Abstract
The protocol for obtaining electrically sealed membrane vesicles from E. coli cells is presented. Proton pumps such as Complex I, quinol oxidase, and ATPase are active in the obtained vesicles. Quality of the preparation was tested by monitoring the electric potential generated by these pumps.
Keywords: Escherichia coli, Membrane vesicles, Proton pumping
Background
Studying of membrane enzymes often requires embedding them into the natural lipid environment. Inside-out, everted membrane vesicles allowed to explore effects of different substrates, inhibitors and other ligands on the operation of these enzymes. Functioning proton pumps, such as NADH: ubiquinone oxidoreductase Type 1 (Complex I), quinol oxidase, and ATPase also requires good electrical sealing of the vesicles. This protocol provides sufficient results in preparation of such vesicles for studying these enzymes (e.g., Euro et al., 2008 ; Belevich et al., 2011 ).
Materials and Reagents
Glass test tubes (approximately 10 x 90 mm)
E. coli MWC215 (SmR ndh::CmR) or GR70N (wild type or mutated) cells grown under aeration
Luria Broth or other medium suitable for aerobic growth of E. coli culture
Antibiotics, as required for particular E. coli strain
Lysozyme
Ethylenediaminetetraacetic acid (EDTA)
Magnesium sulfate (MgSO4)
Dithiothreitol (DTT)
Phenylmethylsulfonyl fluoride (PMSF)
Ethanol
Liquid N2
Electric potential-sensitive probe Oxonol VI (Sigma-Aldrich, catalog number: 75926)
Ammonium sulfate, (NH4)2SO4
Decylubiquinone (Sigma-Aldrich, catalog number: D7911)
Potassium cyanide (KCN)
Nicotinamide adenine dinucleotide reduced (NADH)
Rolliniastatin
Ubiquinone 1 (Sigma-Aldrich, catalog number: C7956)
Adenosine triphosphate (ATP)
-
Aurovertin B (Sigma-Aldrich, catalog number: A5297)
Note: This product has been discontinued.
Tris-HCl
Sucrose
HEPES
Potassium hydroxide (KOH)
Potassium chloride (KCl)
Magnesium chloride (MgCl2)
3-(N-morpholino)propanesulfonic acid (MOPS)
1,3-bis(tris(hydroxymethyl)methylamino)propane (BTP)
The medium (see Recipes)
Buffer A (see Recipes)
Buffer B (see Recipes)
Buffer C (see Recipes)
Buffer D (see Recipes)
Equipment
200 ml Erlenmeyer flasks
2 L Erlenmeyer flasks
Shaker, such as Sertomat R/Sertomat HR or other suitable
Centrifuges, such as (Thermo Fisher Scientific, model: Sorvall RC 5C Plus)
Rotors (Thermo Fisher Scientific, Thermo ScientificTM, models: SS-34, SLA 3000; Beckman Coulter, model: Ti70)
Sonicator (Emerson, model: Branson Sonifier Cell Disruptor B15) or other with the tip diameter 10-12 mm
UV/VIS spectrophotometer (i.e., Ocean Optics, model: HR2000)
Procedure
-
Cells
Inoculate the cells into 5-100 ml (dependently on how much you want to grow next day) LB (+ antibiotics dependently on the strain) to get overnight culture (RT, orbital shaker).
Inoculate overnight culture 1:50 to LB (with antibiotics if necessary) in 50 ml to 200 ml Erlenmeyer flasks or 700 ml LB in 2 L Erlenmeyer flasks and grow in orbital shaker at 37 °C, 220-230 rpm for ~4-5 h (A600 ~2.5 for wild type). Follow the growth: cells must be harvested in the late exponential phase. Do not overgrow, otherwise the cell wall would be thicker and there could be problem with cell disintegration.
Pellet cells at 7,500 x g (8,000 rpm) x 10 min in SS-34 or 8,000 rpm x 20 min in SLA 3000 or other appropriate rotor (10-20 min at approximately 8,000 x g).
-
Spheroplasts
Suspend cell pellet at RT in 10 ml (2 x 50 ml culture) or 100 ml (3 x 700 ml culture) in buffer A (see Recipe 1).
-
Add 100 μg/ml lysozyme (10 mg/ml stock solution) and incubate at RT until cells become osmosensitive (don’t treat the cells longer than necessary), approx. 5-10 min. On that purpose perform the osmosensitivity test. Place 50 μl cells to 1 ml 10 mM EDTA, pH 7.0 in the glass tube. Visually seen lysis (the suspension is getting transparent) should occur in 1-2 min. The cells without lysozyme are not disrupted under these conditions.
