Abstract
Brain predicted age difference, or BrainPAD, compares chronological age to an age estimate derived by applying machine learning (ML) to MRI brain data. BrainPAD studies in youth have been relatively limited, often using only a single MRI modality or a single ML algorithm. Here, we use multimodal MRI with a stacked ensemble ML approach that iteratively applies several ML algorithms (AutoML). Eligible participants in the Healthy Brain Network (N = 489) were split into training and test sets. Morphometry estimates, white matter connectomes, or both were entered into AutoML to develop BrainPAD models. The best model was then applied to a held‐out evaluation dataset, and associations with psychometrics were estimated. Models using morphometry and connectomes together had a mean absolute error of 1.18 years, outperforming models using a single MRI modality. Lower BrainPAD values were associated with more symptoms on the CBCL (p corr = .012) and lower functioning on the Children's Global Assessment Scale (p corr = .012). Higher BrainPAD values were associated with better performance on the Flanker task (p corr = .008). Brain age prediction was more accurate using ComBat‐harmonized brain data (MAE = 0.26). Associations with psychometric measures remained consistent after ComBat harmonization, though only the association with CGAS reached statistical significance in the reduced sample. Our findings suggest that BrainPAD scores derived from unharmonized multimodal MRI data using an ensemble ML approach may offer a clinically relevant indicator of psychiatric and cognitive functioning in youth.
Keywords: biomarkers, brain age, connectome, diffusion tensor imaging, machine learning
Our study demonstrates that multimodal brain imaging coupled with novel, rigorous machine learning methods has the potential to improve the accuracy of brain age estimation. BrainPAD (or the difference between chronological age and estimated brain age) shows promise as a general measure of risk for psychopathology and cognitive impairment in youth, with some evidence for distinct associations between particular domains and decreased versus increased brain age.
1. INTRODUCTION
Human neuromaturation is the complex process that governs the formation and refinement of the structures and connections of the central nervous system from conception through early adulthood. Whereas the mechanisms underlying neuromaturation—such as neural migration, myelination, and synaptic pruning—are relatively conserved across individuals (Tamnes et al., 2017), the rates at which these processes unfold are heterogeneous (Foulkes & Blakemore, 2018). Deviations in the rate of neuromaturation can lead to differences in brain structure, connectivity, and function, with potential implications for the etiology and phenomenology of cognitive impairments and psychopathology (Shahab et al., 2019). These links, however, between neuromaturation, cognition, and psychiatric outcomes remain relatively poorly characterized in youth, despite adolescence being the period when many serious psychiatric conditions emerge and neuromaturation is accelerated.
Altered rates of neuromaturation can be estimated by deviations between an individual's chronological age relative to his or her “brain age.” Brain age and its utility as a marker of disease have been examined in adults across neurologic and psychiatric disorders. Generally, having an “older” brain (i.e., brain age exceeds chronological age) is associated with disease. To name some examples, patients with Alzheimer's disease have been shown to have older brains than healthy controls (Gaser et al., 2013). Increased white matter lesion load in patients with multiple sclerosis has been associated advanced brain age (Høgestøl et al., 2019). In the ENIGMA study of 2,533 participants, major depressive disorder was associated with older brain age (Han, Dinga, Hahn, et al., 2019). Adults with schizophrenia have also been found to have advanced brain ages relative to both healthy adults and adults with bipolar disorder (Nenadic, Dietzek, Langbein, Sauer, & Gaser, 2017). Finally, in a sample of 45,615 individuals between the ages of 3–96, increased brain aging was also associated with several disorders, including schizophrenia, multiple sclerosis, and bipolar spectrum disorder (Kaufmann, van der Meer, Doan, et al., 2019).
While the contribution of brain age to psychopathology continues to be explored in adults, there are fewer studies in children and adolescents. This is a notable omission for three reasons. First, peak incidence for most major psychiatric disorders including depression, anxiety disorders, schizophrenia, and substance use disorders all occur during adolescence, the same period that neuromaturation is at an apex (Walker, Sabuwalla, & Huot, 2004). Second, the research to date has revealed a generally more complex relationship between brain age and psychopathology in youth. Third, preliminary work applying machine learning (ML) algorithms to neuroimaging data in children and adolescents suggest this approach has promise for prognostication (Franke & Gaser, 2019).
In contrast to adults, advanced brain age in youth is not uniformly associated with negative outcomes. For example, older brain age in children, rather than being linked to diminished cognitive states, as in adults, was associated with increased processing speed (Boyle, Jollans, Rueda‐Delgado, et al., 2021; Erus et al., 2015). Among adolescents, “younger” compared to older functional brain states have been associated with adverse symptoms, such as increased risk‐taking behavior (Rudolph, Miranda‐Dominguez, Cohen, et al., 2017). Conversely, a study utilizing structural MRI data found that teenage participants at high‐risk for psychosis were more likely to become psychotic if they had older brain ages relative to “normal” brain ages (Chung et al., 2018). In sum, in youth the associations between brain age, psychiatric symptoms, and neurocognitive functioning may vary across psychiatric and neurocognitive domains.
Previous brain age estimation tools have used brain morphology and functional imaging data, but only a select few have incorporated white matter connectomes, much less brain morphology and white matter connectomes together (Brown et al., 2012; Erus et al., 2015). Studies using MRI‐derived whole‐brain connectomes suggest that atypical development of the connectome is associated with long‐term difficulties with emotion, cognition, and behavior in infants and adolescents (Kaufmann et al., 2017). For example, white matter connectomes at birth were predictive of cognitive performance at age 2 in both full‐term and preterm infants (Girault et al., 2019). Among adolescents with attentional problems, temporoparietal tracts were found to have lower fractional anisotropy (Tymofiyeva, Gano, Trevino, et al., 2018). Thus, the inclusion of white matter connectomes is likely important to studies characterizing the relationship between brain development and psychiatric symptoms, functioning, and cognition in youth.
Previous studies using ML to predict brain age have often employed a single ML algorithm, such as Support Vector Regression (Franke, Luders, May, Wilke, & Gaser, 2012; Schnack et al., 2016), Relevance Vector Regression (Franke et al., 2012), or convolutional neural networks (Cole et al., 2017). Employing a pipeline that systematically tests multiple ML algorithms, and chooses the most accurate one, could improve accuracy, particularly if there is validation with a held‐out sample to diminish the risk of overfitting (Acion et al., 2017). In addition, ensemble learning that combines several individual ML algorithms could further improve accuracy (Wolpert, 1992).
Here, we use an automated ML pipeline that incorporates multiple ML algorithms, including an ensemble method, to estimate brain age from morphometry estimates and whole‐brain white matter connectomes obtained from participants in the Healthy Brain Network (HBN), a large community‐based cohort study of children and adolescents. We then apply the best ML model to a held‐out evaluation dataset of participants with and without a high likelihood for psychopathology to identify associations between deviations in brain age, cognitive functioning, and psychiatric symptoms. To our knowledge, this is the first study to use morphometry, white matter connectomes, and an ensemble ML algorithm to look at broad measures of symptoms and functioning in children and adolescents.
