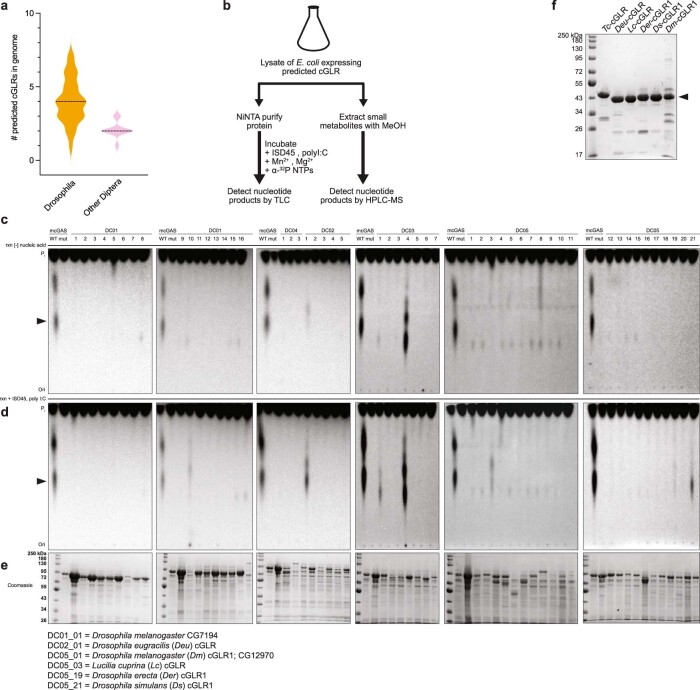
Extended Data Fig. 2. Forward biochemical screen of predicted cGLRs in Diptera.
a, Violin plot showing the number of predicted cGLRs in Diptera genomes. Drosophila genomes (n = 31 species) have a median of four predicted cGLRs in contrast to a median of two predicted cGLRs in other dipteran insects (n = 11 species). b, Schematic of the in vitro screen of predicted cGLRs in the order Diptera. Fifty-three sequences were selected representing each clade in the phylogeny in Fig. 2a. Following recombinant protein expression in E. coli, lysates were split into two samples for parallel TLC analysis of in vitro enzymatic activity and HPLC-MS analysis of lysate nucleotide metabolites.c, d, Purified cGLR proteins were incubated overnight at 37 °C with α32P-radiolabelled nucleotides, a mixture of Mn2+ and Mg2+, and the 45-bp immunostimulatory DNA ISD45 or the synthetic dsRNA analogue poly I:C as potential nucleic acid ligands, and reactions were visualized by PEI-cellulose TLC. Wild-type (WT) and catalytically inactive mouse cGAS enzymes were used as controls for each sample set. Note that mouse cGAS exhibits dsDNA-independent activity in the presence of Mn2+ (ref. 51). Predicted Diptera cGLRs are grouped by clade (DC01–05) and numbered within each clade. Ligand-dependent activity was identified for DC02_01, DC05_03, DC05_19 and DC05_21; species listed below. We observed ligand-independent activity for two enzymes in clade 3. Data represent n = 2 independent experiments. e, SDS–PAGE and Coomassie stain analysis of NiNTA-purified cGLR protein fractions used for the biochemical screen. f, SDS–PAGE and Coomassie stain analysis of final cGLR proteins used for biochemical studies, which were purified by NiNTA-affinity, ion-exchange chromatography and size-exclusion chromatography.