Abstract
The rodents of hystricomorpha and sciuromorpha suborders exhibit remarkably lower incidence of cancer. The underlying genetic basis remains obscure. We report a convergent evolutionary split of human 3p21.31, a locus hosting a large number of tumour-suppressor genes (TSGs) and frequently deleted in several tumour types, in hystrico- and sciuromorphs. Analysis of 34 vertebrate genomes revealed that the synteny of 3p21.31 cluster is functionally and evolutionarily constrained in most placental mammals, but exhibit large genomic interruptions independently in hystricomorphs and sciuromorphs, owing to relaxation of underlying constraints. Hystrico- and sciuromorphs, therefore, escape from pro-tumorigenic co-deletion of several TSGs in cis. The split 3p21.31 sub-clusters gained proximity to proto-oncogene clusters from elsewhere, which might further nullify pro-tumorigenic impact of copy number variations due to co-deletion or co-amplification of genes with opposing effects. The split of 3p21.31 locus coincided with the accelerated rate of its gene expression and the body mass evolution of ancestral hystrico- and sciuromorphs. The genes near breakpoints were associated with the traits specific to hystrico- and sciuromorphs, implying adaptive significance. We conclude that the convergently evolved chromosomal interruptions of evolutionarily constrained 3p21.31 cluster might have impacted evolution of cancer resistance, body mass variation and ecological adaptations in hystrico- and sciuromorphs.
Subject terms: Cancer, Genome, Cancer
Introduction
Unlike myomorphs (mouse, rat, etc.), hystricomorphs (naked-mole rat, capybara, etc.) and sciuromorphs (squirrels, marmots, etc.) exhibit extraordinary diversity in body mass, longevity and ecological habitat1–12. One of the common traits of rodents of hystrico- and sciuromorpha suborders, which gained considerable attention recently, is their apparent resistance to cancer13–16. It has been proposed that the large body mass and long lifespan are the evolutionary forces that innovate cancer resistance in mammals13,16,17. Accelerated cell proliferation to gain large body mass might lead to cancer18–20. To minimise the risk of cancer, many rodents with large body mass have evolved with repressed telomerase activity to allow replicative senescence that suppresses cancer13,16. Similarly, to have a longer lifespan, a species may need to delay cancer incidence beyond its reproductive age. Long-living mammals, therefore, evolved with efficient cancer-resistance mechanisms13,16,21,22. The cancer resistance of hystricomorphs is particularly well-studied in naked-mole rat (NMR). Tian et al.14 has shown that NMRs contain high-molecular-mass Hyaluronan (HMM-HA), a component of the extracellular matrix, unlike other mammals which have low-molecular-mass HA (LMM-HA). This was presumably an evolutionary selection for highly elastic skin adapted to the underground habitat. Interaction of HMM-HA with the CD44 receptor induces p16, which in turn mediates early contact inhibition, an anticancer mechanism that arrests cell division when cells reach a certain density, in NMR14. HMM-HA also inhibits the process of metastasis23,24. The genome of NMR exhibits remarkable epigenetic stability and efficient DNA repair when compared with that of other mammals, which eventually contributes to cancer resistance25,26. Capybara, the largest extant rodent, exhibits cancer resistance through the expansion of genes associated with T-cell-mediated tumour suppression27. The Kurloff cells in guinea pig are shown to have anti-leukemic activities28. Squirrels also exhibit cancer resistance despite having high telomerase activity13. However, the underlying mechanisms are not known. Among other cancer-resistant mammals, genomes of elephants and whales have shown expansions in the copy number of tumour-suppressor genes29,30. Bats gained cancer resistance through an efficient genome maintenance mechanism31. These observations imply that cancer resistance had evolved independently in phylogenetically distant mammals and that multiple mechanisms can converge to endow cancer resistance to a species.