2. METHODS AND MATERIALS
2.1. Data source
We used neuroimaging and psychometric data from the HBN collected between 2015, when the study was initiated, through 2017, when HBN neuroimaging data was made publicly available. All phenotypic information was obtained in accordance with the Data Usage Agreement required by the HBN. Briefly, the HBN recruited a community‐based sample of healthy and nonhealthy boys and girls between ages 5 and 21 in multiple sites in New York City. Participants were excluded from the HBN if they had immediate safety concerns, medical conditions that would confound neuroimaging research, or their symptoms interfered with the participation in the study. Detailed inclusion and exclusion criteria are available elsewhere (Alexander et al., 2017).
2.2. MRI scanning protocols
Participants in the HBN study were scanned at the following three sites. The protocols for each site were as follows. Staten Island: mobile scanner, 1.5T Siemens Avanto, T1: 176 slices, resolution (mm) 1.0 × 1.0 × 1.0, TR 2,730 ms, TE 1.64 ms, multiband acceleration off; Diffusion Kurtosis imaging: 72 slices, resolution (mm) 2.0 × 2.0 × 2.0, TR 3,110 ms, TE 76.2 ms, multiband acceleration 3. Rutgers University Brain Imaging Center: Siemens 3T Tim Trio, T1: 224 slices, resolution (mm) 0.8 × 0.8 × 0.8, TR 2,500 ms, TE 3.15 ms, multiband acceleration off; Diffusion Kurtosis imaging: 72 slices, resolution (mm) 1.8 × 1.8 × 1.8, TR 3,320 ms, TE 100.2 ms, multiband acceleration 3, 64 directions, b = 0, 1,000, 2,000). And Citigroup Cornell Brain Imaging Center: Siemens 3T Prisma, T1: 224 slices, resolution (mm) 0.8 × 0.8 × 0.8, TR 2,500 ms, TE 3.15 ms, multiband acceleration off; Diffusion Kurtosis imaging: 81 slices, resolution (mm) 1.8 × 1.8 × 1.8, TR 3,320 ms, TE 100.2 ms, multiband acceleration 3, 64 directions, b = 0, 1,000, 2,000. Additional scan parameters are listed elsewhere (Protocol HBN, 2017).
2.3. Quality assessment
To maximize the potential clinical utility of our brain age prediction model, we aimed to include all participants, regardless of neuroimaging data quality. In addition, quality‐related artifacts might contain meaningful information. Therefore, no manual edits were performed on the FreeSurfer reconstructions. Rather than excluding participants with low quality data, we examined to what degree image quality changed the relationship between predicted age from the optimal ML model and chronological age. MRIQC was used to estimate the signal to noise ratio for the structural images, and FSL was used to obtain the temporal signal to noise ratio for the diffusion‐weighted imaging (DWI) data (Andersson & Sotiropoulos, 2016; Esteban et al., 2017). Relative motion parameters were obtained using eddy_cuda9.1 as part of the Mrtrix DWI pipeline. All quality metrics were normalized by site. Structural signal to noise ratios were normalized by site and T1 image used.
First, we examined whether the quality measures were independently correlated with chronological age (p <.20). Metrics associated with chronological age were then included in a linear model (LM), with chronological age as the outcome, and predicted age and the relevant quality metrics as predictors. Finally, we calculated the percentage change in the coefficient of predicted age in that model compared to a model without quality metrics. The greater the percentage change, the more imaging quality, rather than brain features, were driving the model's performance.
2.4. Brain morphometry
FreeSurfer v6.0 ‐(https://surfer.nmr.mgh.harvard.edu/) was used to generate 678 morphometric estimates from structural MRI (sMRI) data, including thickness, volumes, surface, and mean curvature, for each participant. The Desikan–Killiany atlas was used (Desikan, Segonne, Fischl, et al., 2006). In short, structural image processing with FreeSurfer included motion correction, removal of nonbrain tissue, Talairach transformation, segmentation, intensity normalization, tessellation of the gray matter/white matter boundary, topology correction, and surface deformation. Deformation procedures used both intensity and continuity information to produce representation of cortical thickness. The maps produced were not restricted to the voxel resolution of the original scans and were thus capable of detecting submillimeter differences. Cortical thickness measures have been validated against histological analysis and manual measurements (Fischl et al., 2002; Salat et al., 2005). Further information about this process is provided elsewhere (Dale, Fischl, & Sereno, 1999; Desikan et al., 2006; Fischl et al., 2002; Fischl & Dale, 2000).
Morphometric data were not normalized for estimated total intracranial volume (eTIV), given our goal of accurate brain age estimation. Instead, eTIVs were included in the dataset supplied to the ML pipeline. We then examined whether the optimal ML model did more than rely on eTIV data (see below).
2.5. White matter connectomes
To obtain accurate brain phenotypic estimates, we used an individualized connectome approach, rather than population‐based regions of interest or tracts of interest methods. We used the MrtrixMRI analysis pipeline to preprocess diffusion MRI (dMRI), estimate whole‐brain white matter tracts, and generate individualized connectome features. Generated features included number of estimated streamlines within a given connection, a commonly used measure of fiber connection strength (Cha et al., 2015; Cha et al., 2016), and fractional anisotropy (FA) from the diffusion tensor model. dMRI images were de‐noised (Veraart et al., 2016), motion corrected (Andersson & Sotiropoulos, 2016), and then processed through the Advanced Normalization Tools (ANTs) pipeline using the N4 algorithm (Tustison et al., 2010). Probabilistic tractography was performed using second‐order integration over fiber orientation distributions with a whole‐brain streamline count of 20 million (Tournier, Calamante, & Connelly, 2010).
To discard potential false positive streamlines and improve biological plausibility, initial tractograms were filtered using spherical‐deconvolution informed filtering with a final streamline count target of 10 million (2:1 ratio). Using the filtered tractogram, an 84 × 84 whole‐brain connectome matrix was generated for each participant using the T1‐based parcellation and segmentation from FreeSurfer, which was then registered and warped to the participant's diffusion MRI (b0 images) using ANTs. With this approach, each participant's white matter connectome estimates were constrained by his or her own neuroanatomy. A total of 7,140 connectome estimates were obtained, weighted by streamline count and FA. Computation was done on supercomputers at Argonne Leadership Computing Facility Theta and Texas Advanced Computing Center Stampede2.
2.6. Harmonization of brain data
ComBat (short for Combating batch effects when combining Batches) can be used to reduce “scanner effects” in processed structural and diffusion imaging data (Fortin et al., 2017; Fortin et al., 2018). ComBat harmonizes the mean and variance of brain measures across scanners using a modified linear mixed effects model. However, ComBat harmonization can also attenuate the relationship between brain data and other measures (Chen et al., 2020). For example, if participants imaged on one scanner are younger than participants imaged on another, ComBat might misattribute age‐related differences in brain volume to a scanner effect. To protect these relationships, variables of interest should be included in the ComBat model. In addition, ComBat does not account for variation in the correlation of brain data and other measures across scanners. This might limit the effectiveness of some ML algorithms (Chen et al., 2020). Finally, ComBat cannot handle missing covariate data; subjects with any missing covariates are dropped.
