Abstract
While the activation of the transcription factor interferon regulatory factor 5 (IRF5) is critical for the induction of innate immune responses, it also contributes to the pathogenesis of the autoimmune disease systemic lupus erythematosus (SLE). IRF5 phosphorylation is a hallmark of its activation in the Toll-like receptor (TLR) pathway, where active IRF5 induces type I interferon and proinflammatory cytokine genes. By using the phosphate-binding molecule Phos-tag, without either radioisotopes or phospho-specific antibodies, the protocol described here enables detection of the phosphorylation of both human and murine IRF5, as well as that of other proteins.
Keywords: IRF5, Phosphorylation, Innate immunity, TLR, SLE, Phos-tag, Immunoblot, SDS-PAGE
Background
In the TLR-MyD88 pathway, IRF5 is activated through post-translational modifications such as ubiquitination and phosphorylation, and then active IRF5 translocates into the nucleus and induces its target genes ( Takaoka et al., 2005 ; Balkhi et al., 2008 ; Tamura et al., 2008 ; Hayden and Ghosh, 2014). Regarding the activation status of IRF5 in SLE, it has been reported that IRF5 accumulates in the nucleus in monocytes of SLE patients ( Stone et al., 2012 ). Furthermore, we recently showed in an SLE murine model that IRF5 hyperactivation (e.g., elevated phosphorylation) leads to the development of an SLE-like disease ( Ban et al., 2016 ). Therefore, analyzing the activation status of IRF5 is important for studying SLE as well as innate immune responses. Phosphorylation is central to the activation of IRF5, as numerous studies have revealed the functional phosphorylation sites of IRF5 by site-directed mutagenesis and/or mass spectrometry ( Barnes et al., 2002 ; Lin et al., 2005 ; Chen et al., 2008 ; Chang Foreman et al., 2012 ; Lopez- Pelaez et al., 2014 ; Ren et al., 2014 ). However, antibodies specific for these phosphorylation sites are not commercially available. In addition, phosphorylated IRF5 is normally not separated from non-phosphorylated IRF5 using standard SDS-PAGE. We thus utilized the functional molecule Phos-tag, which binds specifically to the phosphate group via metal ions ( Kinoshita et al., 2006 ). Without using radioisotopes or phospho-specific antibodies, this protocol enables the detection of multiple phosphorylations of the IRF5 protein as up-shifted bands in the resulting immunoblot analysis (Figure 1). This protocol can be applied for detecting the phosphorylation of other proteins if a specific antibody for the total protein of the target protein is available.
Figure 1. Schematic of Phos-tag immunoblot analysis.
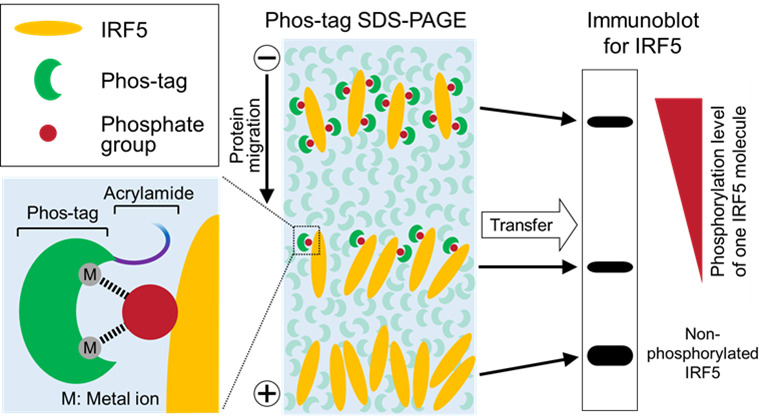
Phos-tag binds specifically to a phosphate group on the target protein via metal ions, such as Zn2+ or Mn2+. Non-phosphorylated and phosphorylated forms of the target protein (IRF5 in this figure) are separated by SDS-PAGE using acrylamide conjugated with Phos-tag, and then detected by immunoblot analysis using an appropriate specific antibody. The mobility shift of the phosphorylated protein is caused by trapping of its phosphate groups by the polyacrylamide gel-conjugated Phos-tag. Thus, multiple phosphorylations of IRF5 appear as different up-shifted bands, whose mobility shift increases with the phosphorylation level of each IRF5 molecule.
