide (Fisher Scientific, catalog number: BP1408-1)
TEMED (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 17919)
Ammonium persulfate (Acros Organics, catalog number: 327081000)
Acetonitrile (Fisher Scientific, catalog number: A9561)
Formic acid (Fisher Scientific, catalog number: A117-50)
Boric acid (Fisher Scientific, catalog number: A73-500)
Calcium nitrate tetrahydrate (Fisher Scientific, catalog number: C109-500)
Magnesium sulphate anhydrous (Fisher Scientific, catalog number: M65-500)
Potassium nitrate (Fisher Scientific, catalog number: P263-500)
Potassium chloride (Fisher Scientific, catalog number: P217-500)
Dithiothreitol (DTT) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0861)
Ethylenediaminetetraacetic acid, disodium salt dihydrate (EDTA) (Fisher Scientific, catalog number: S311-100)
Tris base (Fisher Scientific, catalog number: BP152-500)
Methanol (Fisher Scientific, catalog number: A456-1)
Acetic acid (Fisher Scientific, catalog number: A507-P500)
Double distilled water
Coomassie Brilliant Blue R-250 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 20278)
Trypsin solution sequencing grade (Roche Diagnostics, catalog number: 11418475001)
Ammonium bicarbonate (Fisher Scientific, catalog number: A643-500)
Calcium chloride (Fisher Scientific, catalog number: C70-500)
Trifluoroacetic acid (TFA) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28904)
Germination solution (see Recipes)
Homogenization buffer (see Recipes)
Staining solution (see Recipes)
Destaining solution (see Recipes)
Trypsin buffer (see Recipes)
Equipment
Vortex (e.g., Fisher Scientific, model: Fisher ScientificTM Analog Vortex Mix, catalog number: 02-215-365)
Centrifuge (e.g., Eppendorf, model: 5424 R)
Ultracentrifuge (e.g., Thermo Fisher Scientific, model: Sorvall® DiscoveryTM 90 SE)
Daisy BBs (Daisy, model: 24, catalog number: 980024-001)
Tissuelyser II (QIAGEN, model: Tissuelyser II, catalog number: 85300)
Speed-Vac Labogene ApS (LabGeneTM, model: ScanVac CoolSafe 110-4, catalog number: H01130032)
Pipette
Sonicator (BANDELIN electronic, model: Sonorex RK 100, catalog number: 301.00044253.024)
Precolumn (Eksigent, Redwood City, CA, USA)
Ascentis column (Sigma-Aldrich, Supelco, model: Ascentis® Express Peptide ES-C18 HPLC Column, catalog number: 53552-U), dimension 15 x 100 μm, pore size 2.7 μm
Orbitrap LTQ XL mass spectrometer (Thermo Fisher Scientific, model: LTQ XLTM)
Software
Proteome Discoverer 1.3 (Thermo, Germany) or other software like MaxQuant (http://www.maxquant.org/), Protmax (http://www.univie.ac.at/mosys/software.html) and MASCOT (http://www.matrixscience.com/)
PROMEX (http://promex.pph.univie.ac.at/promex/) or other software like ProteomeXchange (http://www.proteomexchange.org/)
Procedure
-
Harvesting of mature pollen grains
-
Grow plants (e.g., tomato cv. Moneymaker–MM hereafter and cv. Red setter–RS hereafter) under controlled greenhouse conditions (for tomato: 24 °C/20 °C, day/night; 60% humidity ( Fragkostefanakis et al., 2015 ).
Note: Growth conditions need to be adapted for the plant species or cultivar in use.
Harvest anthers (> 10 mm length for MM and RS) from flower buds after anthesis as they contain mature pollen grains (preferable time is 0-72 h post anthesis).
Make a horizontal cut on the collected anther and put four anthers in one 1.7 ml microtube containing 500 µl of germination solution.
Squeeze anther tissue with slight mechanical stress using pipette tips and vortex for 15-20 sec to release the mature pollen from anther locules.
Further, pass germination solution containing isolated mature pollen grains through cheesecloth to remove anther debris.
Centrifuge the collected pollen grains at 10,000 × g for 15 min at 4 °C.
Remove the supernatant, wash the pelleted pollen grains with 200 µl of germination solution and centrifuge at 10,000 × g for 1 min at 4 °C.
Remove the supernatant and repeat the washing step A7.
Remove the supernatant, immerse the collected pollen in liquid nitrogen and store at -80 °C till further usage.
-
-
Isolation of membrane-enriched fraction
Add metal beads (Daisy BB) that help to homogenize the tissue to microtubes and grind the collected mature pollen grains with TissueLyser II (at 25/sec for 1 min).
Add 10-20 ml homogenisation buffer containing sodium chloride (NaCl, 750 mM) and protease inhibitor cocktail to the ground mature pollen, re-suspend and leave it on ice for 30 min.