Copy number variation (CNV) is one of the common mechanisms leading to the gain or loss of relative abundance of proto-oncogenes (POGs) and tumour-suppressor genes (TSGs)32. Amplification of one or both copies of a POG can cause cancer32. Though both copies of a TSG need to be deleted to remove its tumour-suppressor impact, deletion of either copy predisposes the individuals to cancer32. POGs and TSGs can be found in individual gene clusters, likely constrained by concurrent chromatin states and coordinated transcription of co-positioned genes33. Clustering of POGs or TSGs accompanies a cost of increased susceptibility to cancer since several oncogenes can be co-amplified causing cancer or several tumour-suppressor genes can be co-deleted to predispose the individuals to cancer34,35. Examples include co-amplification of POGs at 8p11–1236, at 14q13.337–40 and co-deletion of TSGs at 8p2241 and at 3p21.3142–44 regions. On the contrary, many other POGs exhibit linear proximity to TSGs under an evolutionary constraint to nullify tumorigenic effect through co-amplification or co-deletion of oncogenes and tumour-suppressor genes45. We hypothesise that certain genomic rearrangements of POG and TSG clusters can endow cancer resistance to some mammals. Indeed, the widespread importance of genomic rearrangements in various ecological and evolutionary processes has been recognised46. Inversions alone can promote speciation either by generating large genetic barriers needed for reproductive isolation or by suppressing recombination to protect co-adapted alleles47–49. Apart from their role in speciation events50, genomic rearrangements are non-trivially linked to divergence of gene expression and of phenotypes51,52. It is, therefore, tenable to assess the relative genome organisation of cancer susceptible and cancer-resistant mammalian species in order to explore mechanisms of cancer resistance. Through analysis of 31 placental mammalian and 3 non-mammalian vertebrate genomes, we showed that TSG-enriched 3p21.31 locus of human was interrupted by large genomic inserts in hystrico- and sciuromorphs. The interruptions of the 3p21.31 locus were independent and did not share breakpoints in hystrico- and sciuromorphs. The locus remained syntenic in myomorphs as well as in other mammals. 3p21.31 is deleted in most cancers, and with a very high frequency in lung carcinoma, renal cell carcinoma, breast cancer and uterine cervix carcinoma43. The evolutionary split of this cluster can, therefore, endow cancer resistance by escaping the co-deletion of several TSGs in cis. We further inferred functional and evolutionary implications of our observations in terms of (i) evolutionary constraints that kept the tight synteny of 3p21.31 cluster throughout mammalia, and (ii) relaxed constraints on 3p21.31, body mass evolution and ecological adaptations in hystrico- and sciuromorphs.
Results
Genomic rearrangements and cancer resistance in naked-mole rat
We first identified the instances of genomic rearrangements between a cancer susceptible (human) and a cancer-resistant (NMR) mammal (Fig. 1a–c and S1a, b). Since genome assemblies of NMR are available as collections of large scaffolds and not as chromosomes, we only focussed on cis (intra-chromosomal) alterations in this study. We plotted intergenic distances between consecutive human genes against the distances between corresponding NMR genes. We focussed on values scoring high either on x axis (Human) or on y axis (NMR). We observed that the most meaningful changes were associated with human chromosome 3 mapping to NMR scaffold JH602043.1 (Fig. 1a). The highest value along y axis (NMR) marked the split of the 3p21.31 locus from a breakpoint flanked by NME6 and PLXNB1 genes. The second highest value along y axis involved chromosomal repositioning of only one gene (OSBPL11) and did not involve large genomic rearrangements. This gene was adjacent to a region annotated as ‘assembly exceptions’, implying the possibility of erroneous sequence mapping (Supplementary Fig. 1a). Similarly, the highest value along x axis involved repositioning of a single pseudogene OR9H1P, which might have resulted from differential retrotransposition in human and NMR (Supplementary Fig. 1b). Therefore, we ignored the events of single-gene repositioning and focussed only on 3p21.31 split.