Materials and Reagents
Tips (BM Equipment, catalog numbers: BMT-10G and BMT-200; Corning, catalog number: 4846)
Disposable pipette (Greiner Bio One International, catalog numbers: 606160 and 607160)
Microcentrifuge tube (Greiner Bio One International, catalog number: 616201)
Filter paper (GE Healthcare, catalog number: 3030-6461)
Polyvinylidene fluoride (PVDF) membrane (EMD Millipore, catalog number: IPVH00010)
Bio-Rad Protein Assay (Bio-Rad Laboratories, catalog number: 5000006)
Bovine serum albumin (BSA) (Wako Pure Chemical Industries, catalog number: 017-23294)
Lambda protein phosphatase (λPPase) (New England Biolabs, catalog number: P0753)
Calf intestinal alkaline phosphatase (CIAP) (Takara Bio, catalog number: 2250)
Manganese(II) chloride tetrahydrate (MnCl2.4H2O) (Wako Pure Chemical Industries, catalog number: 136-15301)
Bromophenol blue (BPB) (Wako Pure Chemical Industries, catalog number: 029-02912)
Marker III (Wako Pure Chemical Industries, catalog number: 230-02461)
Anti-IRF5 antibody (Abcam, catalog number: ab21689)
MaxBlot solution 1 (MEDICAL & BIOLOGICAL LABORATORIES, catalog number: 8455-100)
Horseradish peroxidase-conjugated anti-rabbit IgG antibody (GE Healthcare, catalog number: NA934)
ECL Prime (GE Healthcare, catalog number: RPN2236)
Immunostar® LD (Wako Pure Chemical Industries, catalog number: 292-69903)
Sodium chloride (NaCl) (Wako Pure Chemical Industries, catalog number: 191-01665)
Na2HPO4.12H2O (Wako Pure Chemical Industries, catalog number: 196-02835)
Potassium chloride (KCl) (Wako Pure Chemical Industries, catalog number: 163-03545)
Potassium dihydrogen phosphate (KH2PO4) (Wako Pure Chemical Industries, catalog number: 169-04245)
NP-40 (Nacalai Tesque, catalog number: 25223-75)
Sodium deoxycholate (Nacalai Tesque, catalog number: 10712-12)
SDS (Nacalai Tesque, catalog number: 08933-05)
cOmplete protease inhibitor cocktail tablets (Roche Diagnostics, catalog number: 11836170001)
PhosSTOP phosphatase inhibitor cocktail tablets (Roche Diagnostics, catalog number: 04906837001)
Glycerol (Nacalai Tesque, catalog number: 17018-25)
2-mercaptoethanol (Wako Pure Chemical Industries, catalog number: 131-14572)
Tween 20 (Nacalai Tesque, catalog number: 28353-85)
Skim milk (Morinaga Milk Industry)
Acrylamide (Nacalai Tesque, catalog number: 00809-85)
N,N’-methylenebisacrylamide (bis) (Wako Pure Chemical Industries, catalog number: 138-06032)
Phos-tag acrylamide (Wako Pure Chemical Industries, catalog number: AAL-107)
APS (Wako Pure Chemical Industries, catalog number: 016-08021)
TEMED (Wako Pure Chemical Industries, catalog number: 205-06313)
Glycine (Nacalai Tesque, catalog number: 17109-35)
Tris (Nacalai Tesque, catalog number: 35406-91)
MOPS (Dojindo, catalog number: 343-01805)
Sodium bisulfite (Nacalai Tesque, catalog number: 31219-55)
Methanol (Wako Pure Chemical Industries, catalog number: 139-01827)
EDTA.2Na (Dojindo, catalog number: 345-01865)
7.5% Phos-tag precast gel (Wako Pure Chemical Industries, catalog number: 192-17381)
Phosphate-buffered saline (PBS, 1x) (see Recipes)
EDTA-free lysis buffer (see Recipes)
1% NP-40 TBS buffer (see Recipes)
6x sample buffer (see Recipes)
Tris-buffered saline containing Tween 20 (TBS-T) (see Recipes)
