Dear Editor,
The serotonin 5-HT1 receptor subtypes, including 5-HT1A, 5-HT1B, 5-HT1D, 5-HT1E, and 5-HT1F, are G protein-coupled receptors (GPCRs) that respond to the endogenous neurotransmitter serotonin and couple preferentially to the Gi/o family of G proteins.1 Drugs targeting 5-HT1 receptors are used to treat migraine, depression, and schizophrenia.2 Clinical use of traditional anti-migraine drugs, triptans, caused side effects arising from therapeutic vasoconstrictive actions when targeting 5-HT1B/1D receptors.3 The requirement of new anti-migraine drugs without vasoconstrictive effects led to the development of lasmiditan, a highly selective 5-HT1F receptor agonist with minimized on-target side effects.4
Migraine is one of the most common diseases worldwide and, importantly, a major cause of lost work productivity.3 The selective 5-HT1B/D agonists, triptans, are currently used for a first-line acute treatment of moderate-to-severe migraine attacks. Triptans bind mostly to 5-HT1B/D receptors within cerebral blood vessels, leading to vasoconstriction. Unfortunately, a large percentage of patients are not satisfied with current acute migraine treatments, because 5-HT1B/D receptors are also present on coronary and limb arteries and triptans may cause acute coronary syndromes in patients with or without cardiovascular disease.3,5
Lasmiditan, a potent and selective agonist for the 5-HT1F receptor, has recently been approved for treatment of acute migraine.6 Lasmiditan does not have vasoconstriction effects and may be a safer and more effective option for patients refractory to treatment with triptans and for patients with cardiovascular disease.6 Lasmiditan has a pyridinyl-piperidine scaffold, which is structurally different from the indole derivatives of triptans. In addition, lasmiditan is able to penetrate the blood-brain barrier to act on receptor located in the brain, thus enhancing its action on receptor sites in central nervous system (CNS).6 To better understand the structural basis of lasmiditan selectivity and activation of the 5-HT1F, we determined the structure of the 5-HT1F in complex with lasmiditan and Gi1 at a resolution of 3.4 Å by single-particle cryo-EM. The structure reveals the mechanism of 5-HT1F-selective activation and provides a template for the rational design of anti-migraine drugs.
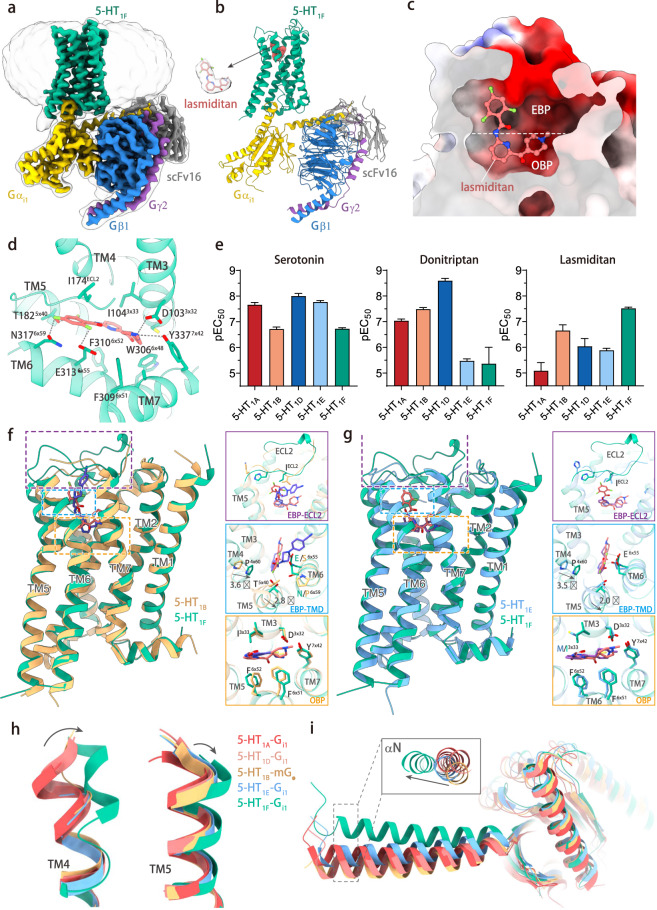
For single-particle cryo-EM structural studies, we prepared the lasmiditan-bound 5-HT1F–Gi complex, which were met with technical challenges of low expression levels and unstable formation of the receptor–G protein complex. Despite these difficulties and after many attempts, we were able to prepare homogenous sample for cryo-EM analysis. The structure was determined at a global resolution of 3.4 Å (Supplementary information, Fig. S1). The lasmiditan-bound 5-HT1F–Gi complex EM density maps are sufficiently clear to define the position of the 5-HT1F receptor, the Gi heterotrimer, scFv16, and the bound ligand lasmiditan. The overall structure of 5-HT1F consists of a canonical transmembrane domain (TMD) of seven transmembrane helices (TM1–7), a short intracellular loop 2 (ICL2) helix, and an amphipathic helix H8 (Fig. 1a, b). The active 5-HT1F receptor shares a similar overall conformation with other active 5-HT1 receptors,7 while a complete backbone structure for extracellular loop 