Abstract
Introduction:
Cancer neoantigens represent important targets of cancer immunotherapy. The goal of cancer neoantigen vaccines is to induce neoantigen-specific immune responses and antitumor immunity, while minimizing the potential for autoimmune toxicity. Advances in sequencing technologies, neoantigen prediction algorithms and other technologies have dramatically improved the ability to identify and prioritize cancer neoantigens. Unfortunately, results from preclinical studies and early phase clinical trials highlight important challenges to the successful clinical translation of neoantigen cancer vaccines.
Areas covered
In this review, we provide an overview of current strategies for the identification and prioritization of cancer neoantigens with a particular emphasis on the two most common strategies used for neoantigen identification: (1) direct identification of peptide ligands eluted from peptide-MHC complexes, and (2) next-generation sequencing combined with neoantigen prediction algorithms. We highlight the limitations of current neoantigen prediction pipelines, and discuss broader challenges associated with cancer neoantigen vaccines including tumor purity/heterogeneity and the immunosuppressive tumor microenvironment.
Expert Opinion
Despite current limitations, neoantigen prediction is likely to improve rapidly based on advances in sequencing, machine-learning, and information sharing. The successful development of robust cancer neoantigen prediction strategies is likely to have significant impact, with the potential to facilitate cancer neoantigen vaccine design.
Keywords: Cancer neoantigen, Cancer immunotherapy, Neoantigen vaccine, Sequencing, Epitope prediction, MHC class I, Binding affinity, Immune checkpoint inhibition
1. Introduction
Genetic alterations are common in cancer, including single nucleotide variants, frameshift insertions and deletions, aberrant splicing, and complex structural alterations. These genetic alterations often result in mutated proteins with novel amino acid sequences. Mutated proteins that are recognized by the immune system are known as cancer neoantigens. Investigators first established the ability of cancer neoantigens to induce immune responses in preclinical models and human cancer patients over two decades ago [1–3].
The development of next generation sequencing technologies has revolutionized the ability to identify and study cancer neoantigens. There is now emerging evidence to suggest that cancer neoantigens are important targets of both endogenous antitumor immune responses and cancer immunotherapies (reviewed in [4]). For example, increased tumor mutation burden (widely considered a surrogate for neoantigen load) is associated with greater numbers of tumor infiltrating lymphocytes and improved survival [5, 6]. Tumor mutation burden is also associated with response to immune checkpoint inhibition [7, 8]. Of particular note, there is now evidence from preclinical studies [9, 10], and early phase clinical trials [11–13] to suggest that targeting neoantigens with neoantigen vaccines and/or adoptive cell therapies can successfully induce antitumor immune responses and potentially improved clinical outcomes. Thus, successful development and validation of strategies and/or technologies for the accurate identification of cancer neoantigens is likely to have a significant impact.
Although neoantigen identification strategies are based on state-of-the-art technologies, only a small proportion of neoantigens selected by next-generation sequencing and in silico neoantigen prediction algorithms induce an immune response in preclinical models and in human translational studies [14, 15]. It is likely that limitations associated with current neoantigen identification strategies contribute to the low response rate. In this review, we describe the strategies that are commonly used to identify and prioritize cancer neoantigens, with a particular focus on the limitations that preclude accurate and robust selection. We also discuss the broader challenges that may limit the success of cancer neoantigen vaccines. To identify important contributions, we performed a systematic literature review using the PubMed, Embase, Web of Science, and Cochrane library databases. We identified publications focused on cancer neoantigens and neoantigen vaccines. The focus of this review on the limitations associated with cancer neoantigen identification and clinical translation is unique, and provides an important perspective on this nascent field of investigation.
1.1. Strategies for neoantigen identification
1.1.1. Direct identification of cancer neoantigens
Currently, two strategies are commonly used for neoantigen identification: (1) direct identification of peptide ligands eluted from peptide-MHC complexes, and (2) next-generation sequencing combined with neoantigen prediction algorithms. Cancer neoantigens are presented by the immune system in association with MHC class I and II molecules. The direct identification strategy for cancer neoantigen identification involves mass spectrometry analysis of MHC ligandomes eluted from peptide-MHC complexes [9, 16–18] (Figure 1). This strategy is typically performed in parallel with exome sequencing of tumor and normal tissues. Genetic alterations identified by exome sequencing are used to generate a patient-specific tumor mutation database. Sequenced peptides from LC-MS/MS are compared to the tumor mutation database to identify cancer neoantigens present in the ligandome [19]. As the direct identification strategy identifies neoantigens that are being presented by MHC molecules, variations in antigen presentation presented by MHC molecules, variations in antigen presentation pathways, such as proteolytic cleavage in the proteasome/immunoproteasome are captured through this approach. In addition, cancer neoantigens derived from post-translational modifications can be recognized [20, 21].
Figure 1:
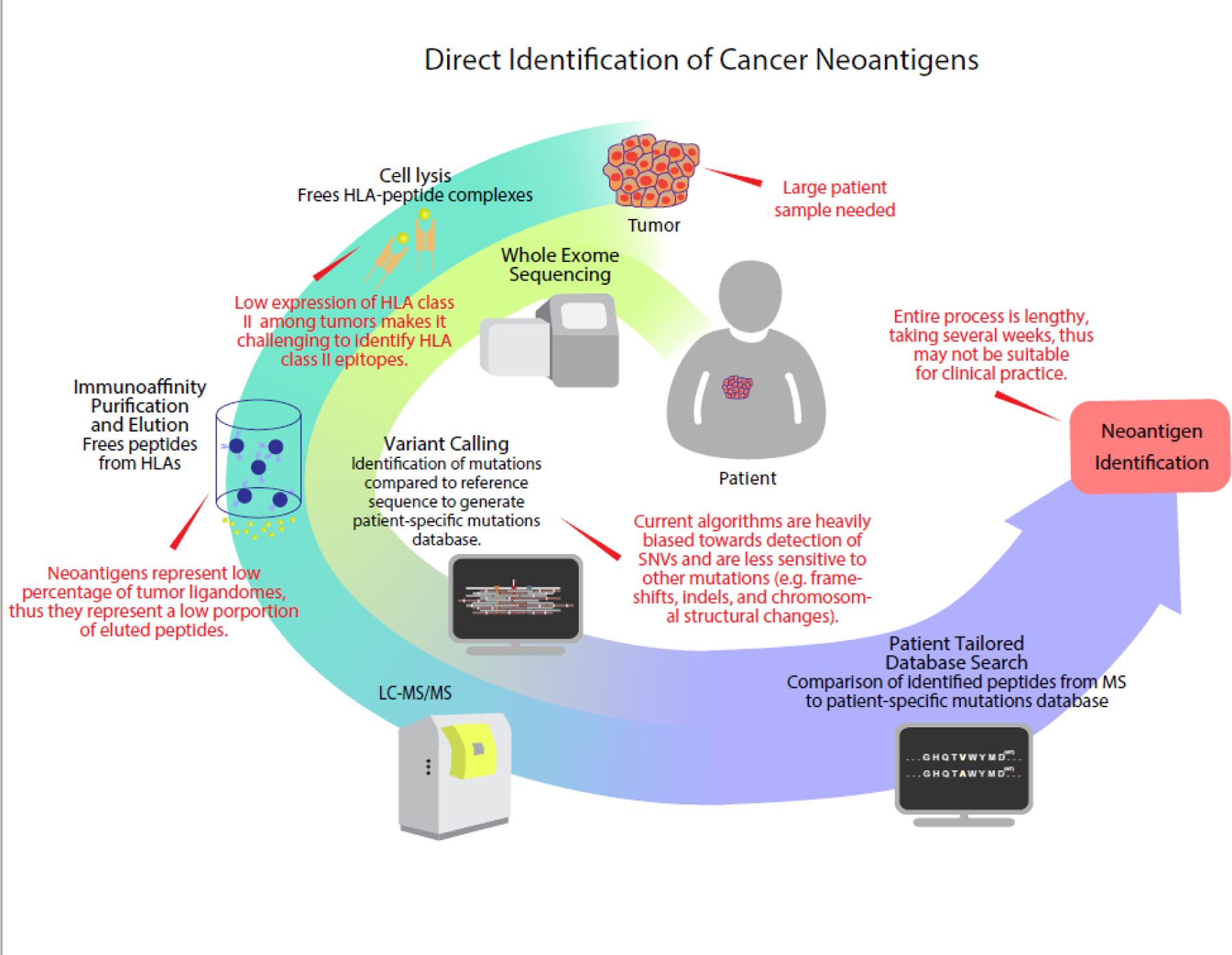
Direct identification of cancer neoantigens. HLA epitopes are eluted from tumor cells in parallel to whole exome sequencing of the tumor. Variant calling of WES data is performed against reference sequence to generate a patient specific mutational database. Eluted ligands, sequenced by MS, are compared to this database to identify neoantigen candidates.