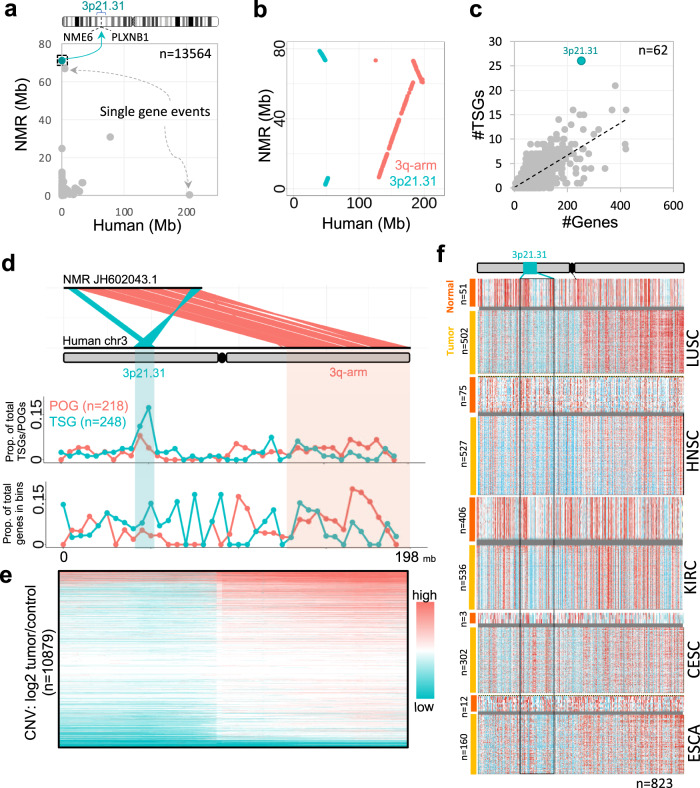
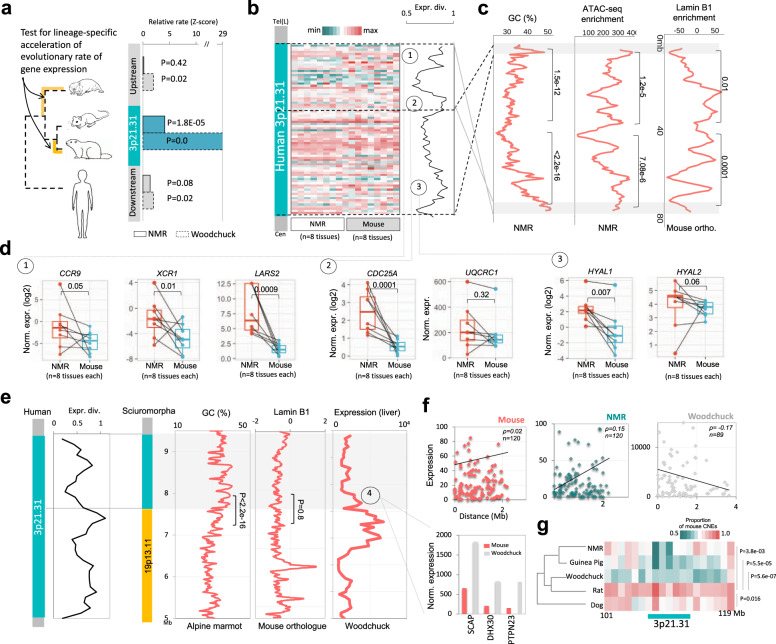
Fig. 1. Genomic rearrangements and tumour-suppressor properties of 3p21.31.