Centrifuge the homogenised mixture at 100,000 × g for 60 min at 4 °C.
-
Re-suspend the pellet in Laemmli buffer and quantify the protein concentration.
Note: The supernatant is enriched in soluble proteins and can also be subjected to shotgun proteomics protocol, if needed.
-
Shotgun proteomics
Load 40 µg of total proteins onto an SDS-PAGE (12.5% with maximum thickness 1 mm) and run the gel at 80 V constant for 1.5 cm in resolving gel (Valledor and Weckwerth, 2014). Laemmli buffer is used as loading buffer.
Stain the gel with methanol:acetic acid:water:Coomassie Brilliant Blue R-250 (40:10:50:0.001) for 30 min and de-stain it overnight in methanol:water (40:60). Afterwards, slice the thick stacked protein band from the remaining gel and chop it into smaller pieces with a sharp blade. Immerse the chopped gel pieces into double distilled water until all samples are processed.
Afterwards, remove water and add 1 ml of sodium bicarbonate (25 mM) and incubate at 37 °C for 15 min. Discard supernatant and add 1 ml of 25 mM sodium bicarbonate and 50% acetonitrile. Incubate the samples at 37 °C for 15 min. Repeat this step.
-
Remove the supernatant and dehydrate gel pieces in 300 μl 100% acetonitrile for 5 min at room temperature. Discard supernatant and dry out in a Speed-Vac for 5 min.
Note: Gel pieces should have a ‘transparent-plastic’ look. If the gel-pieces are still translucent, repeat the step.
-
Digest the chopped gel pieces with 50 µl trypsin solution (10 ng/μl) and incubate for 15 min at 37 °C. If required, add more trypsin solution until the gel pieces are completely rehydrated. Incubate for 14-16 h at 37 °C and avoid shaking during incubation. Afterwards, stop the reaction by adding 5 μl of 1.0% trifluoroacetic acid (pH should be < 4 after addition of TFA).
Note: Add 1 ml of trypsin buffer to dilute trypsin to 10 ng/μl. Incubate on ice for 10 min and then pipette up and down to completely re-suspend the pellet. After dilution, trypsin can be stored at -20 °C for one month. It is highly recommended to store it in aliquots (Valledor and Weckwerth, 2014).
For peptide extraction, add 150 µl of 50% acetonitrile (v/v) and 1% formic acid (v/v) into the microtube containing the gel pieces and incubate for 5 min at room temperature followed by sonication for 3 min (low intensity ultrasound bath).
Spin microtubes (1,000 × g for 1 min) and collect the supernatant in a new microtube ‘A’. Repeat step C6 and transfer the supernatant to microtube ‘A’.
Add 100 µl of 90% acetonitrile (v/v) and 1% formic acid (v/v) to the microtube containing the gel pieces and incubate for 5 min at room temperature. Transfer the supernatant to microtube ‘A’.
Dry the samples in Speed-Vac (microtube A). For long-term storage of extracted peptides -80 °C is preferred.
Afterwards, proceed with peptide desalting using spec 96 well C18, Agilent (Valledor and Weckwerth, 2014).
Dissolve peptides in 4% (v/v) acetonitrile, 0.1% (v/v) formic acid.
Inject digested peptides into a one-dimensional nanoflow LC-MS/MS system that has a precolumn.
Elute the peptides with an Ascentis column during 80 min and a gradient ranging from 5% to 50% (v/v) acetonitrile, 0.1% (v/v) formic acid and perform mass spectral analysis on an Orbitrap LTQ XL mass spectrometer with a controlled flow rate of 500 nl per minute and specific tune settings for MS as follows: spray voltage set to 1.8 kV; temperature of the heated transfer capillary set to 180 °C.
After each full MS scan, perform ten MS/MS scans, and dynamically select ten most abundant peptide molecular ions, with a dynamic exclusion window set to 90 sec.
Omit ions with a +1 or unidentified charge state in the full MS from MS/MS analysis.
Data analysis
-
Shotgun proteome analysis
Search the obtained raw data (in form of mass spectra) with SEQUEST algorithm present in Proteome Discoverer version 1.3. For this, set identification confidence to 5% FDR (false discovery rate) and the variable modifications to acetylation of N-terminus and oxidation of methionine, with a mass tolerance of 10 ppm for the parent ion and 0.8 Da for the fragment ion.
-
Match peptides to the genome sequence of the respective plant species, tomato in this case (Sol Genomics Network), and consider a significant hit when the peptide confidence is at medium or high and an Xcorr threshold is established at 1 per charge (2 for +2 ions, 3 for +3 ions, etc.).
Note: Choose high thresholds to minimize false detection of proteins.
-
Prediction of membrane proteins
First, use a database like ARAMEMNON for prediction of membrane proteins ( Schwacke et al., 2003 ). ARAMEMNON considers 20 and 11 different databases (or algorithms) to predict alpha-helical and beta-barrel proteins, respectively.