Here, we use the ComBat package for R to produce two additional versions of the training, test, and evaluation datasets (Fortin, 2019). First, we harmonized the brain data for all the eligible participants and included age in the ComBat model. We report the type and accuracy of the most accurate age prediction models developed using both the unharmonized and age‐harmonized datasets. Because no age data were missing, there was no reduction in sample size and the same participants comprised the training and test datasets for the unharmonized and age‐harmonized datasets. Second, we harmonized the brain data from all the eligible participants and included both age and the four exploratory outcome measures (described below, Section 2.8), in the ComBat model. This resulted in a reduction in our sample size due to missingness. However, the number of remaining participants in the training and test set preserved the desired 80%/20% split. Therefore, no participants had to be re‐assigned; all remaining participants retained their original training, test, or evaluation group assignment. We report the type and accuracy of the most accurate age predictions model developed using this age‐outcome‐harmonized dataset. We also report the associations between BrainPAD values (defined below) and the four exploratory outcome measures. Additional details on our use of ComBat are included in the Supporting Information.
2.7. Brain age model development and comparison
We first divided the HBN dataset into two groups based on the Child Behavior Checklist (CBCL) Total Problem T‐Score. The CBCL is a well validated, parent report measure of emotional, behavioral, and social difficulties in children and adolescents, and Total Problem T‐scores of 60 or greater are associated with an increased likelihood of psychopathology (Warnick, Bracken, & Kasl, 2008). To train our brain age prediction on a group of likely typically developing children, we used a cutoff of 60, such that participants scoring 60 or above were in one group and below 60 in another.
We then further divided the group with CBCL Total Problem T‐scores below 60 into a training dataset (n = 215), to develop the age prediction models, and a testing dataset (n = 48), to assess their accuracy, using a random 80%/20% split. Lastly, we created an evaluation dataset (n = 249) that consisted of participants with a high likelihood for psychopathology (those with CBCL Total Problem T‐Scores of 60 or greater) and a low likelihood for psychopathology (participants from the testing dataset, all of whom had CBCL Total Problem T‐Scores less than 60). By pooling participants with low and high likelihood for psychopathology into our evaluation dataset, we aimed to determine whether observed relationships between brain age and outcome measures were robust across the full range of CBCL T scores, and not limited to youth with or without psychopathology. Participants without age, sex, race, or ethnicity data were excluded from the evaluation dataset (Figure 1).
FIGURE 1.
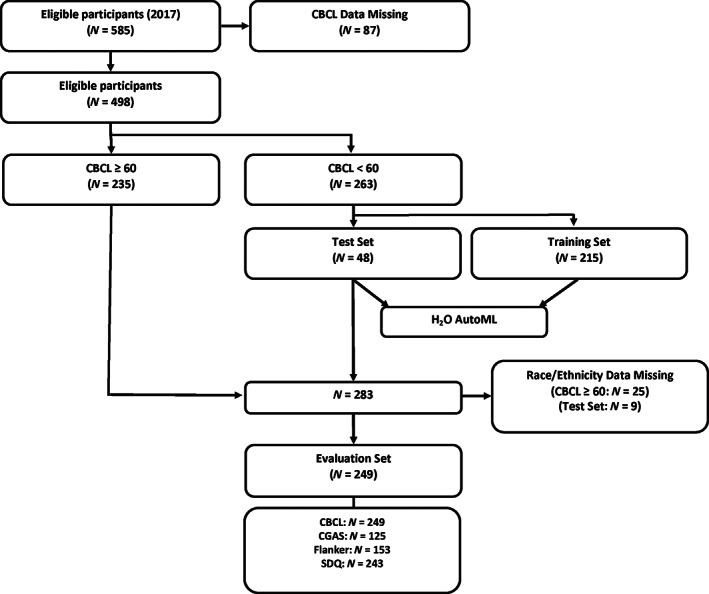
Participant selection workflow. In brief, of 498 participants with CBCL‐Parent report data available, 263 with a CBCL score <60 were used for modeling with H2O's AutoML function using an 80%/20% train/test split. For statistical analysis, test set participants and participants with CBCL ≥60 with race/ethnicity data available were combined for a total of 249 participants
We used H2O AutoML, an open‐source automatic ML pipeline (H2O AutoML, ), to develop and select three brain age prediction models, one for each MRI modality—morphometry (sMRI alone) and connectomes (dMRI alone)—and a third based on both modalities combined (sMRI and dMRI combined; The H2O.ai Team, 2015). As noted above, we did this using the unharmonized brain data, and also the age‐ and age‐outcome‐harmonized brain data. H2O AutoML features multiple ML algorithms and performs scalable, automated model training and hyper‐parameter tuning. Features with near‐zero variance were removed. Random grid search was performed for hyperparameter optimization by H2O. The AutoML pipeline includes Gradient Boosting Method, Generalized Linear Model, Random Forest, “Deep Learning” Multi‐Layer Perceptrons, and Stacked Ensemble ML (SEML). The list of the hyperparameters optimized for each algorithm in the pipeline is available elsewhere (https://docs.h2o.ai/h2o/latest-stable/h2o-docs/automl.html).
The pipeline generated, optimized, and tested a series of brain age prediction models using the MRI modalities and algorithms listed above. Each model was developed using five‐fold cross‐validation, followed by subsequent testing with the held‐out testing data set, producing a ranked list of models. Maximum runtime for AutoML was set to 5 hr. k‐fold cross‐validation and the use of a held‐out dataset to test model accuracy reduce the risk of over‐fitting. Model accuracy was measured using the mean residual deviance (MRD) because it is more robust to non‐normally distributed data. We selected the models with the lowest MRD for each modality (sMRI alone and dMRI alone), and their combination (sMRI and dMRI combined), for further assessment. The mean absolute error (MAE), which is measured in years and is more easily interpreted, was also calculated for all models.
Since eTIV and FA are correlated with age (Schmithorst, Wilke, Dardzinski, & Holland, 2002; Takao, Hayashi, & Ohtomo, 2012), we verified that the optimal ML model produced more accurate results than simpler models relying on these data alone. We regressed age on global FA (mean FA across all white matter tracts), eTIV, and their combination. Parallel to the development of the ML models, this LM was first developed using the training dataset, and then applied to the testing dataset. To compare performance with the ML models, we also calculated the MAE for this model in the testing dataset.
2.8. Exploratory analyses: outcome measures
In addition to the CBCL, we used the following outcome measures: Children's Global Assessment Scale (CGAS), Flanker Uncorrected Standard Score, and Strengths and Difficulties Questionnaire (SDQ) total difficulties score. The CGAS is a clinically administered assessment of global functioning, with higher scores indicating better functioning. The Flanker task is a measure of sustained attention, with higher scores indicating better performance. The SDQ, like the CBCL, is a general measure of behavioral and emotional symptoms in children, but with fewer questions (25 questions) than the CBCL (118 questions). While there is evidence that the CBCL and SDQ are similar measures (Goodman & Scott, 1999; Warnick et al., 2008), there is some evidence that the CBCL better discriminates between community and referred populations (Dang, Nguyen, & Weiss, 2017), and is more sensitive (Warnick et al., 2008), while the SDQ is more specific (Warnick et al., 2008).