Blocking solution (see Recipes)
-
Handmade Phos-tag acrylamide gel (see Recipes)
Separating (lower) gel
Stacking (upper) gel
Tris-glycine running buffer (see Recipes)
Tris-MOPS running buffer (see Recipes)
Transfer buffer (see Recipes)
Equipment
Refrigerated mini-centrifuge (TOMY SEIKO, model: KITMAN-24)
Microplate reader with 595 nm wavelength available (Tecan, model: F039300REMOTER)
Heat block (Major Science, model: MD-02N)
Power supply (Bio-Rad Laboratories, catalog number: 1645070)
Gel tank (NIHON EIDO, model: NA-1012)
Horizontal shaker (YAMATO SCIENTIFIC, model: MK200D)
Semi-dry blotter (NIHON EIDO, model: NA-1512)
Blotting roller (Bio-Rad Laboratories, catalog number: 1651279)
Luminescent image analyzer (GE Healthcare, model: ImageQuant LAS 4000 mini)
Procedure
-
Cell lysate preparation
-
Harvest 1-5 x 106 cells by centrifugation (300 × g, 5 min, 4 °C).
Note: IRF5 phosphorylation occurs in specific cell types and conditions. For example: Bone-marrow-derived dendritic cells (BMDCs) stimulated with CpG-B oligodeoxynucleotide (ODN) (Ban et al., 2016). BMDCs are obtained by culturing mouse bone marrow cells with 100 ng/ml of the Flt3 ligand. After 9 days of culture, BMDCs are collected and seeded at 5 x 106 cells/well in a 6-well culture plate. They are then stimulated with 0.15 μM CpG-B ODN for 3 or 6 h (Figure 2). Other examples of appropriate cell models include HEK293T cells transfected with IRF5 and MyD88 (Ban et al., 2016) or IRF5 and IKKβ (Lopez-Pelaez et al., 2014; Ren et al., 2014), THP-1 cells stimulated with a lipopolysaccharide (LPS) (Ren et al., 2014), and Gen2.2 cells stimulated with CL097 (Lopez-Pelaez et al., 2014).
Remove the supernatant, tap the tube to loosen the cell pellet, add 5 ml of ice-cold PBS, and mix the cells by inverting the tube five times.
Repellet the cells by centrifugation.
Remove the supernatant, quickly centrifuge the sample, and carefully remove all residual supernatant without disturbing the cell pellet.
-
Add 50 μl of ice-cold EDTA-free lysis buffer to the cell pellet and then vigorously pipette up and down 20 times.
Note: EDTA may prevent the binding of the Phos-tag to the target protein phosphate groups by chelating metal ions; thus, the lysis buffer should be EDTA-free.
(Optional) For phosphatase treatment to validate detected phosphorylation, 1% NP-40 TBS buffer is used instead of EDTA-free lysis buffer.
Place the tube on ice for 30 min.
-
Centrifuge the sample at 15,000 × g, for 15 min at 4 °C and then transfer the supernatant (cell lysate) to a clean microcentrifuge tube on ice without transferring the cellular debris at the bottom.
Note: The cell lysate can be stored at -80 °C.
-
-
SDS-PAGE
(Optional) If you prepare handmade Phos-tag acrylamide gels rather than using the precast gels, see Recipes.
Quantify the protein concentration of the cell lysate. For the Bio-Rad Protein Assay, dilute the cell lysate by 20-fold with distilled water, add 5 μl of diluted cell lysate to 245 μl of 1x Bio-Rad Protein Assay, mix well by vortexing, and measure absorbance at 595 nm. Use a dilution series of bovine serum albumin (BSA) aqueous solution to construct a standard curve.
