Abstract
Glutathione is one of the major antioxidant defense components present in cells. It is predominantly present as reduced glutathione (GSH) and converted into oxidized glutathione (GSSG) while reducing the free radicals like hydroxyl ions (OH-). For the measurement of GSH and GSSG, o-phthalaldehyde (OPT) has been used as a fluorescent reagent. O-phthalaldehyde has an ability to react specifically with GSH at pH 8 and GSSG at pH 12 respectively. N-ethylmaleimide (NEM) has been used to prevent auto-oxidation of GSH during measurement of GSSG in the present protocol. The original protocol by Hissin and Hilf was developed for glutathione estimation in Rat liver tissue. The present protocol has been standardized following Hissin and Hilf (1976) for the estimation of glutathione in cultured microglial cell lysate but it can also be used for other mammalian cell lysate. In our lab same protocol has been used for the estimation of glutathione in the whole cell lysate of murine neuroblastoma cell, N2a.
Keywords: Glutathione, Antioxidant, Free radical, Microglia (N9), o-Phthalaldehyde, N-ethylmaleimide
Background
This method was published by Hissin and Hilf in analytical biochemistry way back in 1976 (Hissin and Hilf, 1976). There are methods available to detect GSH accurately however; due to readily oxidative conversion of GSH into GSSG most of the methods give an overestimate of GSSG. Cohen and Lyle (1966) solved the problem by using NEM to prevent oxidative conversion of GSH into GSSG and also preventing GSH to react with OPT during GSSG estimation (Figures 1 and 2). We have used this simple and reliable method to detect GSH and GSSG in our experimental system (microglial cell lysate). The main advantage of this protocol is that, it does not involve sophisticated instrument like high performance liquid chromatography (HPLC) which also needs sufficient expertise to handle as compared to plate reader which is more commonly available and easy to operate ( Rahman et al., 2006 ).
Figure 1. Schematic of chemical reaction during GSH estimation in the N9 cell lysate.
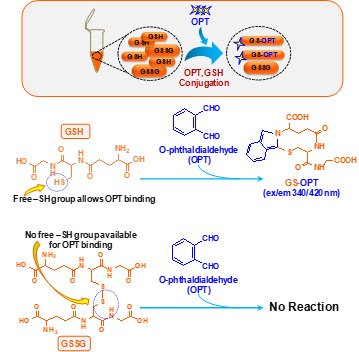
Figure 2. Schematic of chemical reaction during GSSG estimation in the N9 cell lysate.
Materials and Reagents
Pipette tips (Corning, Axygen®, catalog number: T-1005-WB-C-L)
0.22 μm filter
6 well culture plate (SRL LIFE SCIENCES, catalog number: 30006)
Culture flask
1.5 ml centrifuge tube (Corning, Axygen®, catalog number: MCT-150-R)
0.5 ml tube (Corning, Axygen®, catalog number: 14-222-292)
96 well plate (SPL life sciences, catalog number: 30096)
Disposable plastic cell scraper (SRL LIFE SCIENCES, catalog number: 90020)
Cell line: In the present protocol mouse microglial cell line, N9 has been used, which was kindly gifted by Dr. Anirban Basu, National Brain Research Centre (NBRC), India ( Singh et al., 2016 ). N9 cell line was developed by retroviral transfection of primary microglia cells with the v-myc or v-mil oncogenes of the avian retrovirus MH2. Cells were cultured in DMEM/F12 medium supplemented with 10% FBS and 1% penicillin-streptomycin in a 5% CO2 incubator at 37 °C
DMEM/F12 (Sigma-Aldrich, catalog number: 56498C)
Sodium bicarbonate
Double distilled water
Fetal bovine serum (FBS) (Genetix Biotech, Cell cloneTM, catalog number: CCS-500-SA-U)
Penicillin-streptomycin (Pen-Strep) (Thermo Fisher Scientific, GibcoTM, catalog number: 10378016)
Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340)
Bradford reagent (Bio-Rad Laboratories, catalog number: 5000006)
Tri-chloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T6399)
o-Phthalaldehyde (Sigma-Aldrich, catalog number: P1378)
N-ethylmaleimide (Sigma-Aldrich, catalog number: E3876)
Glutathione reduced (GSH) (Sigma-Aldrich, catalog number: G4251)
Glutathione oxidized (GSSG) (Sigma-Aldrich, catalog number: G4376)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Dipotassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P3786)
Potassium phosphate dibasic trihydrate (K2HPO4.3H2O)
-
EDTA disodium salt (Sigma-Aldrich, catalog number: E5513)
Note: This product has been discontinued.