2 (ECL2) is visible, which is partly missing in other 5-HT1 structures due to the flexibility. The cryo-EM map includes well-defined features for amino acids forming the agonist-binding pocket and clear density for lasmiditan in 5-HT1F (Fig. 1b). We found that negatively-charged amino acids in the ligand-binding pocket of 5-HT1F are primarily responsible for the affinity of lasmiditan (Fig. 1c, d). In the orthosteric binding pocket (OBP), the primary amine on methylpiperidine group of lasmiditan forms a canonical charge interaction with D1033x32 of 5-HT1F (Fig. 1d), which simultaneously forms a hydrogen bond with Y3377x42, supporting a stable interaction between the ligand and the receptor (Fig. 1d). Mutational studies showed that these residues are critical for lasmiditan binding (Supplementary information, Fig. S2). The interactions between D3x32 of the receptor and the primary amine of agonists as well as the supportive Y7x42 are conserved in aminergic GPCRs.8 In addition, the methylpiperidine group of lasmiditan forms hydrophobic interactions with F3096x51 in TM6 of 5-HT1F (Fig. 1d), and mutations in F3096x51 cause a nearly 100-fold reduction in lasmiditan affinity (Supplementary information, Fig. S2). Meanwhile, the aromatic pyridine scaffold of lasmiditan is sandwiched between I1043x33 and F3106×52, forming a hydrophobic interaction core (Fig. 1d). F3106x52A mutation simultaneously eliminated 5-HT1F-G protein coupling signals and lasmiditan affinity, and I1043×33A mutation also caused a nearly 60-fold reduction in lasmiditan affinity, suggesting that these hydrophobic interactions are crucial for lasmiditan-induced 5-HT1F activation (Supplementary information, Fig. S2). I3x33 of 5-HT1F is 3.5 Å away from the aromatic ring of lasmiditan, which provides a stronger hydrophobic interaction than V3x33 of 5-HT1A and M3x33 of 5-HT1E. In the extended binding pocket (EBP), the trifluorobenzene group of lasmiditan forms additional hydrophobic interactions with I174ECL2 and P1584x60, and forms hydrogen bonds with residue E3136x55, N3176x59, T1825x40, and H176ECL2 of 5-HT1F (Fig. 1d). These structural observations are also confirmed by mutation experiments (Supplementary information, Fig. S2).
Fig. 1. Structure of lasmiditan–5-HT1F–Gi1 complex.
a Cryo-EM map of the 5-HT1F–Gi complex. b Structural model of the 5-HT1F–Gi complex. The ligand model is shown on left side of the complex with surrounding density map. c Electrostatic surface representation of lasmiditan-binding pocket of 5-HT1F. d The binding mode of lasmiditan in the ligand-binding pocket of 5-HT1F. e Gi recruitment assay using NanoBiT for wild-type 5-HT1A, 5-HT1B, 5-HT1D, 5-HT1E, and 5-HT1F induced by serotonin, donitriptan and lasmiditan. f Structural comparison of lasmiditan-bound 5-HT1F and donitriptan-bound 5-HT1B (PDB code: 6G79). g Structural comparison of lasmiditan-bound 5-HT1F and BRL54443-bound 5-HT1E (PDB code: 7E33). h Structure comparison focuses on extracellular ends of TM4 (left) and TM5 (right) among 5-HT1A (red, PDB code: 7E2Y), 5-HT1B (tan, PDB code: 6G79), 5-HT1D (yellow, PDB code: 7E32), 5-HT1E (blue, PDB code: 7E33) and 5-HT1F (green). i Comparison of the Gα conformation among the structures of Gi/o-coupled 5-HT1A (red, PDB code: 7E2Y), 5-HT1B (tan, PDB code: 6G79), 5-HT1D (yellow, PDB code: 7E32), 5-HT1E (blue, PDB code: 7E33) and 5-HT1F (green).