The development of modern high-throughput mass spectrometry techniques has greatly enhanced the depth to which the ligandome can be surveyed. Bassani-Sternberg et al. published a manuscript highlighting the impact of high throughput mass spectrometry techniques on neoantigen identification [19]. The authors assembled the ligandomes from human melanomas, analyzing to a depth of 95,500 ligands. Eleven ligands were derived from neoantigens, and 4 were proven to be immunogenic in T-cell validation assays [19]. In this proof-of-concept study, the authors clearly demonstrated that it is possible to use high-throughput mass spectrometry techniques to identify cancer neoantigens.
However, this approach does have limitations. The process of identifying eluted ligands is resource and labor intensive, and the entire process can take weeks to months, limiting clinical translation [22]. The accuracy of the direct identification strategy is also highly dependent on the depth of analysis. Only a few peptide/MHC complexes are required to induce an immune response. As a result, the direct identification strategy has been limited, until recently, to use in cell lines and other preclinical models [23]. For instance, in the study by Bassani-Sternberg et al., the amount of tissue and analytical depth required to identify fewer than a dozen neoantigens in tumors known to have high mutation burden highlights how labor-intensive this process is, and the challenge of using this strategy in clinical translation [19]. While improvements have been made in starting tumor material, current immunoprecipitation protocols still require 5 × 107 to 1 × 109 cells, thus precluding the use of most needle aspirations and biopsies [24]. This is an area of active investigation, suggesting that improved technologies may enhance the translational potential.
1.1.2. Sequencing-based identification of cancer neoantigens
Given the limitations of the direct identification strategy, most studies targeting cancer neoantigens have relied on next-generation sequencing and in silico prediction algorithms (Figure 2). Next-generation sequencing technologies have greatly reduced the cost and time required for whole-exome and RNA-sequencing. Comparison of tumor/normal whole exome sequences and tumor RNA sequences is used to identify genetic alterations present only in cancer cells. Neoantigen identification and prioritization is then conducted in silico, generally using algorithms derived based on vast databases of MHC ligandomes assembled by in vitro binding assays and/or high-throughput mass spectrometry [25]. Early epitope prediction algorithms, such as NetMHC and Pickpocket, are trained on in vitro peptide binding affinity for specific MHC alleles, which has been shown to be correlated with immunogenicity [9, 26, 27]. NetMHC uses an artificial neural network while Pickpocket uses position-specific weight matrices [28, 29]. Segal et al. were the first to demonstrate such an approach can be used to identify cancer neoantigens derived from somatic mutations found in breast and colorectal cancer; post hoc validation revealed individual breast and colorectal cancers contain 10 and 7 HLA-A*0201-restricted cancer neoantigens, respectively [30].
Figure 2:
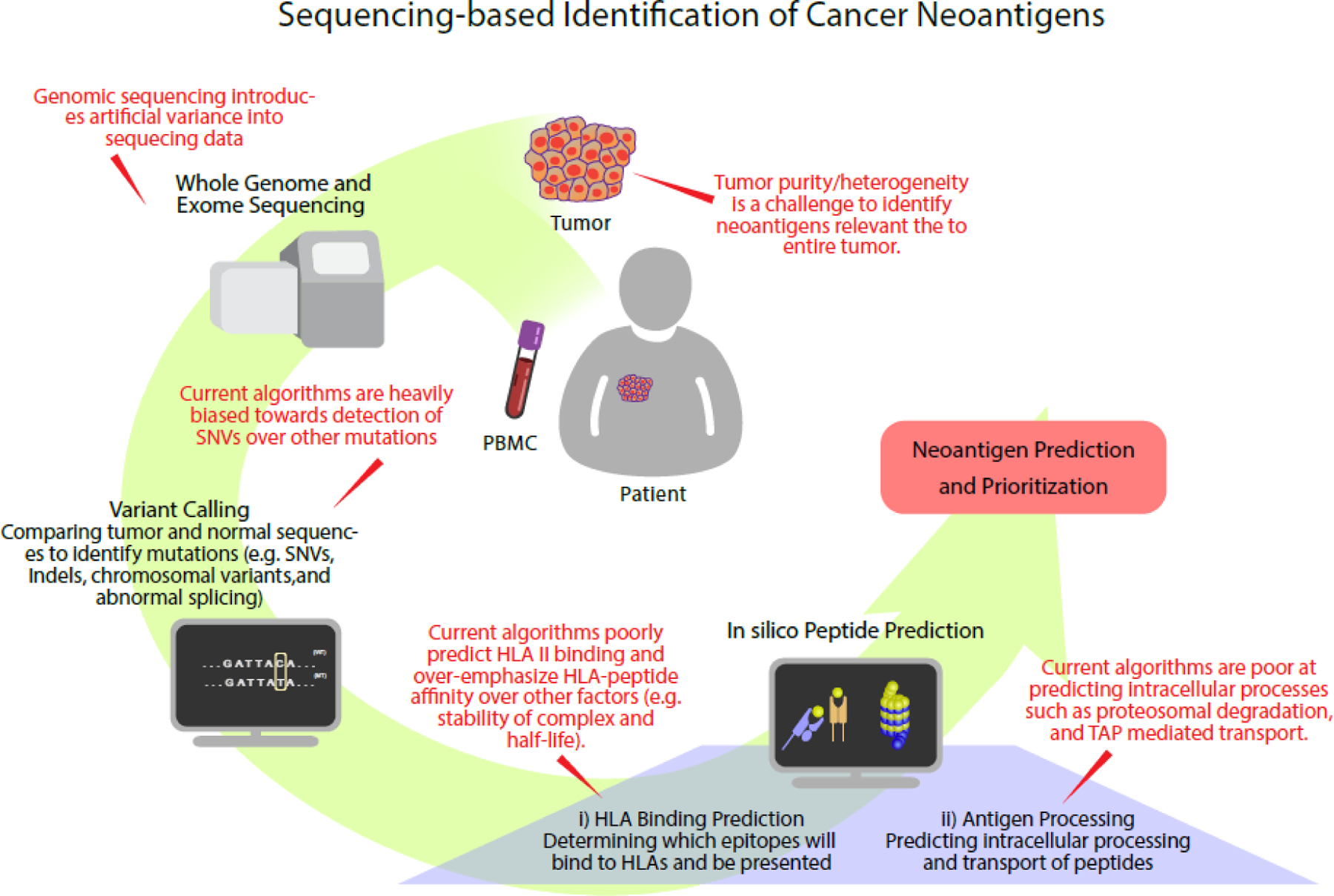
Sequencing-based identification of cancer neoantigens. Tumor and patient genomes are sequenced and variant calling is performed between the two sequences. Identified mutations are processed by neoantigen prediction algorithms based on known rules of HLA binding and antigen processing to predict neoantigen candidates.