If the plant species of your interest is not listed in ARAMEMNON, you can predict Arabidopsis co-orthologs of the detected protein by well-established approaches ( Simm et al., 2015 ).
The detected proteins should be verified by a second program like THMMM (http://www.cbs.dtu.dk/services/TMHMM/), which predicts transmembrane alpha helices based on hidden Markov models ( Krogh et al., 2001 ).
Recipes
-
Germination solution
2 mM boric acid
2 mM calcium nitrate tetrahydrate
2 mM magnesium sulphate anhydrous
1 mM potassium nitrate
For preparing 100 ml mix:
0.012 g of boric acid
0.047 g of calcium nitrate tetrahydrate
0.024 g of magnesium sulphate anhydrous
0.010 g of potassium nitrate
-
Homogenization buffer
100 mM potassium chloride
5 mM DTT
1 mM EDTA
50 mM Tris pH 7.2
750 mM sodium chloride
For preparing 100 ml mix:
0.745 g of potassium chloride
0.077 g of DTT
0.029 g of EDTA
0.605 g of Tris pH 7.2
Note: Always add sodium chloride in the last place as per the homogenization buffer; for 20 ml homogenization buffer, add 0.876 g of sodium chloride.
-
Staining solution
methanol:acetic acid:water:Coomassie Brilliant Blue R-250 (40:10:50:0.001)
-
Destaining solution
Methanol and water (40:60)
-
Trypsin buffer
25 mM sodium bicarbonate
10 % (v/v) acetonitrile
5 mM calcium chloride
For preparing 100 ml mix:
0.21 g of sodium bicarbonate
100 μl of pure acetonitrile
645 μl of water
Note: Always add calcium in the last place as per the trypsin buffer; for 10 ml of trypsin buffer, add 0.005 g of calcium chloride.
Acknowledgments
This protocol is adapted from ( Paul et al., 2016a ). This is an article of the SPOT-ITN consortium funded by Marie-Curie (EU Grant Agreement Number 289220) to E.S.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Chaturvedi P., Ghatak A., and Weckwerth W.(2016). Pollen proteomics: from stress physiology to developmental priming. Plant Reprod 29(1-2): 119-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi P., Ischebeck T., Egelhofer V., Lichtscheidl I. and Weckwerth W.(2013). Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J Proteome Res 12(11): 4892-4903. [DOI] [PubMed] [Google Scholar]
- 3.Fragkostefanakis S., Simm S., Paul P., Bublak D., Scharf K. D. and Schleiff E.(2015). Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis . Plant Cell Environ 38(4): 693-709. [DOI] [PubMed] [Google Scholar]
- 4.Krogh A., Larsson B., von Heijne G. and Sonnhammer E. L.(2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305(3): 567-580. [DOI] [PubMed] [Google Scholar]
- 5.Paul P., Chaturvedi P., Selymesi M., Ghatak A., Mesihovic A., Scharf K. D., Weckwerth W., Simm S. and Schleiff E.(2016). The membrane proteome of male gametophyte in Solanum lycopersicum . J Proteomics 131: 48-60. [DOI] [PubMed] [Google Scholar]
- 6.Paul P., Roth S. and Schleiff E.(2016). Importance of organellar proteins, protein translocation and vesicle transport routes for pollen development and function. Plant Reprod 29(1-2): 53-65. [DOI] [PubMed] [Google Scholar]
- 7.Paul P., Simm S., Blaumeiser A., Scharf K. D., Fragkostefanakis S., Mirus O. and Schleiff E.(2013). The protein translocation systems in plants- composition and variability on the example of Solanum lycopersicum . BMC Genomics 14: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul P., Simm S., Mirus O., Scharf K. D., Fragkostefanakis S. and Schleiff E.(2014). The complexity of vesicle transport factors in plants examined by orthology search. PLoS One 9(5): e97745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pertl H., Schulze W. X. and Obermeyer G.(2009). The pollen organelle membrane proteome reveals highly spatial-temporal dynamics during germination and tube growth of lily pollen. J Proteome Res 8(11): 5142-5152. [DOI] [PubMed] [Google Scholar]
- 10.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flugge U. I. and Kunze R.(2003). t>ARAMEMNON, a novel database for Arabidopsis integral membrane proteins . Plant Physiol 131(1): 16-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simm S., Fragkostefanakis S., Paul P., Keller M., Einloft J., Scharf K. D. and Schleiff E.(2015). t>Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum . Bioinform Biol Insights 9: 1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valledor L. and Weckwerth W.(2014). An improved detergent-compatible gel-fractionation LC-LTQ-Orbitrap-MS workflow for plant and microbial proteomics. Methods Mol Biol 1072: 347-358. [DOI] [PubMed] [Google Scholar]