-
In a clean microcentrifuge tube, prepare 1-3 μg/μl cell lysate (10-30 μg/10 μl/well) by diluting with EDTA-free lysis buffer, and add 6x sample buffer (2 μl/well).
(Optional) For phosphatase treatment, 10 μg of the cell lysate prepared by using 1% NP-40 TBS buffer is incubated with the reaction mixture containing 1x NEBuffer for PMP (supplied with λPPase), 1 mM MnCl2, and 400 U of λPPase for 60 min at 30 °C. It is then further incubated with 30 U of CIAP for 30 min at 37 °C. The reaction is stopped by addition of 6x sample buffer.
Heat the samples at 100 °C for 5 min and then cool the samples to room temperature (20-24 °C).
-
Set up the handmade or precast gel in a gel tank by following the manufacturer instructions, and attach the leads to the power supply. Use running buffer in both the upper and lower chamber buffers.
Note: Tris-glycine running buffer can be used for both the handmade gel (containing Mn2+), and the precast gel (containing Zn2+), while Tris-MOPS running buffer can be used only for the precast gel. Both precast and handmade gels were successfully employed to detect IRF5 phosphorylation in the Tris-glycine running buffer. For other proteins, it will be necessary to empirically determine which combination of gel (metal ion) and running buffer is optimal for phosphorylation detection.
-
Apply the samples into the wells (12 μl/well) and run the gels as follows:
Handmade: Constant-voltage condition of 80 V for approximately 40 min (until bromophenol blue [BPB] dye reaches the separating gel), and then at 120 V for 200 min at room temperature
-
Precast: Constant-current condition of 15 mA/gel for 30 min, and then 30 mA/gel for 100 min at room temperature
Note: The above are optimized conditions for the separation of phosphorylated IRF5, where the position of the BPB dye and running time are used as a guide for electrophoresis. For other proteins, first run the gels until the BPB dye reaches the bottom of the separating gel, and increase the running time if better separation is required. The WIDE VIEW prestained protein size marker III (WAKO) can be used to evaluate the transfer efficiency, although notably, it does not identify the actual molecular weight of proteins. To avoid band distortion in the adjacent lanes, at least one blank lane should be left between this marker and the analyzed samples.
-
Transfer and blocking
During the run, prepare blocking solution (see Recipes) and immerse the filter papers in transfer buffer.
When the run is complete, remove the gels from the apparatus and cut off the stacking gel.
-
Incubate the gels in EDTA-containing transfer buffer (25 ml/gel) for 10 min with gentle shaking (60 rpm) at room temperature. Discard the buffer.
Note: Since metal ions in the gel interfere with the transfer, EDTA-containing transfer buffer is used to remove them from the gel after SDS-PAGE.
Repeat the incubation twice (three times in total).
-
Incubate the gels once in EDTA-free transfer buffer (25 ml/gel) for 10 min with gentle shaking at room temperature.
Note: Since EDTA in the transfer buffer also interferes with the transfer, EDTA-free transfer buffer is used to remove EDTA from the gel after removal of the metal ions.
Briefly pre-wet the PVDF membrane in methanol until the membrane is completely wet, discard methanol, add transfer buffer, and incubate for at least 10 min with gentle shaking (60 rpm) at room temperature.
Stack 5 filter papers, the PVDF membrane, gel, and 5 filter papers in this order from the anode (+) to the cathode (-) of the semi-dry blotter. Gently remove any air bubbles with the blotting roller.
Run at 100 mA/gel (2 mA of current per cm2 of gel) for 60 min at room temperature.
When the run is complete, remove the membrane from the stack and incubate the membrane in blocking solution for 30 min with gentle shaking at room temperature.
-
Antigen-antibody reaction and ECL detection
-
Prepare primary antibody solution (anti-IRF5 antibody diluted by 3,000-fold with MaxBlot solution 1).