Lasmiditan is a new-generation 5-HT receptor agonist with high affinity and selectivity to 5-HT1F (Ki = 2 nM) over other serotonin receptors (Ki > 500 nM),4 and this selectivity is also confirmed by our NanoBiT G protein recruitment assays (Fig. 1e; Supplementary information, Fig. S3a). Comparison between the structure of 5-HT1F bound to lasmiditan and other 5-HT1 structures7,9 uncovers the structural basis of selectivity for lasmiditan (Fig. 1f–h). Structual comparison of 5-HT1F and 5-HT1B shows that the ligand–receptor interaction is basically conserved in OBP, but different in EBP, which are formed by TM4/5/6, and ECL2. For the TM4/5, the residues that interact with the trifluorobenzene group of lasmiditan in 5-HT1F are highly conserved in 5-HT1B (Fig. 1f). However, the conformation of TM4/5 shows significant changes between 5-HT1F and 5-HT1B. On the extracellular side, TM4 shifts 3.6 Å, and TM5 shifts 2.8 Å in 5-HT1F away from those in 5-HT1B (Fig. 1f). For the TM6, the E6x55 and N6x59 of 5-HT1F form hydrogen bonds with lasmiditan, while the corresponding residues are S6x55 and P6x59 in 5-HT1B, which cannot establish the corresponding interactions. For the ECL2, the region that interacts with lasmiditan shows different conformations between 5-HT1F and 5-HT1B (Fig. 1f).
5-HT1E is the receptor with the highest homology to 5-HT1F. The structure of 5-HT1E that we previously reported reveals the mechanism of selectivity of the 5-HT1E/1F-selective ligand BRL54443 to 5-HT1E. Lasmiditan is only selective to 5-HT1F, rather than to 5-HT1E. Structural comparison of 5-HT1F and 5-HT1E provides an opportunity to uncover the mechanism of selectivity of lasmiditan to 5-HT1F. Although 5-HT1F and 5-HT1E have relatively conserved residues for ligand binding both in OBP and EBP, the conformations of TM4, TM5, and ECL2 show significant differences. Among them, the extracellular ends of TM4 shifted by 3.5 Å and TM5 shifted by 2.0 Å, and the backbone of ECL2 shows a different conformation (Fig. 1g). These changes are similar in comparison with 5-HT1B (Fig. 1f). We further compared the 5-HT1F structure with those of other 5-HT1 subfamily receptors; the results showed that the conformation of the TM4–TM5–ECL2 region is relatively conserved in 5-HT1A, 5-HT1B, 5-HT1D, and 5-HT1E, but not in 5-HT1F (Fig. 1h; Supplementary information, Fig. S3b–d). To confirm the roles of ECL2 in ligand selectivity, we replaced the ECL2 of 5-HT1F with that of other 5-HT1 receptors and tested the receptor activation. The result shows that the lasmiditan-induced activation was significantly affected (Supplementary information, Fig. S3e). The importance of EBP for lasmiditan binding and the different shape of EBP of 5-HT1F from other 5-HT receptors determines the high selectivity of lasmiditan to 5-HT1F.