2.9. Exploratory analyses: calculation of brain predicted age difference (BrainPAD)
We calculated brain predicted age difference (BrainPAD) (Cole, Marioni, Harris, & Deary, 2019; Franke et al., 2012), to capture the difference between the predicted and chronological ages for each participant, with BrainPAD equal to the predicted age minus the chronological age. BrainPAD was therefore positive when the predicted age was greater than the chronological age and negative when the predicted age was less than the chronological age. We calculated BrainPAD scores using the most accurate models built from the unharmonized brain data and the age‐outcome‐harmonized brain data.
2.10. Statistical analyses
Descriptive statistics were generated for the training, testing, and evaluation datasets. We used analysis of variance (ANOVA) tests to evaluate differences in the training, testing, and evaluation groups. LMs were used to estimate the association between BrainPAD values and the outcome measures, controlling for age, sex, race, ethnicity, and scanner (site). These analyses were therefore restricted to participants with those data. A separate LM was used for each outcome measure. Plotting and modeling were performed using Rstudio version 1.2.5033 and the R Stats package version 4.0.4 (Team RC, 2012). In these analyses, we used the CBCL Total Raw score instead of the Total T‐score, as the T‐score is already adjusted for age and sex. Throughout, p‐values of less than or equal to .05 were considered statistically significant. We report both the nominal and Bonferroni corrected p‐values for the four LMs.
3. RESULTS
3.1. Participant selection and demographics
Workflow for participant selection and analysis is outlined in Figure 1. In brief, of the 585 HBN participants initially eligible for this study, 87 did not have CBCL data and were excluded. The age range among the remaining 498 participants with CBCL data was 5–18. Among the remaining 498 participants, 263 had CBCL Total T‐scores less than 60, and 235 had T‐scores equal to or greater than 60. Eighty percent of the participants with T‐scores less than 60 (n = 215) were included in the training dataset, and 20% (n = 48) in the testing dataset. For the evaluation dataset (n = 249), only participants with race and ethnicity data were included. The evaluation set consisted of 39 participants from the testing set and 210 participants whose CBCL scores were equal to or greater than 60. This same workflow is described for the age‐outcome‐harmonized dataset in the Supporting Information. Demographic information for the participants in the training, testing, and evaluation datasets are displayed in Table 1. Note that while groups differ on clinical markers, the distributions of age, sex, race, and ethnicity are comparable across the three groups.
TABLE 1.
Healthy Brain Network participant demographics
| Training set (N = 215) | Test set (N = 48) | Evaluation set (N = 249) | p‐value | |
|---|---|---|---|---|
| Age | 1.79 (±2.98) | 10.65 (±3.42) | 10.97 (±3.29) | .74 |
| Sex | .90 | |||
| Male | 133 (61.86%) | 29 (60.42%) | 149 (59.84%) | |
| Female | 82 (38.14%) | 19 (39.58%) | 100 (40.16%) | |
| Race | .86 | |||
| White/Caucasian | 93 (43.26%) | 21 (43.75%) | 127 (51.00%) | |
| Black/African‐American | 34 (15.81%) | 5 (10.42%) | 42 (16.87%) | |
| Hispanic | 19 (8.84%) | 5 (10.42%) | 23 (9.24%) | |
| Asian | 6 (2.79%) | 0 (0.00%) | 8 (3.21%) | |
| Indian | 6 (2.79%) | 1 (2.08%) | 1 (0.40%) | |
| Native‐American Indian | 0 (.00%) | 0 (0.00%) | 1 (0.40%) | |
| Two or more races | 32 (14.88%) | 8 (16.67%) | 38 (15.26%) | |
| Other race | 4 (1.86%) | 0 (0.00%) | 6 (2.41%) | |
| Unknown | 2 (.93%) | 0 (0.00%) | 3 (1.20%) | |
| Missing | 19 (8.84%) | 8 (16.67%) | 0 (0.00%) | |
| Site | .77 | |||
| CBIC | 21 (9.77%) | 3 (6.25%) | 27 (10.84%) | |
| RU | 81 (37.67%) | 22 (45.83%) | 101 (40.56%) | |
| SI | 113 (52.56%) | 23 (47.92%) | 121 (48.59%) | |
| Ethnicity | .94 | |||
| White/Caucasian | 127 (59.07%) | 25 (52.08%) | 167 (67.07%) | |
| Black/African‐American | 48 (22.33%) | 12 (25.00%) | 61 (24.50%) | |
| Hispanic | 13 (6.05%) | 4 (8.33%) | 15 (6.02%) | |
| Asian | 4 (1.86%) | 1 (2.08%) | 6 (2.41%) | |
| Missing | 23 (1.70%) | 6 (12.50%) | 0 (0.00%) | |
| CBCL total raw score | 2.22 (±1.09) | 19.08 (±10.91) | 57.68 (±25.81) | <.0001 |
| CGAS total score | <.0001 | |||
| Mean (SD) | 67.91 (±11.18) | 64.74 (±9.80) | 61.08 (±10.19) | |
| Missing | 116 (53.95%) | 25 (52.08%) | 124 (49.80%) | |
| Flanker uncorrected standard score | .46 | |||
| Mean (SD) | 87.39 (±13.91) | 83.94 (±17.89) | 86.04 (±15.71) | |
| Missing | 67 (31.16%) | 16 (33.33%) | 96 (38.55%) | |
| SDQ difficulties total score | <.0001 | |||
| Mean (SD) | 8.81 (±4.69) | 8.12 (±4.57) | 16.38 (±6.32) | |
| Missing | 7 (3.26%) | 0 (0%) | 6 (2.41%) | |
Note: Between group differences were assessed for the Training Set, Test Set, and Evaluation Set using ANOVA. No significant differences were found for age, sex, race, ethnicity, or site (scanner).
3.2. Brain age prediction accuracy
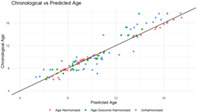
Table 2 shows the performance of the most accurate age prediction models using morphometry (sMRI), connectomes (dMRI), and combined MRI modalities, with the reported accuracy obtained from the testing dataset. Using the unharmonized brain data, a SEML‐based model, applied to the combined MRI modalities, was the most accurate overall (MAE = 1.180, MRD = 1.962). A SEML‐based model was also the most accurate among the tested models using only connectomes. However, with morphometry data alone, the Deep Learning algorithm performed best. Notably, all ML models were more accurate than the LMs using eTIV alone (MAE = 2.591 years), global FA alone (MAE = 2.858 years), or their combination (MAE = 2.582 years). Figure 2 shows a scatter plot of the predicted and chronological ages using the SEML‐based multimodal model. The ranked list of the top 5 tested models for the uni‐ and multimodal approaches is provided in Tables S1a–c.
TABLE 2.
