Abstract
Seedless vascular plants, including ferns and lycophytes, produce spores to initiate the gametophyte stage and to complete sexual reproduction. Approximately 10% of them are apomictic through the production of genomic unreduced spores. Being able to measure the spore nuclear DNA content is therefore important to infer their reproduction mode. Here we present a protocol of spore flow cytometry that allows an efficient determination of the reproductive modes of seedless vascular plants.
Keywords: Apomixis, Bead-vortex, Fern, Flow cytometry, Lycophyte, Spore
Background
In seedless vascular plants, sporogenesis features, such as meiotic chromosome counts, were traditionally used to infer nuclear DNA content as well as reproductive modes. However, these approaches are time-consuming, or can only provide indirect evidence. An efficient and reliable method to estimate spore nuclear DNA content of these plants had not been established until Kuo et al. (2017) . Herein, we describe a protocol using flow cytometry to evaluate spore genome sizes of these plants based on the work of Kuo et al. (2017) .
Materials and Reagents
Pipette tips (10, 100, and 1,000 μl)
50-ml tube
1.7-ml tubes with caps
2.0-ml tubes with caps
2.3-mm stainless steel beads (Bio Spec Products, catalog number: 11079123ss)
30-μmnylon meshes (Sysmex, CellTrics®, catalog number: 04-0042-2316)
20-μmnylon meshes (Sysmex, CellTrics®, catalog number: 04-0042-2315)
10-μm nylon meshes (Sysmex, CellTrics®, catalog number: 04-0042-2314)
Glass Petri dish (Corning, PYREX®, catalog number: 423790)
Leaf tissue of C-value standard (e.g., Nicotiana tabacum L. ‘Xanthi’; 2C = 10.04 pg, Johnston et al., 1999 )
Spores of ferns or lycophytes (kept by dry storage and under < 4 °C)
PVP-40 (Sigma-Aldrich, catalog number: PVP40)
2-mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
RNaseA solution (10 mg/ml in ddH2O) (Sigma-Aldrich, catalog number: R5000-100MG)
Triton X-100
Sodium sulfite (Na2SO3)
Tris-HCl (pH 7.5)
Propidium iodide
-
Backmen stock buffer (see Recipes)*
*Note: LB01 buffer (Doležel et al., 2007) or GPB buffer (Loureiro et al., 2007) can be alternatively used depending on plant material properties.
PI solution (see Recipes)
Equipment
Pipette (10, 100, and 1,000 μl)
Vortex (Scientific Industries, model: Vortex-Genie 2)
Razors and razor pen
-
Flow cytometer (BD, BD Biosciences, model: FACScan)*
*Note: FACScan with a 15 microW blue argon ion laser of an emission wave length of 488 nm.
Software
BD FACSCan system (BD Biosciences, Franklin Lake, NJ, USA)
Procedure
-
Prepare buffer for use
Allocate appropriate amount of Backmen stock buffer to a 50-ml tube based on an estimation of 1-1.5 ml per sample.
Add 0.04 g PVP-40, 5 μl 2-mercaptoethanol, 1 μl RNase per ml of buffer.
-
Extract spore nuclei by bead-vortexing
-
For each sample, weigh ~0.007 g spores into a 1.7-ml tube. Green spores that usually have thin spore walls, 4 times the amount of spores are recommended ( Kuo et al., 2017 )*.
*Note: For bead-vortexing, the detailed process can be seen in the video supplied in Kuo et al. (2017): http://onlinelibrary.wiley.com/store/10.1111/nph.14291/asset/supinfo/nph14291-sup-0002-VideoS1.mov?v=1&s=dae6f0590d33413f1444bc0add857d285fb73cd6
Add 16 stainless steel beads into each 1.7-ml tube.
Add 250 μl of buffer into each 1.7-ml tube.
Vortex these tubes at a speed of 1,900 rpm for 1 min. For green spores, a speed of 3,200 rpm and a vortex duration of 0.5 min are recommended ( Kuo et al., 2017 ).
Filter bead-vortexed samples into 2.0-ml tubes through nylon meshes. The size of nylon mesh is selected based on spore sizes to prevent spore being filtered through the mesh.
Add additional buffer to the samples, and ensure each of filtered spore nuclei solutions is greater than 500 μl in volume.
-
-
Extract standard nuclei by chopping leaf tissue
Add 500 μl of buffer to a glass Petri dish.
Add a (~400 mm2) piece of young leaf to the Petri dish, and chop it with a razor on ice until most tissue slices are less than 1 mm in size.
Filter the chopped sample into a 2.0-ml tube through a 30-μm nylon mesh.
Add additional buffer to the sample, and ensure that the filtered leaf nuclei solution is greater than 500 μl in volume or more depending on need.