a Scatter plot of all intergenic distances between consecutive genes of human vs. that of corresponding NMR orthologues. The box on the y axis highlights the genes (NME6 and PLXNB1) that were proximal in human, but distant in NMR, and marks the large genomic split of 3p21.31 locus. The events of single-gene repositioning are marked by grey arrows. Details of these events are given in Supplementary Fig. 1a, b. b Dot plot between human chr3 and NMR JH602043.1 scaffold. Sea-green coloured dots represent 3p21.31 locus, while the 3q arm of chr3 is shown in red. c Number of tumour-suppressor genes as a function of the total number of genes in each chromosomal band of human genome. The 3p21.31 cluster is marked in sea-green colour. d Upper panel: linear view of genomic rearrangements between human chr3 and NMR JH602043.1. Middle and lower panels: density of proto-oncogenes and tumour-suppressor genes along human chr3, normalised by the total number of TSGs and POGs (upper panel), and by the total number of genes in respective chromosomal bands (lower panel). e Copy number variations along human chr3 in TCGA Pan-cancer dataset (n = 10,879). Each horizontal line represents one instance of CNV, each row represents one patient, and the colour grade scales to the tumour-to-control ratio of genomic DNA in TCGA Pan-Cancer dataset. f Gene expression (RNA-seq data normalised to TCGA cohort) of tumour and normal tissues along chromosome 3. Data shown in panels e and f were directly obtained from UCSC Xena browser. LUSC lung squamous cell carcinoma, HNSC head–neck squamous cell carcinoma, KIRC kidney renal clear cell carcinoma, CESC cervical squamous cell carcinoma and endocervical adenocarcinoma, ESCA oesophageal carcinoma.
Upon closer inspection of human chr3 and NMR JH602043.1 through dot plot, we found that human 3p21.31 locus and 3q arm were engaged in a complex rearrangement in NMR. In particular, human 3p21.31 locus was interrupted by 71 Mb region from 3q arm in NMR (Fig. 1b). We observed the same through analysis of another genome assembly53 of NMR (GCA_014060925.1, Supplementary Table 2 ), suggesting that the split of 3p21.31 locus in NMR was not a genome assembly artefact.
The 3p21.31 locus was highly enriched with TSGs (n = 25, Fig. 1c, d), while 3q arm was relatively enriched with POGs (Fig. 1d). This was coherent with the reports that 3p21.31 is a dominant tumour-suppressor locus and 3q26 and 3q29 are OncCassettes54–56. To further demonstrate tumour suppressing potential of 3p21.31, we browsed TCGA Pan-cancer data using UCSC Xena browser. The map of copy number variations (CNVs) confirmed that human 3p-arm exhibited more instances of deletions and, on the contrary, 3q arm had more of amplifications (Fig. 1e and S1c). Concomitantly, the locus also exhibited a greater frequency of somatic point mutations in TCGA Pan-cancer cohort (Supplementary Fig. 1d). By mapping gene expression data for various tumour types (TCGA cohort), we observed that genes on 3p-arm were mostly downregulated and those on 3q arm were upregulated in tumour samples when compared with normal (Fig. 1f). Clustering of TSGs at 3p21.31 locus allows their co-deletion or co-silencing, and subsequent predisposition to cancer42–44,57. The 25 known TSGs of 3p21.31 were split into sub-clusters of 16 and 9 TSGs by a 71 Mb insert from 3q arm in NMR. The extreme distance constraint would not have allowed the co-deletion (or co-silencing) of all TSGs of 3p21.31 in NMR, and, therefore, might have contributed to cancer resistance.
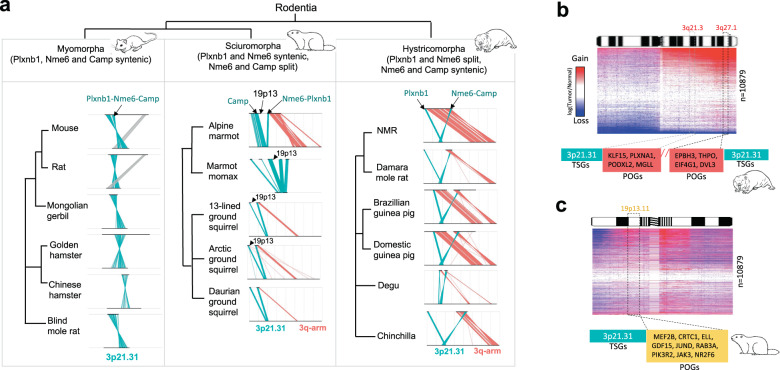
We further tested whether the observed rearrangement was specific to NMR. While we observed the synteny of 3p21.31 cluster in myomorphs, all analysed hystricomorphs exhibited the same rearrangement as in NMR, highlighting that the rearrangement occurred in the common ancestor of hystricomorphs (Fig. 2a and Supplementary Table 2 ). Interestingly, sciuromorphs exhibited an independent split of 3p21.31 from a different breakpoint, and involved an insert from 19p13.11 locus (Fig. 2a). While the precise length of this insert could not be determined due to fragmented genome assemblies, it was at least 8 Mb in alpine marmot. Importantly, like in NMR, split 3p21.31 also gained proximity to a known onco-cluster on 19p13.11 in alpine marmot, harbouring at least nine well-known POGs, and frequently amplified in Pan-cancer samples (Fig. 2b, c). The gained proximity to the POGs from 3q arm in hystricomorphs and from 19p13.11 in sciuromorphs might have allowed nullification of tumorigenic effects though co-deletion (or co-silencing) or co-amplification (or co-activation) of co-positioned TSG and POG clusters, as proposed elsewhere45.