Brain age prediction accuracy across all models
| Model | MAE (years) | MRD | Optimal algorithm |
|---|---|---|---|
| Morphometry + WM Connectomes | 1.1801 | 1.962 | SEML—Family |
| WM alone | 1.3494 | 2.634 | SEML—Family |
| Morphometry alone | 1.578 | 4.301 | Deep learning (MLP) |
| Age‐harmonized—Morphometry + WM | 0.261 | 0.128 | SEML—Family |
| Age‐harmonized—WM alone | 0.332 | 0.185 | SEML—Family |
| Age‐harmonized—Morphometry alone | 1.438 | 3.964 | SEML—Family |
| Outcome‐harmonized—Morphometry + WM | 0.880 | 1.270 | SEML—Family |
| Outcome‐harmonized—WM alone | 0.776 | 1.246 | SEML—Family |
| Outcome‐harmonized—Morphometry alone | 1.801 | 4.152 | XGBoost |
| Global FA + eTIV | 2.582 | — | — |
| Global FA alone | 2.858 | — | — |
| eTIV alone | 2.591 | — | — |
Note: SEML Models using both morphometry and white matter connectomes performed best when using the unharmonized brain data (top) and age‐harmonized brain data (middle). A SEML model using only the white matter connectomes performed best when using the outcome‐harmonized brain data (middle). All ML models outperformed linear models using FA and/or eTIV (bottom).
Abbreviations: FA, fractional anisotropy; MAE, mean absolute error; MLP, multi‐layer perceptron; MRD, mean residual deviance; SEML, stacked ensemble machine learning, WM, white matter.
FIGURE 2.
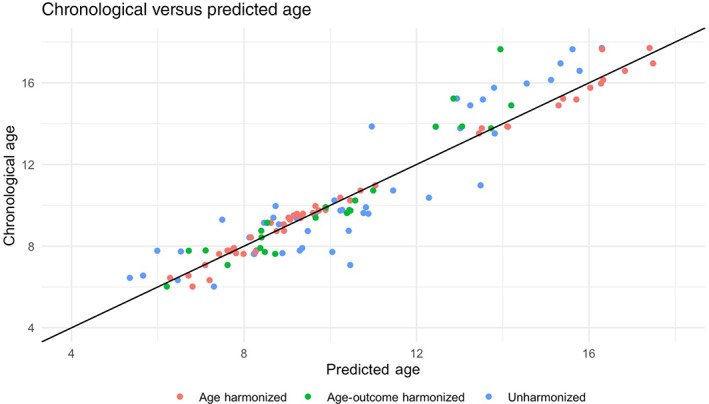
Scatterplot of predicted age versus chronological age. Scatterplot depicting the relationship between predicted and chronological age for the participants in the held‐out test sets for the best age models built using the unharmonized (N = 48), age‐harmonized (N = 48), and age‐outcome‐harmonized (N = 23) brain data
Results for the age prediction model developed using age‐harmonized and age‐outcome‐harmonized brain data (dMRI, sMRI, and combined) are given in Tables S2a–c and S3a–c, respectively. For the age‐harmonized brain data, a SEML‐based model using both MRI modalities was again the most accurate (MAE = 0.26 years). For the age‐outcome‐harmonized brain data, however, a SEML‐based model using only the dMRI data was the most accurate (MAE = 0.78). We therefore used the dMRI‐only model in the exploratory analyses with the age‐outcome‐harmonized dataset.
Regarding imaging quality, all three quality metrics—signal to noise ratio from the structural images, temporal signal to noise ratio, and relative motion from the diffusion data—were found to be associated with chronological age (p <.05 in all cases). Specifically, lower signal to noise ratios, and more motion, were associated with younger age. For the model built using unharmonized brain data, adding these terms to a model of chronological age regressed on predicted age resulted in only a 2% change in the coefficient of predicted age (Tables S4a–e).
3.3. BrainPAD, risk of psychopathology, functioning, and cognition
The relationship between brain age, as measured by the best model built from the unharmonized brain data, and the outcome measures was assessed using the evaluation data set (n = 249 after removing participants with missing race or ethnicity data). Note that, positive BrainPAD values imply a brain age that exceeds chronological age, while negative scores imply a brain age below the chronological age. Also, note that higher CBCL scores imply more symptoms, whereas lower CGAS scores imply worse functioning (and typically more symptoms). Lower BrainPAD values were significantly associated with more symptoms on the CBCL (β = −3.305, p = .003, p corr = .012) and worse functioning on the CGAS (β = 1.853, p = .003, p corr = .012), adjusting for age, sex, race, ethnicity, and scanner. Higher BrainPAD values were also significantly associated with better performance on the Flanker (β = 2.224, p = .002, p corr = .008). There was no apparent association between BrainPAD value and SDQ score (β = −.495, p = .072, p corr = .288). The strength and direction of these associations was consistent in the age‐outcome‐harmonized evaluation dataset (n = 125), though only the association with CGAS remained statistically significant after Bonferroni correction (CBCL β = −3.709, p = .155, p corr = .620; CGAS β = 2.799, p = .010, p corr = .040; Flanker β = 2.264, p = .051, p corr = .204; and SDQ β = −1.148, p = .091, p corr = .364). CBCL and SDQ scores were correlated in this sample (Pearson's r = .73, p = <.001).
Scatter plots of BrainPAD values (developed using the unharmonized brain data) and the four exploratory outcome measures, along with regression lines, are shown in Figure 3. The plotted data are adjusted for age, sex, race, ethnicity, and scanner. The plots demonstrate the inverse associations between BrainPAD and CBCL (Figure 3a, top left) and Flanker scores (Figure 3c, bottom left), and the positive association with CGAS scores (Figure 3b, top right). A relatively flat line is observed for the relationship between BrainPAD and SDQ score (Figure 3d, bottom left).
FIGURE 3.
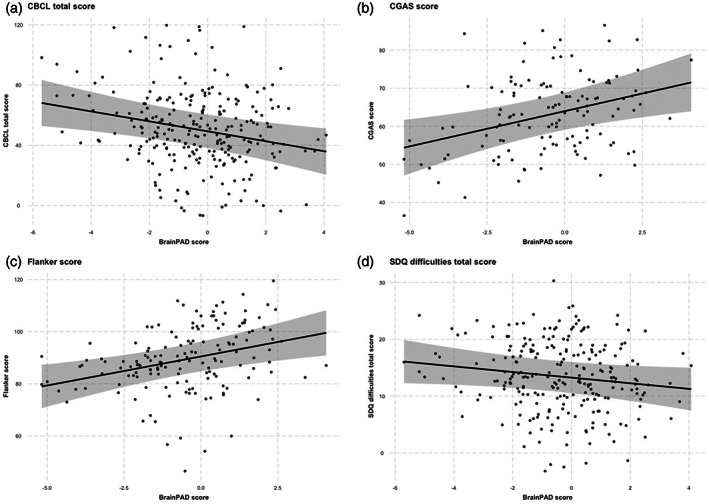
Scatterplot of BrainPAD versus outcome measures. Scatterplots depicting the relationship between BrainPAD and (a) CBCL, (b) CGAS, (c) Flanker, and (d) SDQ scores among participants in the evaluation dataset (N = 249). BrainPAD scores are adjusted for age, sex, race, ethnicity, and site (scanner)
4. DISCUSSION
In this study, we predicted brain age in youth with and without a high likelihood for psychopathology by applying an automated ML pipeline to MRI‐derived morphometry and white matter connectomes. For the unharmonized and age‐harmonized brain data, we found that morphometry and white matter connectomes together yielded the most accurate brain age prediction, compared to using either modality alone, consistent with prior studies comparing uni‐ and multimodal approaches (Brown et al., 2012; Erus et al., 2015). Also of note, the SEML method, which combines multiple ML algorithms, was the best predictor of brain age. This suggests that integrating multiple ML algorithms might be helpful for accurately modeling the complex relationship between large‐scale, multimodal brain data, and phenotypic measurements, like age.