100% ethanol (Merck, catalog number: 1009831011)
Methanol (SRL Laboratories, catalog number. 65524)
0.1 M potassium phosphate EDTA buffer (KPE buffer) (see Recipes)
50% trichloroacetic acid (see Recipes)
o-Phthaldehyde solution (10 mg/ml) (see Recipes)
0.4 M N-ethylmaleimide (see Recipes)
0.1 N sodium hydroxide (see Recipes)
Equipment
1 ml and 200 µl pipettes (Eppendorf)
Table top centrifuge (Sigma-zentrifuges, model: Sigma 3-18KS)
Microplate reader (BMG LABTECH, model: FLUOstar Omega)
CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Steri-CycleTM CO2 Incubators)
Sonicator (Sonics & Materials, model: VC 505)
Software
Mars data analysis software, ver. 1.01 (Data analysis software for Microplate reader)
Procedure
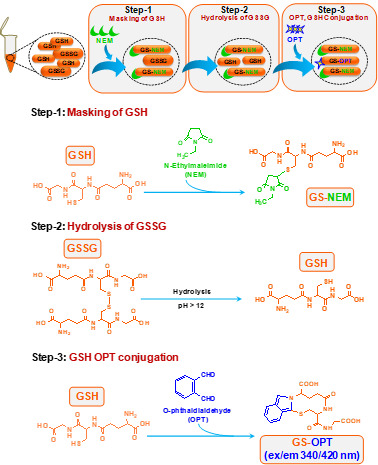
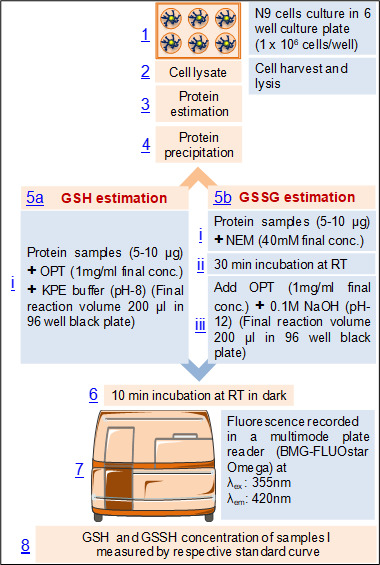
Flow diagram of the procedure (Figure 3)
Figure 3. Flow diagram of the procedure for the estimation of glutathione in mouse microglial cell line, N9.
-
Media preparation and cell culture
For 1 L medium, 12 g DMEM/F12, 2.44 g sodium bicarbonate is dissolved in 890 ml of double distilled water and then 100 ml FBS along with 10 ml Pen-Strep is added. Finally media is filtered through 0.22 μm filter.
N9 cells are seeded at a density of 1 x 106/2 ml in a 6-well culture plate and grown for 16-18 h in complete culture medium (DMEM/F12 medium supplemented with 10% FBS and 1% Pen-Strep).
N9 cells are passaged after every 48 h and used till 6-7 passage after thawing.
Note: If any treatment is given, please ensure that dose and duration of treatment is not toxic for cells.
-
Cell harvesting and lysis
Remove culture medium and wash the cells with phosphate buffered saline (PBS). Add ice cold 0.1 M potassium phosphate buffer with EDTA (KPE buffer, 1 ml) to the each well and cells are dislodged from the surface of culture flask with the help of cell scraper and collected in a 1.5 ml centrifuge tube.
Centrifuge the cell suspension at 350 × g for 5 min at 4 °C.
Resulting cell pellet is resuspended in 200 µl KPE buffer with 1% protease inhibitor cocktail (PIC) and lysed by sonication for 5 sec pulse at 25 W two times on ice.
Note: Cell scrapper is preferred instead of enzymatic dissociation to minimize the handling time. Besides, for enzymatic dissociation cells needs incubation in 37 °C whereas ice cold buffer can be used while using scrapper which is desirable during preparing cell lysate to keep structure and function of the protein unaltered.
-
Protein sample preparation and estimation
After sonication, lysed samples are centrifuged at 18,000 × g for 10 min at 4 °C and the resulting supernatants are collected in separate pre-cooled 1.5 ml centrifuge tubes.
Keep 10 µl samples in 0.5 ml tubes for protein estimation by Bradford reagent.
-
Protein precipitation
After protein estimation, 10 µg protein sample is precipitated. Initially 80 µl protein sample is mixed with 20 µl trichloroacetic acid (TCA) (50% stock concentration), vortexed and kept in ice for 10 min. Protein sample with TCA is centrifuged at 9,100 × g for 10 min at 4 °C, the supernatant is transferred into a fresh 1.5 ml centrifuge tube.
-
GSH and GSSG estimation
-
GSH estimation
Mix 10 µl supernatant with equal volume of OPT (1 mg/ml) and 180 µl KPE buffer (pH-8) in a black 96 well plate.
-
GSSG estimation
Transfer 50 µl of the supernatant into a new centrifuge tube, add 0.5 µl N-ethylmaleimide (stock concentration: 4 M) and mix thoroughly.
Incubate for 30 min at room temperature to inhibit GSH.
-
Add 10 µl of this sample, 10 µl OPT and 180 µl 0.1 N NaOH (pH-12) in a black 96 well plate.
Note: Use multichannel pipette to add OPT and KPE buffer mixture in the 96 well plate to minimize the time lapse.