More recently, epitopes identified by MS have greatly outnumbered ones identified in vitro [31]. Dozens of pipelines (e.g. pVAC-Seq, EDGE, and MHCflurry) have since been trained on MS data [32–36]. In addition to having a vastly larger datasets for training, algorithms trained with MS data avoid the circular logic of early in vitro data—only predicted epitopes were tested, thus potentially missing large portions of potential candidates [31]. While early algorithms relied predominantly on linear regression methods such as position weight matrices, neural networks have become the dominant algorithm for prediction (e.g. NetMHCpan, MHCflurry, EDGE) [37–39]. Although neural network based algorithms require more upfront data for learning, they are able to capture the non-linear relationship between peptide sequence and binding affinity that a linear regression model cannot. Benchmarking studies were performed by Zhao et al, whom found prediction accuracy to be improved with incorporation of MS datasets and use of neural networks versus regression-based algorithms [40]. However with machine learning approaches, there is always a concern of model overfitting.
Limitations of the sequencing and in silico prediction approach include biases introduced by sequencing technologies, imperfect algorithms used for variant calling and binding affinity predictions, and the imperfect association between predicted binding and immunogenicity. Accurate identification of immunogenic MHC class II neoantigens is particularly challenging given our relatively limited understanding of class II antigen processing and presentation, and the fact that MHC class II binding is more promiscuous.
1.1.2.1. Tandem minigene approach to test immunogenicity
Tandem minigenes (TMGs) have been used to as a strategy to screen for immunogenic mutations. In this approach, mutations are identified using whole-exome sequencing. Rather than predicting immunogenicity via binding prediction algorithms, minigene constructs are created from mutations flanked by 12 amino acids of normal protein sequence, with each minigene containing 6–24 mutated gene products [41, 42]. These TMGs are transfected into APCs to allow for translation, processing, and MHC presentation, and subsequently co-cultured with various TIL cultures to identify immunogenic neoantigens [43]. This approach has been used to identify neoantigen targets for adoptive T-cell therapy in melanoma and epithelial cancers and have led to durable regressions [42, 44].
1.2. Challenges of neoantigen identification and prioritization
We have identified two strategies for neoantigen identification. In practice, the direct identification strategy is less commonly used based on tissue requirements and other considerations. However, the direct identification strategy has informed sequencing-based strategies for neoantigen identification. In this section, we discuss limitations of sequencing-based strategies for neoantigen identification, but also highlight progress that is being made with direct identification of cancer neoantigens and how this progress is informing sequencing-based strategies for neoantigen identification.
1.2.1. Limitations identifying genetic alterations in cancers
Next-generation sequencing technologies have inherent error rates that can impact neoantigen identification. Artifacts introduced by differences between sample preparation, sequencing alignments, sequencing read length and depth, and library construction may preclude accurate identification of genetic variants, and explain some of the discrepancies detected between different pipelines [45]. Despite the great strides made with next-generation sequencing, challenges remain to neoantigen prediction. One limitation is the relatively short reading frame, requiring genetic material to be read in fragments and later reassembled to acquire the original sequence. In sequences where there are numerous repeats, this lends itself to misassembly of read fragments. Furthermore, while the short reads can accurately capture SNVs and short indels, larger mutations and structural variants are more difficult to detect [46]. Sequencing of FFPE samples, which represent a significant number of specimens due to their use in clinical pathology, is further impeded by damage caused by chemical processing [47, 48]. Low tumor purity and/or tumor heterogeneity in some clinical specimens can influence sequencing readouts [49]. Sequencing errors can introduce false-positive variants. Targeting cancer neoantigens identified based on sequencing errors may induce immune responses, but will not improve antitumor immunity.
Imperfect variant-calling may also influence neoantigen selection [50, 51]. SNVs are readily identified by current variant calling algorithms, and are overrepresented in the predicted neoantigen repertoire. Genetic variants resulting from insertion and deletion (indel) and gene fusion mutations are only now being routinely identified. Additionally, cancer-specific post-translational modifications, which can generate potentially immunogenic amino acid variations, are not evaluated [20, 21]. Therefore, by focusing primarily on SNVs, current sequencing-based algorithms do not adequately evaluate several other classes of cancer neoantigens, potentially reducing the ability to identify immunogenic neoantigens. For instance, in a pan-cancer analysis, Turajlic et al. predicted indel mutations contributed greatly to the overall immunogenic phenotype of certain malignancies. Compared to SNVs, indel neoantigens were predicted to generate epitopes that had higher MHC class I binding affinity and mutant binding specificity [52]. The authors note indel mutations have not been widely incorporated into neoantigen prediction pipelines as current variant calling algorithms do not identify indel variants as accurately and reliably as SNVs [53, 54]. Indel variant calling is still at an early stage and additional advances need to be made to provide the high fidelity required for clinical application.
1.2.2. Limitations in MHC class I neoantigen prediction algorithms
Another limitation in neoantigen identification is the in silico algorithms used to predict candidate neoantigen binding to MHC class I alleles. Most in silico algorithms are based on machine-learning and their performance is highly dependent on the databases on which they are trained. Attempts at designing ab initio algorithms in which predictions are based on structural analysis rather than homology to prior data sets have been disappointing [55–58].
Recently, MHC ligandome compilation has been enhanced by data generated by high-throughput mass spectrometry. Thus, while high-throughput mass spectrometry may have limited utility in direct identification of neoantigens clinically, it has been essential to enhancing our understanding of epitope binding [59]. This technology allows for greater depth of analysis capturing information on thousands of MHC ligands. This provides a richer database for improving and/or developing tools to predict epitope binding. In an analysis by Bassani-Sternberg et al, an unbiased mass spectrometry approach was used to generate ligandomes from cancer cell lines and deconvolute these ligands to their respective MHC class I alleles. A prediction algorithm trained on this database had a higher predictive value than standard approaches, which mostly relied on in vitro binding assays, with improvements most evident for MHC class I alleles that have few known ligands in current databases [60].
High-throughput mass spectrometry has the power to analyze ligands at great depths, thus revealing peptide-specific determinants of MHC-binding that were otherwise missed by standard techniques. Addition of these motifs to training databases has enriched our understanding of antigen processing and presentation. Peptide abundance, peptide length, and the influence of complex allosteric interactions on binding motifs have all been demonstrated to be important determinants of peptide-MHC binding that were inferred from mass spectrometry-generated data [59, 61–63]. MS peptidomics have also identified protein hotspots, or regions within a protein prone to proteasomal cleavage and ligand production, thus adding a new dimension to our understanding of antigen processing [64, 65]. Similarly, information regarding ligands formed from peptide splicing can help us predict de novo ligands that would otherwise be missed by simple analysis of sequencing data [66].