Consistent with work on the accuracy of age prediction models using ComBat‐harmonized brain data, we found smaller MAEs for the unimodal and multimodal models built using age‐harmonized brain data compared to unharmonized brain data (Pomponio et al., 2020). Interestingly, in the smaller, outcome‐harmonized dataset, a SEML model built using the white matter connectomes alone was the most accurate. This might be because white matter connectomes had less outcome‐related variability in our sample than the morphometry measures. In any case, this result reinforces the importance of multimodal datasets, and white matter connectomes in particular, to ML‐based brain age estimation.
All ML models yielded more accurate estimates of age than the LMs that used global FA, eTIV, or their combination. The best SEML model built using the unharmonized data improved the MAE by over 1 year compared to the best LMs suggesting that ML models are doing more than recapitulating known, gross relationships. Furthermore, the relationship between chronological age and predicted age was minimally altered when adjusting for quality metrics, suggesting that neuroimaging quality does not account for our results. Moreover, this raises the possibility that the SEML model built from the unharmonized brain data is sufficiently robust to low quality data to be used in clinical settings.
When applying our brain age prediction model to a held‐out evaluation dataset, we found associations between deviations from typical brain age, as measured by BrainPAD, and a measure of symptoms (CBCL), functioning (CGAS), and neurocognition (Flanker). The overall strength and direction of these associations was largely preserved in the smaller, outcome‐harmonized evaluation dataset, though only the association with CGAS remained statistically significant after correction for multiple comparison.
Our results indicate that BrainPAD is associated with dysfunction irrespective of reporting source (parents and clinicians) and symptom domain (psychiatric symptoms and neurocognitive performance). We did not find an association between BrainPAD and SDQ scores, despite CBCL and SDQ scores being generally well correlated (Goodman & Scott, 1999; Warnick et al., 2008). One possible explanation is that the CBCL better discriminated between participants at high and low risk for psychopathology in this sample. Studies have suggested that the CBCL is a more sensitive measure relative to the SDQ (Warnick et al., 2008), which may explain the more robust relationship between BrainPAD and CBCL than with the SDQ. Taken together, BrainPAD, derived from unharmonized morphometry and white matter connectomes, may provide a useful objective neuromaturity index that correlates with behavior, functioning, and neurocognition in youth.
Overall, we found that negative BrainPAD values were associated with more symptoms and poorer functioning, and that positive BrainPAD values were associated with stronger neurocognitive function. This pattern appears consistent with the epidemiology of psychiatric disorders linked to delayed versus accelerated neurodevelopment. Attentional disorders, which are common overall and particularly in children (Froehlich et al., 2007), have been associated with delayed neurodevelopment (Gallo & Posner, 2016; Rudolph et al., 2017), and possibly contribute to elevated scores on the CBCL and reduced scores on the CGAS in this age group. On the other hand, psychotic disorders, which have been associated with accelerated neurodevelopment (Schnack et al., 2016), might also be associated with elevated CBCL and reduced CGAS scores–however, these disorders are less common overall and markedly less common in children (Courvoisie, Labellarte, & Riddle, 2001), possibly accounting for the absence of associations between positive BrainPAD and symptoms or impairment (i.e., CGAS).
Our finding of an association between BrainPAD and Flanker performance is consistent with a previous report linking accelerated neurodevelopment with improved cognitive performance (Erus et al., 2015). In contrast to studies in adults and teens, increased brain age in our youth sample is associated with better neurocognition, but not with increased risk of psychopathology or poor functioning. Nonetheless, our results do not preclude increased brain age being disadvantageous across other neurocognitive domains, or during other developmental periods (Cole et al., 2018; Gaser et al., 2013; Kaufmann et al., 2019).
Our study has several limitations. First, as is common to ML studies, the resulting model is not easily interpreted, and the use of the SEML algorithm renders the model even less interpretable. Second, our study sample was large enough to develop the model with subsequent testing in a held‐out set but did not have enough participants with specific psychiatric conditions to assess the relationship between neuromaturity and individual DSM‐based disorders. Hence, while we detected a general relationship between decreased brain age and psychiatric symptoms, and increased brain age and cognitive functioning, we are not able to shed light on the particular neuromaturity‐related etiologies of specific disorders, such as schizophrenia (Nenadic et al., 2017; Schnack et al., 2016). Third, this study relies on cross‐sectional data, and therefore does not clarify the direction of the relationship between brain changes, symptoms, and functioning. And fourth, given the amount of missing data, and the limitations of existing brain data harmonization procedures, including ComBat, we were not able to replicate our results precisely in an outcome‐harmonized dataset.
Fortunately, we see ample opportunities to address these shortcomings in future work. Regarding interpretability, methods are currently in development to specifically enhance the interpretability of the SEML algorithm, such as the use of targeted maximum loss‐based estimation (Van der Laan & Rose, 2011). As to the sample size, the HBN has continued to collect neuroimaging data. Future studies could deploy the method described in this article on the updated HBN dataset to establish the replicability of the brain age model and, with sufficient sample size, the relevance of neuromaturity‐related etiologies to certain disorders. The ongoing collection and release of data from the Adolescent Brain and Cognitive Development study (Volkow et al., 2018), for example, is an opportunity to test the brain age model and explore the association with long‐term outcomes on a larger, prospective, longitudinal dataset. Finally, methodological improvements in harmonizing brain data specifically for ML pipelines might further improve accuracy, specificity, and reproducibility.
In conclusion, our study demonstrates that using multimodal brain imaging, including white matter connectome estimates, and novel, rigorous ML methods, such as SEML, has the potential to improve the accuracy of brain age estimation. Furthermore, BrainPAD shows promise as a general measure of risk for psychopathology and cognitive impairment, with some evidence for distinct associations between particular domains and decreased versus increased brain age. Additional work is needed in larger, longitudinal datasets to replicate this approach, clarify causal mechanisms, and make more specific associations between deviations in neuromaturation and specific psychiatric conditions and neurocognitive impairments.
Supporting information
AppendixS1. Supporting Information.
ACKNOWLEDGMENTS
The authors thank the Healthy Brain Network participants and the staff at the Child Mind Institute. Dr. Alex Luna received no financial support for the research, authorship, and/or publication of this article. Dr. Joel Bernanke was supported by Grant No. T32 MH016434‐41, by the AACAP Pilot Research Award for Attention Disorders, and by the Bender‐Fishbein Award in Child Psychiatry Research. Dr. Jiook Cha was supported by Grant No. K01 MH 109836, by the Brain Pool Program, National Research Foundation of Korea Grant No. 2021R1C1C1006503, 2021M3E5D2A01022515, and the BK21 FOUR Program (5199990314123) funded by the Korea government (MSIT). Dr. Jonathan Posner was supported by the Edwin S Webster Foundation, Suzanne Crosby Murphy Endowment under award number R01 MH036197 and UH3 OD023328.