Note: Remember to take control before lysozyme addition! The cells without lysozyme stay intact and should be used for comparison with lysozyme-treated cells by matching of the turbidity of the cell suspension.
Add 4 mM MgSO4 (from 1 M stock solution) and traces of DNase. The suspension viscosity should decrease almost immediately.
Pellet spheroplasts at 7,500 x g (8,000 rpm) x 10 min in SS-34.
Suspend the spheroplasts on ice in 20 ml or 40 ml of cold buffer B (see Recipe 2).
-
Add 2 mM dithiothreitol (DTT) (from 100 mM stock solution), 0.5 mM PMSF (from 100 mM fresh stock solution).
Notes:
PMSF has short life time in water solutions. It is stable only in absolute ethanol kept anaerobically, therefore the best way is to make fresh stock solution in 95% ethanol.
This medium is optional and can be changed dependently on further experiments, however, high [Mg2+], dithiothreitol (DTT) and protease inhibitor PMSF always significantly improve the membrane quality. Buffer B is developed for measurements of intravesicular acidification by proton pumps. For monitoring the electric potential generation buffer C with low salts concentration was used (see below).
-
Membranes (on ice/water bath)
Sonicate spheroplasts at approximately 75 W (watts). For efficient sonication the distance between the sonicator tip and walls of the tube with spheroplasts suspension should not be wider than 6-8 mm. To prevent overheating 50% sonication pulses are used. After approx. 30 sec sonication it is necessary to make 1 min break for cooling. For osmosensitive cells this should be repeated 4-6 times. The cell disruption can be visually controlled.
Pellet undisrupted cells and cell debris at 7,200 x g x 10 min in SS-34 and discard it.
Pellet membranes in Ti70 at 150,000 x g (40,000 rpm) x 40 min or other appropriate rotor at 80,000-150,000 x g for 30-40 min. If the membrane vesicles with larger internal volume are required the sedimentation should proceed shorter or at lower RCF (relative centrifugal force).
Rinse the pellet if necessary and suspend membranes (in minimal volume) of desired buffer B (see Recipe 2).
If the membranes are not used immediately freeze them in aliquots in liquid N2 and store at -80 °C.
The quality of obtained membrane vesicles can be simply tested by monitoring the electric potential (∆ψ) generated by proton pumps using ∆ψ-sensitive probe, Oxonol VI, as shown in Figure1. The value of ∆ψ can be estimated by calibration of absorption changes with imposed diffusion potentials (i.e., Ghelli et al., 1997 ; Pouliquin et al., 1999 ). H+ or K+ diffusion potential could be established in the presence of proton uncoupler such as carbonyl cyanide m-chlorophenyl hydrazine (CCCP) or potassium transporter valinomycin, correspondingly. The value of the diffusion potential can be calculated by the Nernst equation. The detailed description of the procedure can be found in the paper by Ghelli et al., 1997 .
The NADH:ubiquinone reductase activity of membrane-bound Complex I can be measured in buffer C at 30 °C by following NADH oxidation at 340 nm (ε = 6.2 mM-1 cm-1). The NADH:ubiquinone oxidoreductase activity of Complex I in the membrane vesicles prepared from E. coli GR70N is 0.92 ± 0.13 (n = 10) ( Belevich et al., 2011 ).
Figure 1. Following the electric potential generation by three proton pumps in E. coli membranes using ∆ψ .