Our understanding of antigen processing and presentation was further enhanced by a study conducted by Abelin et al., which used mass spectrometry to profile MHC ligandomes in mono-allelic cells expressing single HLA-I alleles. This model enables more accurate class I epitope prediction without need for deconvolution [67]. The authors were able to identify 24,000 HLA class I peptides through this pipeline, providing insights into how protein cleavage and levels of gene expression influence antigen presentation. Machine-learning algorithms trained on this database outperformed the standard. Similarly, Sarkizova et al used mono-allelic cells to profile >185,000 peptides and develop HLAthena, which predicted endogenous HLA class I- associated ligands with a 1.5-fold improved accuracy compared to existing tools, and correctly identified >75% of observed class I presented peptides for 11 patient-derived tumor cell lines [68]. Of note, tumor cells often have aberrant antigen processing and presentation pathways. These pathways are not well understood. High-throughput mass-spectrometry analyses of normal tissue may not accurately predict the factors influencing antigen presentation in tumor cells, which may among themselves have differences in antigen presentation given tumor heterogeneity. Focused peptidomic analyses of malignant tissue are necessary to design robust neoantigen prediction pipelines that can be used to design neoantigen-directed therapies.
Bioinformatic tools, such as NNAlign_MA, have also been developed to deconvolute ligandomes from MS datasets. The typical process for deconvolution requires 1) clustering of peptides, and 2) annotation of a cluster to a specific MHC molecule [69]. Earlier deconvolution tools, such as GibbsCluster and MixMHCp requires prior knowledge of MHC binding motifs [70, 71]. NNAlign_MA combines these two steps with a machine learning to generate a prediction algorithm covering all MHCs in the dataset [69]. By performing the three tasks simultaneously, NNAlign_MA is able to iteratively update peptide clustering, MHC annotation, and prediction algorithms resulting in improved accuracy. This motif deconvolution technique serves as the basis of NetMHCIIpan [72].
Most neoantigen prediction algorithms use predicted binding affinity to MHC class I as a surrogate for immunogenicity, and several studies have indeed identified MHC-peptide binding affinity to be highly correlated with immunogenicity [9, 26]. However, antigen immunogenicity is complicated, and many biologic processes are likely to contribute beyond MHC binding. Delivery of antigen to antigen presenting cells, antigen cleavage and processing by immunoproteasomes, and recognition of peptide-MHC complexes by circulating cognate T cells are just some of the other factors that influence immunogenicity. Furthermore, inherent differences in these processes between the MHC class I and class II pathways further complicate accurate prediction [73]. Each process is highly variable and can impact immunogenicity of a particular protein antigen or neoantigen.
For example, current pipelines do not capture T cell-specific determinants of immunogenicity, such as T cell receptor affinity for peptide-MHC complexes, the prevalence of T cell precursor frequencies (frequencies of T-cells for a given antigen) in circulation and degree of costimulation. Furthermore, aberrant mechanisms of antigen processing and presentation within tumor cells further complicate neoantigen prioritization [74]. Several studies have challenged the predictive accuracy of peptide binding affinity, suggesting other factors, such as predicted peptide-MHC complex stability, are more important determinants of immunogenicity [75]. Duan et al. evaluated the conformational stability of several peptide-MHC class I complexes derived from neoantigens and identified immunogenic neoantigens that were predicted to have low-binding affinity by NetMHC [76]. However, approaches integrating peptide-MHC complex stability with binding are limited given the relative lack of robust tools for prediction of peptide-MHC complex stability, and may not be feasible in a clinical setting [77, 78]. To date, epitope prediction algorithms integrating antigen processing and presentation have only demonstrated modest gains compared to predictions based on binding affinity alone [25].
1.2.3. Limitations in MHC Class II neoantigen prediction algorithms
CD4 T cell responses are an integral part of adaptive immunity. CD4 T cells are essential for the generation of specific, potent, and long-lasting cellular immunity [79, 80]. CD4 T cells may also contribute to reprogramming the tumor microenvironment, promoting antitumor immunity [81]. In a study conducted by Alspach et al. evaluating the mediators of antitumor immunity following checkpoint blockade, the authors found activation of CD4 T-cells was essential for tumor rejection, emphasizing the important role of MHC class II antigens in antitumor immunity [82]. Of note, tumor cells that downregulate MHC class I expression to evade cytotoxic T cells are still susceptible to CD4 T cell-mediated immunity. There is evidence from preclinical studies and early clinical trials that cancer neoantigens may contain MHC class II epitopes. Accurate identification of neoantigens containing class II neoantigens may be critical for optimizing neoantigen vaccines and other therapies targeting cancer neoantigens.
MHC class I and II antigen processing and presentation are fundamentally different. Developing epitope-prediction algorithms for MHC class II epitopes has been challenging. MHC class II peptides are longer and more variable in length than MHC class I peptides, ranging from 11–20 amino acids [83]. MHC class I peptides reliably anchor within the MHC class I binding groove at their N- and C-termini. However, binding of MHC class II peptides is more variable and flanking regions, which do not engage with the peptide binding groove, still contribute to epitope specificity [84]. Furthermore, MHC class II binding is not as dependent on the anchor residues, resulting in more promiscuous peptide binding. While algorithms that prioritize neoantigens based on MHC class II binding affinity have been developed, they are only now being translated into the clinic [85, 86].
Kreiter et al. were one of the first to develop a neoantigen prioritization pipeline to identify MHC class II-restricted neoantigens based on binding affinity. Vaccination with these neoantigens in a preclinical model led to tumor regression [81]. The importance of CD4 T cell-mediated responses is further highlighted by some of the first clinical trials evaluating neoantigen vaccines in melanoma patients. Sahin et al. vaccinated patients with polyepitope RNA vaccines encoding 10 neoantigens predicted to have high class I and/or class II binding affinities and found the majority of immunogenic neoantigens induced exclusively CD4 T cell-mediated responses [11]. A more surprising observation was made by Ott et al., who immunized patients with a multi-peptide vaccine; despite only including neoantigens identified via a class I binding affinity algorithm, most detectable immune responses were generated by CD4 T cells [12]. From these observations, it appears evident that optimization of personalized neoantigen-directed therapy requires inclusion of MHC class II neoantigens.
With the advent of high-throughput mass spectrometry, comprehensive analysis of MHC class II ligands has allowed for the development of more reliable binding prediction algorithms [87, 88]. Similar to studies characterizing class I ligandomes, high-throughput mass spectrometry analyses of class II ligandomes have revealed patterns of antigen processing including locations of cleavage hotspots as important determinants of class II recognition and CD4 T-cell-mediated immunity [65, 89]. In an important study performed by Abelin et al., class II ligandomes were generated from a series of mono-allelic cells. The authors then used this dataset to develop a class II neoantigen prediction algorithm and found higher fidelity than the most commonly used algorithms [90]. The study also identified key aspects of class II ligand presentation that should be incorporated into class II prediction algorithms. For example, they found that class II peptide loading is influenced by individual chaperone protein alleles, such as HLA-DM alleles, and behaviors of professional antigen-presenting cells. The authors discovered that representation of certain genes in the ligandome did not correlate with gene expression levels, suggesting other factors affecting antigen presentation need to be considered in designing class II prediction algorithms. Other investigators have also used machine-learning algorithms trained on MS-generated data to develop class II prediction algorithms. These algorithms have also demonstrated superior predictive value compared to the most commonly used algorithms [88, 91–94]. Taken together, these studies have enhanced our understanding of class II antigen processing and presentation, which had previously remained incompletely understood [85, 95].