Luna, A., Bernanke, J., Kim, K., Aw, N., Dworkin, J. D., Cha, J., & Posner, J. (2021). Maturity of gray matter structures and white matter connectomes, and their relationship with psychiatric symptoms in youth. Human Brain Mapping, 42(14), 4568–4579. 10.1002/hbm.25565
Alex Luna and Joel Bernanke shared first authorship.
Funding information Edwin S Webster Foundation, Grant/Award Numbers: UH3 OD023328, R01 MH036197; National Research Foundation of Korea (NRF), Grant/Award Number: 5199990314123; BK21 Four program, Brain Pool Program, Grant/Award Numbers: 2019H1D3A2A01102270, K01 MH 109836; AACAP Pilot Research Award for Attention Disorders, Grant/Award Number: T32 MH016434‐41
DATA AVAILABILITY STATEMENT
Data were from the Healthy Brain Network (HBN), a large, publicly available dataset from a community‐based cohort of children and adolescents.
REFERENCES
- Acion, L., Kelmansky, D., van der Laan, M., Sahker, E., Jones, D., & Arndt, S. (2017). Use of a machine learning framework to predict substance use disorder treatment success. PLoS One, 12(4), e0175383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, L. M., Escalera, J., Ai, L., Andreotti, C., Febre, K., Mangone, A., … Milham, M. P. (2017). An open resource for transdiagnostic research in pediatric mental health and learning disorders. Scientific Data, 4(1), 170181. 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J. L., & Sotiropoulos, S. N. (2016). An integrated approach to correction for off‐resonance effects and subject movement in diffusion MR imaging. NeuroImage, 125, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, R., Jollans, L., Rueda‐Delgado, L. M., Rizzo, R., Yener, G. G., McMorrow, J. P., … Whelan, R. (2021). Brain‐predicted age difference score is related to specific cognitive functions: a multi‐site replication analysis. Brain imaging and behavior, 15(1), 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T. T., Kuperman, J. M., Chung, Y., Erhart, M., McCabe, C., Hagler, D. J., Jr., … Dale, A. M. (2012). Neuroanatomical assessment of biological maturity. Current Biology, 22(18), 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, J., Fekete, T., Siciliano, F., Biezonski, D., Greenhill, L., Pliszka, S. R., … Posner, J. (2015). Neural correlates of aggression in medication‐naive children with ADHD: Multivariate analysis of morphometry and tractography. Neuropsychopharmacology, 40(7), 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, J., Ide, J. S., Bowman, F. D., Simpson, H. B., Posner, J., & Steinglass, J. E. (2016). Abnormal reward circuitry in anorexia nervosa: A longitudinal, multimodal MRI study. Human Brain Mapping, 37(11), 3835–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A.A., Beer, J.C., Tustison, N.J., Cook, P.A., Shinohara, R.T., & Shou, H. 2020. Removal of scanner effects in covariance improves multivariate pattern analysis in neuroimaging data. bioRxiv. 858415.
- Chung, Y., Addington, J., Bearden, C. E., Cadenhead, K., Cornblatt, B., Mathalon, D. H., … for the North American Prodrome Longitudinal Study (NAPLS) Consortium and the Pediatric Imaging, Neurocognition, and Genetics (PING) Study Consortium . (2018). Use of machine learning to determine deviance in neuroanatomical maturity associated with future psychosis in youths at clinically high risk. JAMA Psychiatry, 75(9), 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, J. H., Marioni, R. E., Harris, S. E., & Deary, I. J. (2019). Brain age and other bodily 'ages': Implications for neuropsychiatry. Molecular Psychiatry, 24(2), 266–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, J. H., Poudel, R. P. K., Tsagkrasoulis, D., Caan, M. W. A., Steves, C., Spector, T. D., & Montana, G. (2017). Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage, 163, 115–124. [DOI] [PubMed] [Google Scholar]
- Cole, J. H., Ritchie, S. J., Bastin, M. E., Valdés Hernández, M. C., Muñoz Maniega, S., Royle, N., … Deary, I. J. (2018). Brain age predicts mortality. Molecular Psychiatry, 23(5), 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvoisie, H., Labellarte, M. J., & Riddle, M. A. (2001). Psychosis in children: Diagnosis and treatment. Dialogues in Clinical Neuroscience, 3(2), 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, A. M., Fischl, B., & Sereno, M. I. (1999). Cortical surface‐based analysis. I. Segmentation and surface reconstruction. NeuroImage, 9(2), 179–194. [DOI] [PubMed] [Google Scholar]
- Dang, H. M., Nguyen, H., & Weiss, B. (2017). Incremental validity of the Child Behavior Checklist (CBCL) and the strengths and difficulties questionnaire (SDQ) in Vietnam. Asian Journal of Psychiatry, 29, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R. S., Ségonne, F., Fischl, B., Quinn, B. T., Dickerson, B. C., Blacker, D., …Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31(3), 968–980. 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Erus, G., Battapady, H., Satterthwaite, T. D., Hakonarson, H., Gur, R. E., Davatzikos, C., & Gur, R. C. (2015). Imaging patterns of brain development and their relationship to cognition. Cerebral Cortex, 25(6), 1676–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban, O., Birman, D., Schaer, M., Koyejo, O. O., Poldrack, R. A., & Gorgolewski, K. J. (2017). MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS One, 12(9), e0184661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B., & Dale, A. M. (2000). Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America, 97(20), 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B., Salat, D. H., Busa, E., Albert, M., Dieterich, M., Haselgrove, C., … Dale, A. M. (2002). Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron, 33(3), 341–355. [DOI] [PubMed] [Google Scholar]
- Fortin, J‐P. (2019). ComBat Harmonization. Available from https://github.com/Jfortin1/ComBatHarmonization.