Note: Using the blocking solution (5% skim milk in TBS-T) to dilute the anti-IRF5 antibody (Abcam) causes the appearance of non-specific bands. For other antibodies, the blocking solution can be used to dilute the primary antibody during Phos-tag immunoblot analysis if it is known to facilitate the appearance of a correct specific (preferably single) band via the standard immunoblot analysis method.
Rinse the membrane with TBS-T and incubate it in primary antibody solution for 90 min at room temperature or overnight at 4 °C with gentle shaking.
Wash the membrane 5 times with TBS-T (5 min incubation for each washing).
Prepare secondary antibody solution (horseradish peroxidase-conjugated anti-rabbit IgG antibody diluted by 20,000-fold with blocking solution).
Incubate the membrane in secondary antibody solution for 60 min at room temperature with gentle shaking.
Wash the membrane 5 times with TBS-T (5 min incubation for each washing).
Incubate the membrane in the working solution of ECL Prime or Immunostar® LD (for high-sensitivity detection) by following the manufacturer’s instructions.
Detect the chemiluminescence using the luminescent image analyzer.
-
Figure 2. Detection of phosphorylated IRF5 by Phos-tag immunoblot analysis.
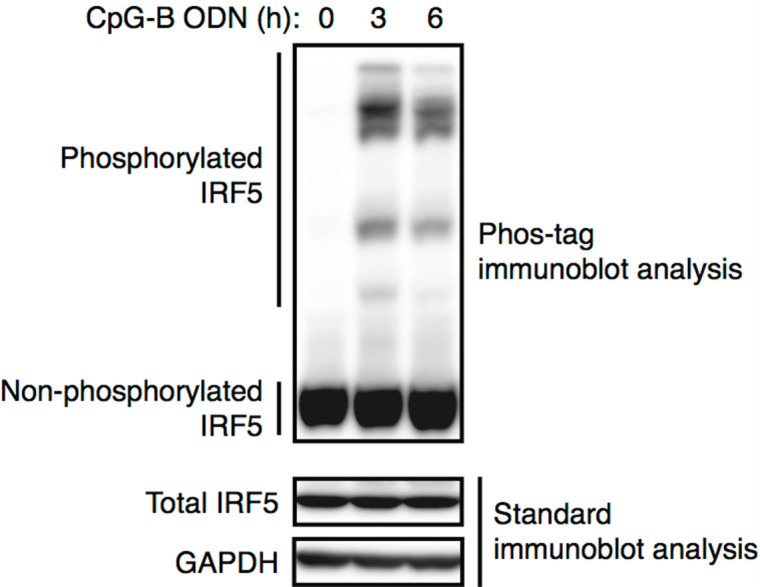
BMDCs were stimulated with 0.15 μM CpG-B oligodeoxynucleotide (ODN) for the indicated times and subjected to Phos-tag (upper panel) or standard (bottom 2 panels) immunoblot analysis using antibodies against IRF5 and GAPDH. GAPDH levels served as a loading control.
Data analysis
As shown in Figure 2, phosphorylated IRF5 proteins are visualized as up-shifted migration bands (cf. Figure S5D of Ban et al., 2016 ). The multiple up-shifted bands indicate differently phosphorylated IRF5 protein species. In BMDCs, a proportion of IRF5 proteins are phosphorylated in response to stimulation with CpG-B ODN (a TLR9 ligand).
Notes:
Protein size cannot be determined by Phos-tag immunoblot analysis since the mobility shift of the phosphorylated protein is caused by trapping of its phosphate groups by the polyacrylamide gel-conjugated Phos-tag (Figure 1), rather than because the size of the phosphorylated protein is enlarged by attachment of the Phos-tag. In addition, it has been previously shown that the actual size of a non-phosphorylated protein can differ from its estimated size as determined by standard SDS-PAGE (Kinoshita et al., 2009). Finally, the number of phosphorylations occurring on a given protein cannot be determined using this protocol because the mobility shift of a given phosphorylated protein is different to that of another protein with the same number of phosphorylations (Kinoshita et al., 2006).