1.3. Additional challenges targeting cancer neoantigens
Even if neoantigen identification and prioritization is improved, there are other challenges that may potentially limit the success of therapies targeting cancer neoantigens. In other words, even if the most immunogenic neoantigens can be identified and prioritized, there may be challenges successfully inducing neoantigen-specific immune responses and/or improving clinical outcomes. For example, cancer patients may have acquired immune system deficiencies resulting in diminished antitumor immunity compared to healthy individuals. Stronen et al. identified a large repertoire of immunogenic neoantigens recognized by T-cells from healthy individuals that were neglected by T-cells from cancer patients with cognate HLA alleles [96]. Tumor-specific properties, such as low tumor mutational burden (TMB), extensive intra-tumor heterogeneity and immunosuppressive tumor microenvironment, may also contribute to resistance to strategies targeting cancer neoantigens. In fact, many tumors appear to have developed microenvironments that confer resistance to immune-based and/or neoantigen-directed therapies.
1.3.1. Malignancies with low mutation burdens
Neoantigens serve as important targets of cancer immunotherapy and TMB is generally correlated with clinical response to immune checkpoint inhibition therapy. Using data from The Cancer Genome Atlas (TCGA), Rooney et al. found that neoantigen load was positively associated with increased cytotoxic activity and improved survival across multiple tumor types [6]. However, mutation burden is highly variable between malignancies [4]. A comprehensive analysis of 27 cancer types conducted by Lawrence et al. identified marked heterogeneity in mutation burden, with median frequency of somatic mutations in cancers ranging from 0.1/megabase to over 100/megabase in melanoma and non-small cell lung cancer (NSCLC) [97, 98]. Initial studies of neoantigen vaccines have been focused on melanoma and NSCLC. However there are studies ongoing in a number of lower TMB cancers with mixed results. Using a preclinical ovarian cancer model known to have a low-to-intermediate mutation rate, Martin et al. designed and administered peptide vaccines encoding 17 neoantigens. The authors were able to detect an immune response to 7 neoantigens, but vaccination did not lead to tumor regression or improved survival [99]. Similarly, Zhang et al. identified neoantigens in human triple negative breast cancer using sequencing and neoantigen prediction algorithms. Neoantigen-specific human CD8 T cells were able to protect immunocompromised mice from tumor challenge with autologous patient-derived xenografts [100].
In order to facilitate the use of neoantigen vaccines and other therapies targeting cancer neoantigens in lower TMB cancers, additional research is needed to understand the biology of cancer neoantigens in these cancers. Cancer neoantigens capable of inducing immune responses are less common in these cancers. However, tumor-infiltrating lymphocytes are still present in many of these cancers, suggesting that other antigens may contribute to antitumor immunity [5]. It is possible that as neoantigen repertoires expand with the incorporation of indel and gene fusion neoantigen predictions, strategies targeting cancer neoantigens may be successful in a broader range of cancers.
1.3.2. Tumor heterogeneity
The accumulation of mutations is one of the factors that contributes to oncogenesis and ultimately cancer invasion and metastasis. Once a malignancy develops, individual cancer cells continue to acquire mutations, resulting in multiple distinct genomic profiles within the tumor. Gerlinger et al. were able to demonstrate this with multiregion sequencing in patients with metastatic renal cell carcinoma. The authors found that more than 2/3 of all somatic mutations were not conserved spatially throughout a tumor, suggesting divergent evolution of tumor cells during tumor progression [101]. This intratumoral heterogeneity can greatly impact neoantigen identification. Neoantigen prioritization is often based on whole-exome sequencing and RNA-seq of a small sample of tumor. In a tumor with extensive heterogeneity, such a sample may not accurately capture the complexity of the neoantigen repertoire. One strategy to mitigate the impact of tumor heterogeneity is multiregion sequencing. This may allow a more comprehensive analysis of the neoantigen repertoire. A second strategy is to target multiple candidate neoantigens with the assumption that generating an immune response to multiple neoantigens will provide coverage of all tumor cells. However, this strategy is limited by the fact that only a small proportion of predicted neoantigens actually induce detectable immune responses.
Of note, several studies have found that immune responses may be dominated by only a few antigens, a phenomenon known as immunodominance [4, 26, 102]. Thus, neoantigen-specific immune responses may be limited to a small number of neoantigens, and strategies focused on incorporating more neoantigen candidates may not be an ideal solution. This phenomenon further underscores the importance of neoantigen prioritization. Conversely, the immunogenic effects of just a few dominant neoantigens may be aided by epitope spreading. In this case, neoantigen-specific antitumor immune responses result in tumor lysis, antigen release, and priming of a broader immune response. Epitope spreading may mitigate resistance associated with tumor heterogeneity and/or antigen loss as well as facilitate antitumor responses to metastatic lesions, as seen in abscopal responses [103].
Cancer therapies often have a significant impact on tumor heterogeneity. Cancer immunoediting is the process by which tumors lose expression of antigens that stimulate antitumor immunity [104, 105]. This phenomenon can be particularly pronounced after strategies targeting cancer neoantigens. Verdegaal et al. demonstrated that advanced melanomas gradually lost expression of cancer neoantigens following adoptive T cell therapies [106]. Preclinical models suggest that tumors that lose expression of cancer neoantigens are more likely to evade antitumor immunity, proliferate, and progress [105].
1.3.3. Immunosuppressive tumor microenvironment
Many cancers develop, grow and metastasize in the context of an immunosuppressive tumor microenvironment. This immunosuppressive tumor microenvironment may be a major barrier to the success of cancer immunotherapies. Multiple mechanisms contribute to the immunosuppressive tumor microenvironment. These mechanisms have been reviewed elsewhere and include expression of immune checkpoint molecules, increased number and/or altered function of regulatory immune cells (such as regulatory T-cells and myeloid-derived suppressor cells), activation of anti-inflammatory pathways, and others [107–109]. Thus, there is considerable interest in combining neoantigen-directed therapies with other cancer immunotherapies. Early clinical trials evaluating neoantigen vaccines demonstrate that immune checkpoint inhibition may enhance the response to neoantigen vaccines [11, 12, 110]. Other strategies include combining neoantigen vaccines with strategies targeting regulatory T cells, tumor-associated macrophages, or myeloid derived suppressor cells [111].
2. Expert opinion
Cancer immunotherapies have the potential to revolutionize cancer therapy. A mechanistic understanding of how cancer immunotherapies target and eradicate tumor cells is essential for optimizing currently available therapies and for the development of new therapies. Cancer neoantigens have emerged as prominent targets of cancer immunotherapies. Specific targeting of cancer neoantigens may avoid central tolerance and minimize autoimmune-related toxicities. Strategies targeting cancer neoantigens attempt to exploit these unique properties of cancer neoantigens to prime and enhance antitumor immune responses. A personalized approach appears mandatory, as the diversity of cancer-associated somatic mutations and HLA polymorphism imply that the neoantigen landscape is almost certain to be unique to an individual. Neoantigen identification has only recently become widely accessible. Advances in sequencing technologies and variant calling algorithms allows for identification of genetic alterations in cancer. In silico neoantigen prediction algorithms are currently the most commonly used tools to identify potentially immunogenic neoantigens. Multiple neoantigen prioritization pipelines have been created based on data generated in different model systems. Each has its own set of advantages and disadvantages, but none so far have been able to definitively identify immunogenic neoantigens with high accuracy.