-
Staining nuclei solutions
Mix spore nuclei and standard leaf nuclei solutions into a 500-μl volume in 2.0-ml tubes.
Add 10 μl PI solution into each of mixed nuclei solutions.
Incubate in the dark at 4 °C for 1 h for staining.
Data analysis
Set up a histogram plot of particle count vs. linear value of relative fluorescence in BD FACSCan system.
After PI staining, measure nuclear DNA content of the samples in BD FACSCan system, and adjust the fluorescence laser voltage to visualize the nuclei peaks on the histogram plot.
Measure > 1,300 particles for each peak, and the coefficient variation for each peak should be lower 5% as the quality criteria suggested by Greilhuber et al. (2007) .
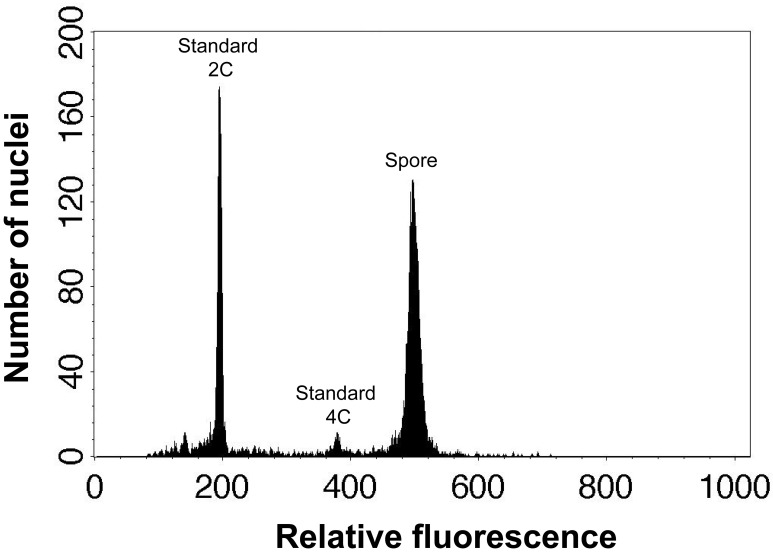
Genome size of spore nuclei (pg or Mbp) = x standard 2C value (Figure 1).
Figure 1. An example of flow cytometric result of spore and standard nuclei.
These peaks are spore nuclei of Dryopteris varia and 2C and 4C leaf nuclei of Nicotiana tabacum L. ‘Xanthi’.
Notes
To dry fertile leaves to release and collect spores, air-drying process under room temperature for 2 to 3 days is recommended. For long-term storage, spore material is better stored in tubes without any solution at a temperature lower than 4 °C.
Recipes
-
Backmen stock buffer ( Ebihara et al., 2005 )
1.0% Triton X-100
50 mM Na2SO3
50 mM Tris-HCl (pH 7.5)
ddH2O (the solvent)
Note: Store at 4 °C up to 1 year.
-
PI solution
2.04 mg/ml propidium iodide
ddH2O (the solvent)
Note: Store in the dark at 4 °C for long-term storage.
Acknowledgments
The bead-vortexing condition to extract spore nuclei of seedless vascular plants is accessed and constructed by Kuo et al. (2017). We thank Fay-Wei Li and three anonymous reviewers for providing comments on the draft of this manuscript.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Doležel J., Greilhuber J. and Suda J.(2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2(9): 2233-2244. [DOI] [PubMed] [Google Scholar]
- 2.Ebihara A., Ishikawa H., Matsumoto S., Lin S. J., Iwatsuki K., Takamiya M., Watano Y. and Ito M.(2005). Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex(Hymenophyllaceae) in Japan and adjacent areas . Am J Bot 92(9): 1535-1547. [DOI] [PubMed] [Google Scholar]
- 3.Greilhuber J., Temsch E. M. and Loureiro J. C. M.(2007). Nuclear DNA Content Measurement. In: Doleel, J., Greilhuber, J. and Suda, J.(Eds.). Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Wiley, pp: 67-102. [Google Scholar]
- 4.Johnston J., Bennett M., Rayburn A., Galbraith D. and Price H.(1999). Reference standards for determination of DNA content of plant nuclei. Am J Bot 86(5): 609-613. [PubMed] [Google Scholar]
- 5.Kuo L. Y., Huang Y. J., Chang J., Chiou W. L. and Huang Y. M.(2017). Evaluating the spore genome sizes of ferns and lycophytes: a flow cytometry approach. New Phytol 213(4):1974-1983. [DOI] [PubMed] [Google Scholar]
- 6.Loureiro J., Rodriguez E., Dolezel J. and Santos C.(2007). Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100(4): 875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]