Fig. 2. Synteny status of 3p21.31 in major rodent clades.
a Orthologous genomic position of human 3p21.31 genes in myomorphs, sciuromorphs, and hystricomorphs. The positions of PLXNB1, NME6 and CAMP genes are highlighted. PLXNB1-NME6 breakage marks the 3p21.31 split in hystricomorphs and NME6-CAMP breakage marks the split in sciuromorphs. b, c Gain of proximities between TSGs from 3p21.31 and the POGs from 3q21.3/3q27.1 in NMR (b), and from 19p13.21 in alpine marmot (c). Heatmaps represent relative gain and loss of CNVs in the PAN-Cancer cohort normalised to control samples.
Constraints on 3p21.31 locus in placental mammals
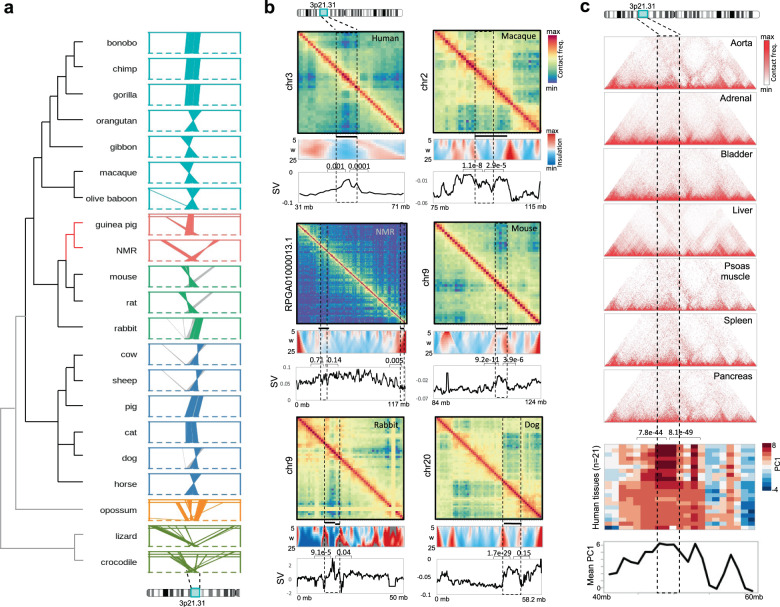
We further extended our analyses to other placental mammals, reptiles and a marsupial. The analysis suggested that 3p21.31 remained syntenic in placental mammals, but not in other terrestrial vertebrates (Fig. 3a), implying that the common ancestor of placental mammals gained the clustering of genes at 3p21.31 locus. We suggest that the selection for terrestrial and mammalian adaptations might underlie the observed gain of pronounced gene clustering at 3p21.31 in placental mammals, as discussed in detail in the ‘Discussion’ section.