Some have interpreted these limitations as reasons to abandon strategies targeting cancer neoantigens [112]. This would be premature. First, neoantigen prioritization pipelines are a relatively new technology, having only emerged within the past 10 years. Pipeline developers are continually modifying these programs to improve prediction accuracy. To date, pipelines have been created by independent groups in parallel, with limited communication between groups. Development of a standardized approach may require extensive information sharing and collaboration [34, 113].
For instance, in 2020 the Tumor Neoantigen Selection Alliance (TESLA) was formed to compare neoantigen prediction algorithms. 25 teams from around the world each used their own unique neoantigen prediction algorithm(s) to identify and prioritize cancer neoantigens. Each group used genomic data provided by the Alliance from the same 6 patient samples (3 melanoma, 3 NSCLC). The immunogenicity of candidate neoantigens was validated by a core laboratory by detection of MHC-restricted T-cells in subject-matched PBMC. The Alliance determined that approximately 50% of immunogenic epitopes are characterized by strong MHC binding affinity, prolonged half-life, high expression, and either low agretopicity or high foreignness. This study highlights the significant differences between groups, and the potential gains that may be realized through further collaboration and efforts to standardize neoantigen prediction pipelines [114].
Second, adoption of immune checkpoint inhibition therapy to treat certain malignancies has revealed the need for accurate neoantigen identification to elucidate mechanisms of action and identify predictive biomarkers of response. Several studies have found immune checkpoint inhibition to enhance the immune responses against cancer neoantigens and, conversely, that the neoantigen repertoire influences response to immune checkpoint inhibition [7, 115, 116]. Thus, accurate identification of neoantigens is not only crucial to targeting cancer neoantigens, but may also identify targets of immune checkpoint inhibition.
Neoantigen prioritization algorithms will benefit from additional research elucidating the mechanisms of antigen processing and presentation, and from advances in sequencing, machine-learning, and increased collaboration. Neoantigen prioritization algorithms are likely to become more standardized, with the ability to optimally identify candidate neoantigens. Strategies targeting cancer neoantigens will continue to evolve with the potential to prime and enhance antitumor immunity.
3. Conclusion
To date, clinical trials targeting shared tumor antigens based on “off-the-shelf” therapeutics have been disappointing. Cancer neoantigens appear to be important targets of cancer immunoediting and cancer immunotherapies, and strategies targeting cancer neoantigens can prime endogenous immunity and enhance antitumor activity. Identification and prioritization of cancer neoantigens has only recently become practical with the development of sequencing and bioinformatics technologies that can identify and validate expression of somatic mutations, and predict immunogenicity in the setting of a particular MHC genotype. Identification of cancer neoantigens by direct analysis of ligands eluted from MHC alleles, while attractive, is currently too laborious to seamlessly translate into clinical applications. Instead, neoantigen identification strategies have evolved to leverage pipelines that combine next-generation sequencing with in silico prediction of MHC binding. While these technologies can readily be translated to a clinical setting, key limitations highlighted in this review need to be addressed prior to optimize success.
ARTICLE HIGHLIGHTS.
Strategies targeting cancer neoantigens, such as neoantigen vaccines, rely on accurate identification of cancer neoantigens.
“Off-the-shelf” immune therapies targeting shared tumor antigens have had limited success, emphasizing the need to target cancer neoantigens. Recent studies demonstrate that cancer neoantigens are important targets of immune checkpoint inhibition, adoptive cell therapy and other cancer immunotherapies.
The two most common strategies to identify cancer neoantigens are: (1) direct identification based on proteomic analysis of ligands eluted from peptide-MHC complexes and (2) indirect identification based on sequencing and bioinformatic pipelines. Since direct identification is currently too cumbersome for widespread use, most clinical trials targeting cancer neoantigens have relied on indirect identification based on sequencing.
Next-generation sequencing is currently in clinical use, facilitating identification of the genetic alterations encoding cancer neoantigens.
While neoantigen vaccines have successfully generated neoantigen-specific immune responses in preclinical models and early phase clinical trials, most candidate neoantigens do not generate immune responses. This suggests that additional study is necessary to improve current neoantigen prediction algorithms.
Neoantigen identification and prioritization pipelines will likely improve in the future, benefitting from insights into the mechanisms of antigen processing and presentation within tumor cells, advances in sequencing and machine-learning technologies, and collaborative efforts.
In addition to improving strategies targeting neoantigens, accurate neoantigen prediction is likely to enhance our mechanistic understanding of cancer immunotherapies.
Acknowledgments
Funding
This manuscript was funded by the Washington University School of Medicine Surgical Oncology Basic Science and Translational Research Training Program grant T32CA009621, and the Washington University School of Medicine Cancer Center Support Grant 2P30CA091842–19 from the National Cancer Institute (NCI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.De Plaen E, et al. , Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum- antigen P91A and identification of the tum- mutation. Proc Natl Acad Sci U S A, 1988. 85(7): p. 2274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monach PA, et al. , A unique tumor antigen produced by a single amino acid substitution. Immunity, 1995. 2(1): p. 45–59. [DOI] [PubMed] [Google Scholar]
- 3.Wolfel T, et al. , A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science, 1995. 269(5228): p. 1281–4. [DOI] [PubMed] [Google Scholar]
- 4.Schumacher TN and Schreiber RD, Neoantigens in cancer immunotherapy. Science, 2015. 348(6230): p. 69–74. [DOI] [PubMed] [Google Scholar]
- 5.Brown SD, et al. , Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res, 2014. 24(5): p. 743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rooney MS, et al. , Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell, 2015. 160(1–2): p. 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Allen EM, et al. , Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 2015. 350(6257): p. 207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubin MM, et al. , Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature, 2014. 515(7528): p. 577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreiter S, et al. , Targeting the tumor mutanome for personalized vaccination therapy. Oncoimmunology, 2012. 1(5): p. 768–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sahin U, et al. , Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature, 2017. 547(7662): p. 222–226. *One of the first clinical trials evaluating personalized neoantigen vaccines in melanoma patients. The authors designed peptide vaccines to incorporate neoantigens selected based on MHC class I and class II binding affinities.
- 12. Ott PA, et al. , An immunogenic personal neoantigen vaccine for patients with melanoma. Nature, 2017. 547(7662): p. 217–221. *One of the first clinical trials evaluating personalized neoantigen vaccines in melanoma patients. The authors designed polyepitope RNA vaccines to incorporate neoantigens selected based on MHC class I affinity, only. However, CD4-mediated immune responses were more prevalent.