The lasmiditan induces activation of 5-HT1F undergoing a canonical conformational rearrangement. Compared to the inactive-state 5-HT1B,10 lasmiditan triggers downward movement of the toggle switch residue W6x48 of 5-HT1F, and then induces the conformational changes in PIF, DRY, and NPxxY motifs (Supplementary information, Fig. S4a–e). These conformational changes further cause an 8 Å outward movement of TM6, which allows the α5 helix of Gαi1 to insert into the intracellular cavity formed by the receptor TMD bundle, a hallmark of GPCR activation (Supplementary information, Fig. S4f). Structural comparison of the Gi-coupled 5-HT1F and the Go-coupled 5-HT1B complexes9 reveals differences in G protein coupling (Supplementary information, Fig. S4g, h). Although the conformations of the main interfaces between receptor and G protein are similar, the G protein conformation shows observable changes. The main body of the Ras-like domain shares a similar conformation, while the N-terminus of αN shift 9.4 Å and the last residue of α5 shift 2.4 Å between Gi and Go (Supplementary information, Fig. S4g). Comparing the 5-HT1F–Gi complex structure with other 5-HT1–Gi/o complex structures,7,9 we found that the αN of 5-HT1F-bound Gi shifts away from other 5-HT1 receptor-bound Gi/o, which suggests that the coupling of 5-HT1F to Gi protein is unique from other 5-HT1 receptors (Fig. 1i).
In summary, in this study, we report the cryo-EM structure of the 5-HT1F–Gi complex bound to a highly selective anti-migraine drug lasmiditan. The structure reveals the binding mode of lasmiditan in 5-HT1F. Comparison of our structure and the previously reported 5-HT1 structures9 provides the basis of the selectivity of lasmiditan to 5-HT1F. The determination for selectivity is mainly attributed to the interaction between the trifluorobenzene group of lasmiditan and the specific EBP of 5-HT1F. Furthermore, our structure reveals a conserved mechanism for activation of 5-HT1F and the unique G protein coupling conformation from that in other 5-HT1–G protein structures.7,9 Together, these results provide a rational template for design of new-generation anti-migraine drugs that selectively target 5-HT1F, therefore avoiding the main disadvantage of cardiovascular side effects associated with the triptan class of anti-migraine drugs.
Supplementary information
Acknowledgements
The cryo-EM data were collected at the Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China). This work was partially supported by the Ministry of Science and Technology of China (2018YFA0507002 to HEX), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB37030103 to HEX), Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX), the National Natural Science Foundation of China (31770796 to YJ), National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (2018ZX09711002-002-002 to YJ).
Author contributions
SH and PX designed the expression constructs, purified the complexes, prepared samples for negative stain and data collection toward the structures, performed functional assay, prepared the figures and manuscript draft. PX evaluated the specimen by negative-stain EM, screened the cryo-EM conditions, prepared the cryo-EM grids, collected cryo-EM images, built the model, and refined the structures. YT, CY, and YZ participated in the NanoBiT G protein recruitment assays. YJ participated in the supervision of SH, PX, YT, CY and YZ, analyzed the structures, and edited the manuscript. HEX conceived and supervised the project, analyzed the structures, and wrote the manuscript with inputs from all authors.
Data availability
The corresponding coordinates and cryo-EM density map have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 7EXD, and in EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-31371.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Sijie Huang, Peiyu Xu, Yangxia Tan.
Contributor Information
Yi Jiang, Email: yijiang@simm.ac.cn.
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00527-4.
References
- 1.McCorvy JD, Roth BL. Pharmacol. Therapeut. 2015;150:129–142. doi: 10.1016/j.pharmthera.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pytliak M, Vargova V, Mechirova V, Felsoci M. Physiol. Res. 2011;60:15–25. doi: 10.33549/physiolres.931903. [DOI] [PubMed] [Google Scholar]
- 3.Negro A, Koverech A, Martelletti P. J. Pain Res. 2018;11:515–526. doi: 10.2147/JPR.S132833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DL, et al. Cephalalgia. 2010;30:1159–1169. doi: 10.1177/0333102410370873. [DOI] [PubMed] [Google Scholar]
- 5.Weder CR, Schneemann M. Orphanet J. Rare Dis. 2019;4:15. doi: 10.1186/1750-1172-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemow DB, et al. J. Headache Pain. 2020;21:71. doi: 10.1186/s10194-020-01132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu P, et al. Nature. 2021;592:469–73. doi: 10.1038/s41586-021-03376-8. [DOI] [PubMed] [Google Scholar]
- 8.Michino M, et al. Pharmacol. Rev. 2015;67:198–213. doi: 10.1124/pr.114.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Nafria J, Nehme R, Edwards PC, Tate CG. Nature. 2018;558:620–623. doi: 10.1038/s41586-018-0241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin W, et al. Cell Discov. 2018;4:12. doi: 10.1038/s41421-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The corresponding coordinates and cryo-EM density map have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 7EXD, and in EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-31371.