Fig. 3. Linear and spatial synteny of 3p21.31 in placental mammals.
a Location of orthologous regions of human 3p21.31 in primates (bonobo, chimp, gorilla, orangutan, gibbon, macaque and olive baboon), rodents (guinea pig, naked-mole rat, mouse and rat), lagomorph (rabbit), artiodactyls (cow, sheep, pig), canines (dog and cat), perissodactyl (horse), marsupial (opossum) and reptiles (lizard and crocodile). Mouse and rat showed an independent split of a small terminal region of 3p21.31 (grey coloured). This region was outside the tumour-suppressor cluster of 25 genes. b Hi-C contact maps (1 Mb resolution), domainograms of insulation scores (200 kb resolution), and leading singular vectors (SV, 200 kb resolution) of 3p21.31 and adjacent regions in the liver of human, macaque, NMR, mouse, dog and rabbit. Black coloured bars above domainograms indicate TAD(s) overlapping with 3p21.31. P values were calculated for the difference of values within 3p21.31 and the adjacent regions of the same length using two-tailed unpaired t tests. c Hi-C contact maps (40 kb resolution), compartment scores (leading principal component as per Schmitt et al.130) of 3p21.31 and adjacent regions across different human tissues. P values for the difference of values within 3p21.31 and the adjacent regions of the same length were calculated using two-tailed unpaired t tests. Effect sizes: 3.35, 2.22 (human); 1.90, 1.27 (monkey); 2.69, 1.59 (mouse); 0.17, 0.74, 1.32 (NMR); 0.91, 0.44 (rabbit); 4.13, 0.28 (dog).
The 3p21.31 locus was spatially converged into one complex in human, rhesus macaque, mouse, dog and rabbit as revealed through the analysis of chromosome conformation datasets (Fig. 3b, c). Interestingly, 3p21.31 locus showed unique spatial seclusion from the rest of the chromosome 3 as inferred from strikingly low inter-loci, and high intra-locus contact frequencies in Hi-C contact maps across different tissues and species (Fig. 3b, c). The spatial seclusion of 3p21.31 locus was also confirmed through insulation scores, plotted as domainograms and the lead singular vectors (SV) of normalised Hi-C contact matrices. The 3p21.31 locus exhibited strong insulation signals at borders and converged to a compartment independent from the rest of the chromosome as marked by higher values of singular vector (Fig. 3b, c and Supplementary Fig. 2). Interestingly, the only other loci that interacted with 3p21.31 on chr3 were the regions marking the human NMR breakpoints on the chr3q arm (Supplementary Fig. 2), implying that the spatial proximity of these loci in the ancestral genome may have mediated the genomic rearrangements, as witnessed elsewhere58. In NMR, as expected, the split 3p21.31 sub-clusters were positioned in separate and distant topological complexes. However, interestingly, one of the split 3p21.31 clusters (left one in Fig. 3b—NMR) was not strongly insulated, and seemingly had spatial mixing with 3q. Since CNVs often span within the range of topological complexes, the above observation may support the possibility of co-deletion or co-amplification of split TSG cluster together with the adjacent POG cluster from 3q, which may nullify pro-tumorigenic potential of CNVs spanning genes of opposing effects, as mentioned earlier.