- 13.Carreno BM, et al. , Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science, 2015. 348(6236): p. 803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The problem with neoantigen prediction. Nature Biotechnology, 2017. 35: p. 97. [DOI] [PubMed] [Google Scholar]
- 15.Vitiello A and Zanetti M, Neoantigen prediction and the need for validation. Nat Biotechnol, 2017. 35(9): p. 815–817. [DOI] [PubMed] [Google Scholar]
- 16.Yadav M, et al. , Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature, 2014. 515(7528): p. 572–6. [DOI] [PubMed] [Google Scholar]
- 17.Kalaora S, et al. , Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget, 2016. 7(5): p. 5110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh-Jasuja H, Emmerich NP, and Rammensee HG, The Tubingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol Immunother, 2004. 53(3): p. 187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bassani-Sternberg M, et al. , Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun, 2016. 7: p. 13404. *First Application of direct neoantigen identification using mass spectrometry in native human tumors
- 20.Mohammed F, et al. , Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol, 2008. 9(11): p. 1236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobbold M, et al. , MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med, 2013. 5(203): p. 203ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creech AL, et al. , The role of mass spectrometry and proteogenomics in the advancement of HLA epitope prediction. 2018. 18(12): p. 1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polyakova A, Kuznetsova K, and Moshkovskii S, Proteogenomics meets cancer immunology: mass spectrometric discovery and analysis of neoantigens. Expert Rev Proteomics, 2015. 12(5): p. 533–41. [DOI] [PubMed] [Google Scholar]
- 24.Purcell AW, Ramarathinam SH, and Ternette N, Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nature Protocols, 2019. 14(6): p. 1687–1707. [DOI] [PubMed] [Google Scholar]
- 25.Backert L and Kohlbacher O, Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med, 2015. 7: p. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yewdell JW and Bennink JR, Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol, 1999. 17: p. 51–88. [DOI] [PubMed] [Google Scholar]
- 27.Richters MM, et al. , Best practices for bioinformatic characterization of neoantigens for clinical utility. 2019. 11(1): p. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreatta M and Nielsen MJB, Gapped sequence alignment using artificial neural networks: application to the MHC class I system. 2016. 32(4): p. 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H, Lund O, and Nielsen M, The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics, 2009. 25(10): p. 1293–1299. **Description of NetMHC, one of the most commonly used epitope-prediction programs. NetMHC infers neoantigen immunogenicity from prediction of peptide-MHC binding affinity.
- 30.Segal NH, et al. , Epitope landscape in breast and colorectal cancer. Cancer Res, 2008. 68(3): p. 889–92. [DOI] [PubMed] [Google Scholar]
- 31. Gfeller D and Bassani-Sternberg M, Predicting Antigen Presentation—What Could We Learn From a Million Peptides? 2018. 9(1716). **Demonstrates in silico neoantigen prioritization pipelines can identify immunogenic tumor-specific mutations.
- 32.Hundal J, et al. , pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med, 2016. 8(1): p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, et al. , TSNAD: an integrated software for cancer somatic mutation and tumour-specific neoantigen detection. R Soc Open Sci, 2017. 4(4): p. 170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bais P, et al. , CloudNeo: a cloud pipeline for identifying patient-specific tumor neoantigens. Bioinformatics, 2017. 33(19): p. 3110–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tappeiner E, et al. , TIminer: NGS data mining pipeline for cancer immunology and immunotherapy. Bioinformatics, 2017. 33(19): p. 3140–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinsteyn A, et al. , Computational Pipeline for the PGV-001 Neoantigen Vaccine Trial. Front Immunol, 2017. 8: p. 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurtz V, et al. , NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol, 2017. 199(9): p. 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Donnell TJ, Rubinsteyn A, and Laserson U, MHCflurry 2.0: Improved Pan-Allele Prediction of MHC Class I-Presented Peptides by Incorporating Antigen Processing. Cell Systems, 2020. 11(1): p. 42–48.e7. [DOI] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, et al. , Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nature Biotechnology, 2019. 37(1): p. 55–63. [DOI] [PubMed] [Google Scholar]
- 40.Zhao W and Sher X.J.P.c.b., Systematically benchmarking peptide-MHC binding predictors: From synthetic to naturally processed epitopes. 2018. 14(11): p. e1006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkhurst M, et al. , Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression. Clin Cancer Res, 2017. 23(10): p. 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y-C, et al. , Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. 2014. 20(13): p. 3401–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran E, et al. , Immunogenicity of somatic mutations in human gastrointestinal cancers. Science, 2015. 350(6266): p. 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran E, et al. , Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science, 2014. 344(6184): p. 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackl H, et al. , Computational genomics tools for dissecting tumour-immune cell interactions. Nat Rev Genet, 2016. 17(8): p. 441–58. [DOI] [PubMed] [Google Scholar]
- 46.van Dijk EL, et al. , The third revolution in sequencing technology. 2018. 34(9): p. 666–681. [DOI] [PubMed] [Google Scholar]
- 47.Yost SE, et al. , Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic Acids Res, 2012. 40(14): p. e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerick M, et al. , Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics, 2011. 4: p. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cibulskis K, et al. , Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol, 2013. 31(3): p. 213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, et al. , The role and challenges of exome sequencing in studies of human diseases. Front Genet, 2013. 4: p. 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertier G, Hetu M, and Joly Y, Unsolved challenges of clinical whole-exome sequencing: a systematic literature review of end-users’ views. BMC Med Genomics, 2016. 9(1): p. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turajlic S, et al. , Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol, 2017. 18(8): p. 1009–1021. [DOI] [PubMed] [Google Scholar]
- 53.Ruark E, et al. , OpEx - a validated, automated pipeline optimised for clinical exome sequence analysis. Sci Rep, 2016. 6: p. 31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munz M, et al. , CSN and CAVA: variant annotation tools for rapid, robust next-generation sequencing analysis in the clinical setting. Genome Med, 2015. 7: p. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altuvia Y and Margalit H, A structure-based approach for prediction of MHC-binding peptides. Methods, 2004. 34(4): p. 454–9. [DOI] [PubMed] [Google Scholar]
- 56.Bordner AJ and Abagyan R, Ab initio prediction of peptide-MHC binding geometry for diverse class I MHC allotypes. Proteins, 2006. 63(3): p. 512–26. [DOI] [PubMed] [Google Scholar]
- 57.Bui HH, et al. , Structural prediction of peptides binding to MHC class I molecules. Proteins, 2006. 63(1): p. 43–52. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, et al. , Limitations of Ab initio predictions of peptide binding to MHC class II molecules. PLoS One, 2010. 5(2): p. e9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bassani-Sternberg M, et al. , Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics, 2015. 14(3): p. 658–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassani-Sternberg M and Gfeller D, Unsupervised HLA Peptidome Deconvolution Improves Ligand Prediction Accuracy and Predicts Cooperative Effects in Peptide-HLA Interactions. 2016. 197(6): p. 2492–9. [DOI] [PubMed] [Google Scholar]
- 61.Trolle T, et al. , The Length Distribution of Class I-Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele-Specific Binding Preference. J Immunol, 2016. 196(4): p. 1480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gfeller D, et al. , The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J Immunol, 2018. 201(12): p. 3705–3716. [DOI] [PubMed] [Google Scholar]
- 63.Bassani-Sternberg M, et al. , Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput Biol, 2017. 13(8): p. e1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jappe EC, et al. , Predicted MHC peptide binding promiscuity explains MHC class I ‘hotspots’ of antigen presentation defined by mass spectrometry eluted ligand data. Immunology, 2018. 154(3): p. 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller M, et al. , ‘Hotspots’ of Antigen Presentation Revealed by Human Leukocyte Antigen Ligandomics for Neoantigen Prioritization. Front Immunol, 2017. 8: p. 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mylonas R, et al. , Estimating the Contribution of Proteasomal Spliced Peptides to the HLA-I Ligandome. Mol Cell Proteomics, 2018. 17(12): p. 2347–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abelin JG, et al. , Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity, 2017. 46(2): p. 315–326. **Utilized mono-allelic cells to generate MHC specific ligandomes using mass spectrometry to improve neoantigen prediction algorithms.