The band shifts are unlikely to be due to increases in the IRF5 molecular weight by ubiquitination, since polyubiquitinated IRF5 is not detectable by standard immunoblot analysis unless MG132 (a proteasome inhibitor that inhibits the degradation of ubiquitin-conjugated proteins) and/or deubiquitinase inhibitors such as N-ethylmaleimide and PR-619 are used.
Phosphatase treatment can be used to confirm that the up-shifted bands are phosphorylated IRF5 (cf. Figure S5A of Ban et al., 2016).
The specificity of the antibody was confirmed by comparing the samples between wild-type and IRF5-deficient cells (cf. Figure S5D of Ban et al., 2016).
Remaining SDS-PAGE samples can be subjected to standard immunoblot analysis to verify the amounts of total IRF5 protein and a loading control protein.
Recipes
-
Phosphate-buffered saline (PBS, 1x)
137 mM NaCl
8.1 mM Na2HPO4.12H2O
2.7 mM KCl
1.5 mM KH2PO4
Note: No pH adjustment is required (pH is approximately 7.4). Store at 4 °C.
-
EDTA-free lysis buffer
50 mM Tris-HCl (pH 7.4)
150 mM NaCl
1% NP-40
0.5% sodium deoxycholate
0.1% SDS
cOmplete protease inhibitor cocktail tablets
PhosSTOP phosphatase inhibitor cocktail tablets
Note: No pH adjustment is required for the EDTA-free lysis buffer. The inhibitor cocktail tablets should be added freshly. The buffer without inhibitors can be stored at 4 °C.
-
1% NP-40 TBS buffer (for phosphatase treatment)
20 mM Tris-HCl (pH 8.0)
150 mM NaCl
1% NP-40
cOmplete protease inhibitor cocktail tablets
Note: No pH adjustment is required for the 1% NP-40 TBS buffer. The inhibitor cocktail tablets should be added freshly. The buffer without the inhibitor cocktail tablets can be stored at 4 °C.
-
6x sample buffer
375 mM Tris-HCl (pH 6.8)
9% SDS
50% glycerol
0.03% BPB
9% 2-mercaptoethanol (2ME)
Note: No pH adjustment is required for the 6x sample buffer. 2ME should be added freshly. The buffer without 2ME can be stored at room temperature. Precipitated SDS in the stored buffer can be resolved by heating to 50 °C.
-
Tris-buffered saline containing Tween 20 (TBS-T)
25 mM Tris-HCl (pH 7.4)
140 mM NaCl
3 mM KCl
0.05% Tween 20
Note: No pH adjustment is required for the TBS-T; store at room temperature.
-
Blocking solution
TBS-T
5% skim milk
Note: Prepare freshly for each experiment.
-
Handmade Phos-tag acrylamide gel (mini size)
Note: Use ddH2O rather than alcohol to overlay the separating gel. The duration of polymerization for the separating gel is approximately 40 min. Use the handmade gel immediately after polymerization, and do not store for later use.
-
Separating (lower) gel
7.5% acrylamide/bis (29:1)
375 mM Tris-HCl (pH 8.8)
0.1% SDS
100 μM MnCl2
50 μM Phos-tag acrylamide
0.1% APS
0.04% TEMED
Note: For proteins other than IRF5, first optimize the concentration of acrylamide/bis (29:1), and then change the concentration of Phos-tag acrylamide as required. Proteins of a size greater and less than 60 kDa should initially be subjected to analysis using 6% and 8% acrylamide/bis (29:1), respectively. The optimal concentration of Phos-tag acrylamide for analysis of a given protein can be selected from a series (20, 50, 100, and 150 μM). The utilized concentration of MnCl2 should be 2-fold higher than that of Phos-tag acrylamide.