- Fortin, J.‐P., Cullen, N., Sheline, Y. I., Taylor, W. D., Aselcioglu, I., Cook, P. A., … Shinohara, R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin, J.‐P., Parker, D., Tunç, B., Watanabe, T., Elliott, M. A., Ruparel, K., … Shinohara, R. T. (2017). Harmonization of multi‐site diffusion tensor imaging data. NeuroImage, 161, 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes, L., & Blakemore, S. J. (2018). Studying individual differences in human adolescent brain development. Nature Neuroscience, 21(3), 315–323. [DOI] [PubMed] [Google Scholar]
- Franke, K., & Gaser, C. (2019). Ten years of BrainAGE as a neuroimaging biomarker of brain aging: What insights have we gained? Frontiers in Neurology, 10, 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, K., Luders, E., May, A., Wilke, M., & Gaser, C. (2012). Brain maturation: Predicting individual BrainAGE in children and adolescents using structural MRI. NeuroImage, 63(3), 1305–1312. [DOI] [PubMed] [Google Scholar]
- Froehlich, T. E., Lanphear, B. P., Epstein, J. N., Barbaresi, W. J., Katusic, S. K., & Kahn, R. S. (2007). Prevalence, recognition, and treatment of attention‐deficit/hyperactivity disorder in a national sample of US children. Archives of Pediatrics & Adolescent Medicine, 161(9), 857–864. [DOI] [PubMed] [Google Scholar]
- Gallo, E. F., & Posner, J. (2016). Moving towards causality in attention‐deficit hyperactivity disorder: Overview of neural and genetic mechanisms. Lancet Psychiatry, 3(6), 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaser, C., Franke, K., Kloppel, S., Koutsouleris, N., Sauer, H., & Alzheimer's Disease Neuroimaging Initiative . (2013). BrainAGE in mild cognitive impaired patients: Predicting the conversion to Alzheimer's disease. PLoS One, 8(6), e67346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault, J. B., Munsell, B. C., Puechmaille, D., Goldman, B. D., Prieto, J. C., Styner, M., & Gilmore, J. H. (2019). White matter connectomes at birth accurately predict cognitive abilities at age 2. NeuroImage, 192, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, R., & Scott, S. (1999). Comparing the strengths and difficulties questionnaire and the child behavior checklist: Is small beautiful? Journal of Abnormal Child Psychology, 27(1), 17–24. [DOI] [PubMed] [Google Scholar]
- Han, L. K. M., Dinga, R., Hahn, T., Ching, C. R. K., Eyler, L. T., Aftanas, L., …Schmaal, L. (2020). Brain aging in major depressive disorder: results from the ENIGMA major depressive disorder working group. Molecular Psychiatry. 10.1038/s41380-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgestøl, E. A., Kaufmann, T., Nygaard, G. O., Beyer, M. K., Sowa, P., Nordvik, J. E., … Westlye, L. T. (2019). Cross‐sectional and longitudinal MRI brain scans reveal accelerated brain aging in multiple sclerosis. Frontiers in Neurology, 10(450). 10.3389/fneur.2019.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, T., Alnæs, D., Doan, N. T., Brandt, C. L., Andreassen, O. A., & Westlye, L. T. (2017). Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nature Neuroscience, 20(4), 513–515. [DOI] [PubMed] [Google Scholar]
- Kaufmann, T., van der Meer, D., Doan, N. T., Schwarz, E., Lund, M. J., Agartz, I., …Westlye, L. T. (2019). Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nature Neuroscience, 22(10), 1617–1623. 10.1038/s41593-019-0471-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenadic, I., Dietzek, M., Langbein, K., Sauer, H., & Gaser, C. (2017). BrainAGE score indicates accelerated brain aging in schizophrenia, but not bipolar disorder. Psychiatry Research: Neuroimaging, 266, 86–89. [DOI] [PubMed] [Google Scholar]
- Pomponio, R., Erus, G., Habes, M., Doshi, J., Srinivasan, D., Mamourian, E., … Davatzikos, C. (2020). Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage, 208, 116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, M. D., Miranda‐Domínguez, O., Cohen, A. O., Breiner, K., Steinberg, L., Bonnie, R. J., …Fair, D. A. (2017). At risk of being risky: The relationship between “brain age” under emotional states and risk preference. Developmental Cognitive Neuroscience, 24, 93–106. 10.1016/j.dcn.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat, D. H., Tuch, D. S., Greve, D. N., van der Kouwe, A. J. W., Hevelone, N. D., Zaleta, A. K., … Dale, A. M. (2005). Age‐related alterations in white matter microstructure measured by diffusion tensor imaging. Neurobiology of Aging, 26(8), 1215–1227. [DOI] [PubMed] [Google Scholar]
- Schmithorst, V. J., Wilke, M., Dardzinski, B. J., & Holland, S. K. (2002). Correlation of white matter diffusivity and anisotropy with age during childhood and adolescence: A cross‐sectional diffusion‐tensor MR imaging study. Radiology, 222(1), 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack, H. G., van Haren, N. E., Nieuwenhuis, M., Hulshoff Pol, H. E., Cahn, W., & Kahn, R. S. (2016). Accelerated brain aging in schizophrenia: A longitudinal pattern recognition study. The American Journal of Psychiatry, 173(6), 607–616. [DOI] [PubMed] [Google Scholar]
- Shahab, S., Mulsant, B. H., Levesque, M. L., Calarco, N., Nazeri, A., Wheeler, A. L., … Voineskos, A. N. (2019). Brain structure, cognition, and brain age in schizophrenia, bipolar disorder, and healthy controls. Neuropsychopharmacology, 44(5), 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao, H., Hayashi, N., & Ohtomo, K. (2012). A longitudinal study of brain volume changes in normal aging. European Journal of Radiology, 81(10), 2801–2804. [DOI] [PubMed] [Google Scholar]
- Tamnes, C. K., Herting, M. M., Goddings, A. L., Meuwese, R., Blakemore, S. J., Dahl, R. E., … Mills, K. L. (2017). Development of the cerebral cortex across adolescence: A multisample study of inter‐related longitudinal changes in cortical volume, surface area, and thickness. The Journal of Neuroscience, 37(12), 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC . (2012). A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Available from https://www.R-project.org [Google Scholar]
- The H2O.ai Team . (2015). H2O: R Interface for H2O. R package version 3.1.0.99999. Available from http://www.h2o.ai.
- Tournier, J. D., Calamante, F., & Connelly, A. (2010). Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. Paper Presented at: Proceedings of the International Society for Magnetic Resonance in Medicine.
- Tustison, N. J., Avants, B. B., Cook, P. A., Yuanjie, Z., Egan, A., Yushkevich, P. A., & Gee, J. C. (2010). N4ITK: Improved N3 bias correction. IEEE Transactions on Medical Imaging, 29(6), 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva, O., Gano, D., Trevino, R. J., Glass, H. C., Flynn, T., Lundy, S. M., …Xu, D. (2018). Aberrant structural brain connectivity in adolescents with attentional problems who were born prematurely. American Journal of Neuroradiology, 39(11), 2140–2147. 10.3174/ajnr.a5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Laan, M. J., & Rose, S. (2011). Targeted learning: Causal inference for observational and experimental data. Springer Series in Statistics, New York, NY: Springer‐Verlag. https://www.springer.com/gp/book/9781441997814. [Google Scholar]
- Veraart, J., Novikov, D. S., Christiaens, D., Ades‐Aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. NeuroImage, 142, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow, N. D., Koob, G. F., Croyle, R. T., Bianchi, D. W., Gordon, J. A., Koroshetz, W. J., … Weiss, S. R. B. (2018). The conception of the ABCD study: From substance use to a broad NIH collaboration. Developmental Cognitive Neuroscience, 32, 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, E. F., Sabuwalla, Z., & Huot, R. (2004). Pubertal neuromaturation, stress sensitivity, and psychopathology. Development and Psychopathology, 16(4), 807–824. [DOI] [PubMed] [Google Scholar]
- Warnick, E. M., Bracken, M. B., & Kasl, S. (2008). Screening efficiency of the child behavior checklist and strengths and difficulties questionnaire: A systematic review. Child and Adolescent Mental Health., 13(3), 140–147. [DOI] [PubMed] [Google Scholar]
- Wolpert, D. H. (1992). Stacked generalization. Neural Networks, 5(2), 241–259. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AppendixS1. Supporting Information.
Data Availability Statement
Data were from the Healthy Brain Network (HBN), a large, publicly available dataset from a community‐based cohort of children and adolescents.