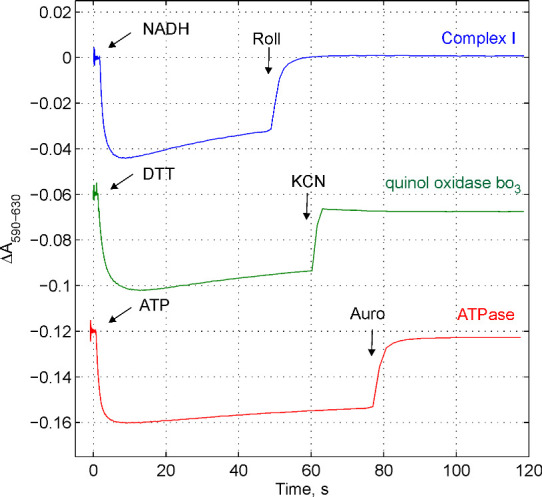 -sensitive probe Oxonol VI. The membrane vesicles were loaded with buffer D. The medium consists of buffer C accomplished with 5 mM (NH4)2SO4 (to convert ∆pH to ∆ψ) and Oxonol VI 2.5 μM (stored in 2-5 mM ethanol or methanol stock solution at -20 °C, stable at least a half of year). The membranes were added at concentration 80 μg/ml. The reaction was followed by the measurements of absorption changes as indicated. To monitor ∆ψ generation by NADH:ubiquinone oxidoreductase, Type I, (Complex I), the medium was accomplished with 100 μM decylubiquinone and 1 mM KCN to block the terminal oxidase. 50 μM NADH was added to start the reaction, ∆ψ was dissipated by Complex I inhibitor, rolliniastatin (Roll) at concentration 1 μM. To monitor ∆ψ generation by quinol oxidase bo3 the medium was accomplished with 50 μM ubiquinone 1. To start the reaction 2 mM dithiothreitol was added, ∆ψ was dissipated by 1 mM KCN addition. To monitor ∆ψ generation by ATPase the medium was accomplished with 1 mM MgSO4. The reaction was initiated by an addition of 1 mM ATP, ∆ψ was dissipated by an addition of ATPase inhibitor, aurovertin B (Auro B) at concentration 10 μM.
-sensitive probe Oxonol VI. The membrane vesicles were loaded with buffer D. The medium consists of buffer C accomplished with 5 mM (NH4)2SO4 (to convert ∆pH to ∆ψ) and Oxonol VI 2.5 μM (stored in 2-5 mM ethanol or methanol stock solution at -20 °C, stable at least a half of year). The membranes were added at concentration 80 μg/ml. The reaction was followed by the measurements of absorption changes as indicated. To monitor ∆ψ generation by NADH:ubiquinone oxidoreductase, Type I, (Complex I), the medium was accomplished with 100 μM decylubiquinone and 1 mM KCN to block the terminal oxidase. 50 μM NADH was added to start the reaction, ∆ψ was dissipated by Complex I inhibitor, rolliniastatin (Roll) at concentration 1 μM. To monitor ∆ψ generation by quinol oxidase bo3 the medium was accomplished with 50 μM ubiquinone 1. To start the reaction 2 mM dithiothreitol was added, ∆ψ was dissipated by 1 mM KCN addition. To monitor ∆ψ generation by ATPase the medium was accomplished with 1 mM MgSO4. The reaction was initiated by an addition of 1 mM ATP, ∆ψ was dissipated by an addition of ATPase inhibitor, aurovertin B (Auro B) at concentration 10 μM.
Notes
This protocol gives highly reproducible results, however, all procedures should be conducted as fast as possible. Breaks (especially after the cells disruption step) may cause an increase in proton leakage through the membrane and decrease in membrane enzymes activity.
This protocol was developed especially for E. coli cells and it should not be used for other bacterial species without modifications.
Recipes
-
Buffer A
200 mM Tris-HCl, pH 8.0
2 mM EDTA
30% sucrose
-
Buffer B
100 mM HEPES-KOH, pH 7.5
100 mM KCl
10 mM MgCl2
-
Buffer C
25 mM HEPES-BTP, pH 7.5
3 mM KCl
-
Buffer D
100 mM MOPS-BTP, pH 7.0
1 mM MgSO4
Acknowledgments
This work was supported by grants from Biocentrum Helsinki, the Sigrid Juselius Foundation, and the Academy of Finland.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Euro L., Belevich G., Verkhovsky M. I., Wikström M. and Verkhovskaya. M. (2008). Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase(Complex I). Biochim Biophys Acta 1777(9): 1166-1172. [DOI] [PubMed] [Google Scholar]
- 2.Belevich G., Knuuti J., Verkhovsky M.I., Wikström M, Verkhovskaya M.(2011). Probing the mechanistic role of the long α-helix in subunit L of respiratory Complex I from Escherichia coli by site-directed mutagenesis . Mol Microbiol 82(5): 1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghelli A., Benelli B. and Esposti M. D.(1997). Measurement of the membrane potential generated by complex I in submitochondrial particles. J Biochem 121(4): 746-755. [DOI] [PubMed] [Google Scholar]
- 4.Pouliquin P., Grouzis J. and Gibrat R.(1999). Electrophysiological study with oxonol VI of passive NO3- transport by isolated plant root plasma membrane . Biophys J 76: 360-373. [DOI] [PMC free article] [PubMed] [Google Scholar]