-
Stacking (upper) gel
4.5% acrylamide/bis (29:1)
125 mM Tris-HCl (pH6.8)
0.1% SDS
0.1% APS
0.1% TEMED
-
-
Tris-glycine running buffer
25 mM Tris
192 mM glycine
0.1% SDS
Note: No pH adjustment is required (pH is 8.1-8.5). Store at room temperature.
-
Tris-MOPS running buffer
100 mM Tris
100 mM MOPS
0.1% SDS
5 mM sodium bisulfite
Note: Sodium bisulfite should be added freshly. No pH adjustment is required (pH is approximately 7.8). The buffer without sodium bisulfite can be stored at 4 °C.
-
Transfer buffer
48.2 mM Tris
38.9 mM glycine
0.037% SDS
20% methanol
10 mM EDTA (included for the first, second, and third incubation of the gel after Phos-tag SDS-PAGE)
Note: No pH adjustment is required (pH is approximately 9.2). Store at room temperature.
Acknowledgments
We thank Dr. Yayoi Kimura at Yokohama City University for her invaluable advice. This protocol was adapted from Wako’s Phos-tag SDS-PAGE protocol. This work was supported by the Fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Science and Technology Agency to T.T.; and Grants-in-Aid (KAKENHI) from the MEXT/Japan Society for the Promotion of Science (Nos. 16K19161 and 25860368 to T.B.).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Balkhi M. Y., Fitzgerald K. A. and Pitha P. M.(2008). Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol 28(24): 7296-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban T., Sato G. R., Nishiyama A., Akiyama A., Takasuna M., Umehara M., Suzuki S., Ichino M., Matsunaga S., Kimura A., Kimura Y., Yanai H., Miyashita S., Kuromitsu J., Tsukahara K., Yoshimatsu K., Endo I., Yamamoto T., Hirano H., Ryo A., Taniguchi T. and Tamura T.(2016). Lyn kinase suppresses the transcriptional activity of IRF5 in the TLR-MyD88 pathway to restrain the development of autoimmunity. Immunity 45(2): 319-332. [DOI] [PubMed] [Google Scholar]
- 3.Barnes B. J., Kellum M. J., Field A. E. and Pitha P. M.(2002). Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol 22(16): 5721-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Foreman H. C., Van Scoy S., Cheng T. F. and Reich N. C.(2012). Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One 7(3): e33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W., Lam S. S., Srinath H., Jiang Z., Correia J. J., Schiffer C. A., Fitzgerald K. A., Lin K. and Royer W. E., Jr (2008). Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol 15(11): 1213-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden M. S. and Ghosh S.(2014). Innate sense of purpose for IKKβ. Proc Natl Acad Sci USA 111: 17348-17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinoshita E., Kinoshita-Kikuta E., Matsubara M., Aoki Y., Ohie S., Mouri Y. and Koike T.(2009). Two-dimensional phosphate-affinity gel electrophoresis for the analysis of phosphoprotein isotypes. Electrophoresis 30(3): 550-559. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita E., Kinoshita-Kikuta E., Takiyama K. and Koike T.(2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5(4): 749-757. [DOI] [PubMed] [Google Scholar]
- 9.Lin R., Yang L., Arguello M., Penafuerte C. and Hiscott J.(2005). A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J Biol Chem 280(4): 3088-3095. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Pelaez M., Lamont D. J., Peggie M., Shpiro N., Gray N. S. and Cohen P.(2014). Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA 111(49): 17432-17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren J., Chen X. and Chen Z. J.(2014). IKKβ is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci USA 111(49): 17438-17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone R. C., Feng D., Deng J., Singh S., Yang L., Fitzgerald-Bocarsly P., Eloranta M. L., Ronnblom L. and Barnes B. J.(2012). Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis Rheum 64(3): 788-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W. and Taniguchi T.(2005). Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434(7030): 243-249. [DOI] [PubMed] [Google Scholar]
- 14.Tamura T., Yanai H., Savitsky D. and Taniguchi T.(2008). The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535-584. [DOI] [PubMed] [Google Scholar]