- 68.Sarkizova S, et al. , A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nature biotechnology, 2020. 38(2): p. 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alvarez B, et al. , NNAlign_MA; MHC Peptidome Deconvolution for Accurate MHC Binding Motif Characterization and Improved T-cell Epitope Predictions. Molecular & Cellular Proteomics, 2019. 18(12): p. 2459–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andreatta M, Alvarez B, and Nielsen M, GibbsCluster: unsupervised clustering and alignment of peptide sequences. Nucleic Acids Res, 2017. 45(W1): p. W458–w463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bassani-Sternberg M and Gfeller D, Unsupervised HLA Peptidome Deconvolution Improves Ligand Prediction Accuracy and Predicts Cooperative Effects in Peptide-HLA Interactions. J Immunol, 2016. 197(6): p. 2492–9. [DOI] [PubMed] [Google Scholar]
- 72.Reynisson B, et al. , Improved Prediction of MHC II Antigen Presentation through Integration and Motif Deconvolution of Mass Spectrometry MHC Eluted Ligand Data. J Proteome Res, 2020. 19(6): p. 2304–2315. [DOI] [PubMed] [Google Scholar]
- 73.Blum JS, Wearsch PA, and Cresswell P, Pathways of antigen processing. Annu Rev Immunol, 2013. 31: p. 443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leone P, et al. , MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst, 2013. 105(16): p. 1172–87. [DOI] [PubMed] [Google Scholar]
- 75.Harndahl M, et al. , Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol, 2012. 42(6): p. 1405–16. [DOI] [PubMed] [Google Scholar]
- 76.Duan F, et al. , Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med, 2014. 211(11): p. 2231–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antes I, Siu SW, and Lengauer T, DynaPred: a structure and sequence based method for the prediction of MHC class I binding peptide sequences and conformations. Bioinformatics, 2006. 22(14): p. e16–24. [DOI] [PubMed] [Google Scholar]
- 78.Yanover C and Bradley P, Large-scale characterization of peptide-MHC binding landscapes with structural simulations. Proc Natl Acad Sci U S A, 2011. 108(17): p. 6981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Godet Y, et al. , Analysis of spontaneous tumor-specific CD4 T-cell immunity in lung cancer using promiscuous HLA-DR telomerase-derived epitopes: potential synergistic effect with chemotherapy response. Clin Cancer Res, 2012. 18(10): p. 2943–53. [DOI] [PubMed] [Google Scholar]
- 80.Fridman WH, et al. , The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer, 2012. 12(4): p. 298–306. [DOI] [PubMed] [Google Scholar]
- 81.Kreiter S, et al. , Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature, 2015. 520(7549): p. 692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alspach E, et al. , MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature, 2019. 574(7780): p. 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Neefjes J, et al. , Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol, 2011. 11(12): p. 823–36. [DOI] [PubMed] [Google Scholar]
- 84.Nielsen M, et al. , MHC class II epitope predictive algorithms. Immunology, 2010. 130(3): p. 319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andreatta M, et al. , Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics, 2015. 67(11–12): p. 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Justesen S, et al. , Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res, 2009. 5: p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen B, et al. , Predicting HLA class II antigen presentation through integrated deep learning. 2019. 37(11): p. 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Racle J, et al. , Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol, 2019. [DOI] [PubMed]
- 89.Godkin AJ, et al. , Naturally processed HLA class II peptides reveal highly conserved immunogenic flanking region sequence preferences that reflect antigen processing rather than peptide-MHC interactions. J Immunol, 2001. 166(11): p. 6720–7. [DOI] [PubMed] [Google Scholar]
- 90. Abelin JG, et al. , Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction. Immunity, 2019. 51(4): p. 766–779.e17. **Used monoallelic cells and mass spectrometry to generate MHC class II ligandomes.
- 91.Chen B, et al. , Predicting HLA class II antigen presentation through integrated deep learning. Nat Biotechnol, 2019. [DOI] [PMC free article] [PubMed]
- 92.Garde C, et al. , Improved peptide-MHC class II interaction prediction through integration of eluted ligand and peptide affinity data. Immunogenetics, 2019. 71(7): p. 445–454. [DOI] [PubMed] [Google Scholar]
- 93.Sofron A, et al. , High-resolution analysis of the murine MHC class II immunopeptidome. Eur J Immunol, 2016. 46(2): p. 319–28. [DOI] [PubMed] [Google Scholar]
- 94.Barra C, et al. , Footprints of antigen processing boost MHC class II natural ligand predictions. Genome Med, 2018. 10(1): p. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andreatta M, et al. , Machine learning reveals a non-canonical mode of peptide binding to MHC class II molecules. Immunology, 2017. 152(2): p. 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stronen E, et al. , Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science, 2016. 352(6291): p. 1337–41. [DOI] [PubMed] [Google Scholar]
- 97.Zhang X, et al. , Personalized cancer vaccines: Targeting the cancer mutanome. Vaccine, 2017. 35(7): p. 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lawrence MS, et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 2013. 499(7457): p. 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin SD, et al. , Low Mutation Burden in Ovarian Cancer May Limit the Utility of Neoantigen-Targeted Vaccines. PLoS One, 2016. 11(5): p. e0155189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, et al. , Breast Cancer Neoantigens Can Induce CD8(+) T-Cell Responses and Antitumor Immunity. Cancer Immunol Res, 2017. 5(7): p. 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gerlinger M, et al. , Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med, 2012. 366(10): p. 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dudley ME and Roopenian DC, Loss of a unique tumor antigen by cytotoxic T lymphocyte immunoselection from a 3-methylcholanthrene-induced mouse sarcoma reveals secondary unique and shared antigens. J Exp Med, 1996. 184(2): p. 441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gulley JL, et al. , Role of Antigen Spread and Distinctive Characteristics of Immunotherapy in Cancer Treatment. Journal of the National Cancer Institute, 2017. 109(4): p. djw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mittal D, et al. , New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol, 2014. 27: p. 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsushita H, et al. , Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature, 2012. 482(7385): p. 400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verdegaal EM, et al. , Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature, 2016. 536(7614): p. 91–5. [DOI] [PubMed] [Google Scholar]
- 107.Rabinovich GA, Gabrilovich D, and Sotomayor EM, Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol, 2007. 25: p. 267–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chanmee T, et al. , Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel), 2014. 6(3): p. 1670–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar V, et al. , The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol, 2016. 37(3): p. 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bassani-Sternberg M, et al. , A Phase Ib Study of the Combination of Personalized Autologous Dendritic Cell Vaccine, Aspirin, and Standard of Care Adjuvant Chemotherapy Followed by Nivolumab for Resected Pancreatic Adenocarcinoma-A Proof of Antigen Discovery Feasibility in Three Patients. Front Immunol, 2019. 10: p. 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gatti-Mays ME, et al. , Cancer vaccines: enhanced immunogenic modulation through therapeutic combinations. 2017. 13(11): p. 2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kissick HT, Is It Possible to Develop Cancer Vaccines to Neoantigens, What Are the Major Challenges, and How Can These Be Overcome? Neoantigens as Vaccine Targets for Cancer. Cold Spring Harb Perspect Biol, 2018. 10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu J, et al. , TSNAdb: A Database for Tumor-specific Neoantigens from Immunogenomics Data Analysis. Genomics Proteomics Bioinformatics, 2018. 16(4): p. 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wells DK, et al. , Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. 2020. [DOI] [PMC free article] [PubMed]
- 115.Snyder A, et al. , Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med, 2014. 371(23): p. 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Luksza M, et al. , A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature, 2017. 551(7681): p. 517–520. *Reports the role of neoantigens as predictive biomarkers of response to checkpoint blockade therapy.