-
Incubate for 10 min in the dark at room temperature.
Measure the fluorescence at λex: 355 nm and λem: 420 nm in a microplate reader.
Data analysis
-
Standards preparation for GSH and GSSG
GSH and GSSG stock solution is prepared at a concentration of 1 mg/ml in KPE buffer and diluted 100 times to 10 µg/ml. 800 µl of 10 µg/ml solution is mixed with 200 µl KPE buffer to make standard GSH and GSSG concentration of 26.4 nmol/ml. From this concentration a series of two fold serial dilutions are (13.2 nmol/ml, 6.6 nmol/ml, 3.3 nmol/ml till 0.103 nmol/ml) prepared in KPE buffer.
-
Calculation of protein concentration in unknown samples
Protein concentrations is determined from standard curve with the formula Y = mX + c (Figures 4A and 4B).
-
Calculation of GSH and GSSG in unknown samples
Concentrations of GSH and GSSG from samples is calculated from their respective standard curve with the formula Y = mX + c (Figures 5A and 5B). This assay can detect GSH from 10 ng to 2 µg and GSSG from 5 ng to 2 µg.
-
Normalization
GSH and GSSG concentrations divided by the protein quantity used in reaction (10 µg protein per sample). Data are represented in terms of µmole/mg protein.
Figure 4. Calculation of protein concentration.
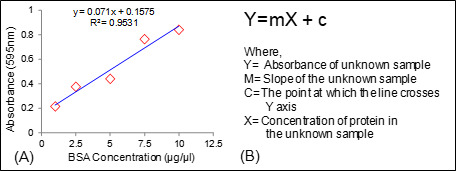
A. Standard curve of BSA; B. Equation used for the calculation of protein concentration in an unknown sample.
Figure 5. Calculation of GSH and GSSG concentration.
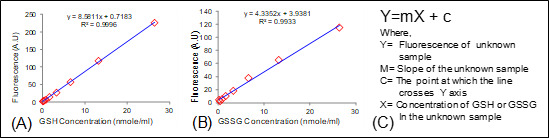
Standard curve of (A) GSH; (B) GSSG and (C) Equation used for the calculation of GSH/GSSG concentration in an unknown sample.
Recipes
-
0.1 M potassium phosphate EDTA buffer (KPE buffer)
Note: KPE buffer is prepared of two different solutions, A and B.
Solution A: dissolving 0.68 g KH2PO4 in 50 ml dH2O
Solution B: dissolving 0.85 g K2HPO4 or 1.14 g K2HPO4.3H2O in 50 ml dH2O
Both the solution A and B can be stored at 4 °C if not using immediately. Just before use 0.1 M KPE buffer is prepared by mixing 8 ml of solution A with 42 ml of solution B and the pH adjusted to 8. Finally 0.16 g of EDTA disodium salt is added to the potassium phosphate buffer and mix well to prepare potassium phosphate EDTA buffer
-
50% trichloroacetic acid (10 ml)
5 g trichloroacetic acid weighed and immediately mixed with dH2O
Adjust the volume to 10ml with dH2O
-
o-Phthaldehyde solution (10 mg/ml)
o-Phthaldehyde solution is prepared by mixing 10 mg orthophthaldehyde with 1 ml methanol
The solution can be stored at 4 °C in dark up to 1 week
-
0.4 M N-ethylmaleimide
50 mg N-ethylmaleimide is dissolved in 1 ml 100% ethanol to make 0.4 M NEM solution. It can be stored at 4 °C up to 1 week
-
0.1 N sodium hydroxide
0.04 g sodium hydroxide pellet is dissolved in 10 ml dH2O to get 0.1 N solution
Acknowledgments
This work has been supported by CSIR-12th 5 year network project, Integrated NextGen Approaches in Health, Disease and Environmental Toxicity (INDEPTH-BSC001); V.S. has been supported by CSIR Senior Research Fellowship. R.G. and M.P. P. have been supported by UGC-Senior Research Fellowship. The authors declare no competing financial interest. The CSIR-IITR manuscript number is 3440. This protocol has been followed and modified from paper entitled “A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem 74(1): 214-226”.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Cohn V. H and Lyle J.(1966). A fluorometric assay for glutathione. Anal Biochem 14(3): 434-40. [DOI] [PubMed] [Google Scholar]
- 2.Hissin P. J. and Hilf R.(1976). A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem. 74(1): 214-26. [DOI] [PubMed] [Google Scholar]
- 3.Rahman I., Kode A. and Biswas S. K.(2006). Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc 1(6): 3159-65. [DOI] [PubMed] [Google Scholar]
- 4.Singh V., Gera R., Kushwaha R., Sharma A. K., Patnaik S. and Ghosh D.(2016). Hijacking microglial glutathione by inorganic arsenic impels bystander death of immature neurons through extracellular cystine/glutamate imbalance. Sci Rep 6: 30601. [DOI] [PMC free article] [PubMed] [Google Scholar]