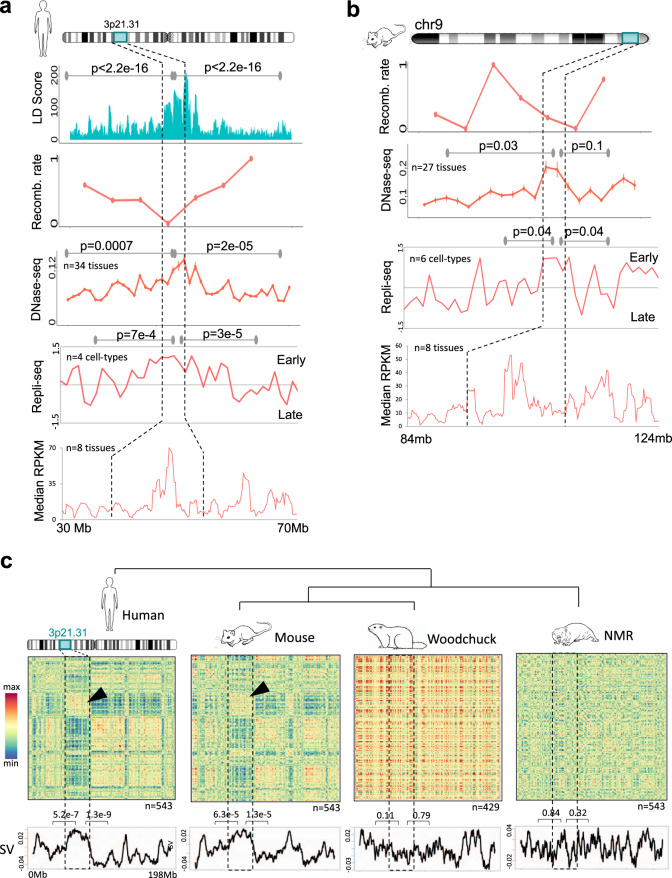
The locus exhibited constitutive early replication, low recombination rate, greater linkage, constitutive open chromatin and higher gene expression (median across eight tissues) in human and mouse, implying selection against expression noise, mutations, and allelic recombination that can break the combinations of beneficial and coordinating tumour-suppressor alleles (Fig. 4a, b). Since co-expression of genes is postulated as one of the potent constraints in the evolution of gene clusters59, we calculated the co-expression matrices using normalised expression data of several different tissues of human, mouse, NMR and woodchuck (‘Methods’). The genes within 3p21.31 were co-expressed in human and mouse, but not in NMR and woodchuck, again in a manner that was independent of the rest of chromosome 3 (Fig. 4c). The linearly and spatially constrained co-regulation of genes within 3p21.31 in mammals may imply selection on coordinated maintenance of transcriptional states of genes. Interestingly, some genes at 3p21.31 locus are known to interact at protein levels60. Their co-expression, therefore, may balance the protein concentrations of interacting partners. Further, the genomic rearrangements of 3p21.31, coincided with the diverged co-expression landscape of genes in hystrico- and sciuromorphs, suggesting the possibility that the relaxed constraint on co-expression might have allowed 3p21.31 to split. The genomic rearrangement of 3p21.3, in turn, may have further facilitated the expression divergence of 3p21.31 genes through the altered genomic neighbourhood.
Fig. 4. Genetic and epigenetic attributes of 3p21.31 in human and mouse.
a, b Linkage disequilibrium (LD) score, recombination rate, chromatin accessibility (across 34 human and 27 mouse tissues), replication timing (across four human and six mouse cell types) and the median expression levels of genes (across eight tissues) in human (a) and in mouse (b). P values for the comparison of values in the 3p21.31 locus and in the adjacent regions were calculated using two-sided Mann–Whitney U tests. Error bars represent 95% confidence intervals. Effect sizes: 0.25 and 0.29 for LD score; 0.63 and 0.41 for DNase-seq (human); 0.66 and 0.16 for DNase-seq (mouse); 0.99 and 1.28 for Repli-seq (human); 0.66 and 1.2 for Repli-seq (mouse). c Co-expression heatmaps of genes from human chromosome 3, and the corresponding orthologues in mouse, woodchuck and naked-mole rat. Below each heatmap is the leading singular vector (SV) of co-expression matrix. High overall SV values at 3p21.31 in human and mouse signify its secluded transcriptional co-regulation, which was largely independent from rest of the chr3. P values were calculated using two-tailed unpaired t-test using the same approach as in Fig. 3b. Effect sizes: 0.82, 1.01 (human); 0.64, 0.71 (mouse); 0.26, 0.04 (woodchuck); 0.02, 0.15 (NMR).
Overall, the conserved synteny in a wide range of phylogenetically distant placental mammals and the genomic attributes thereof highlight the functional constraints on 3p21.31 locus. Indeed, Makino et al. captured the 3p21.31 locus among the 37 highly conserved interacting gene clusters, which exhibit protein–protein interactions of their gene products, implying the constraints on the linkage of favourable alleles60.
Divergence of 3p21.31 in hystrico- and sciuromorphs
We tested if the gene expression within 3p21.31 evolved more rapidly in NMR and woodchuck as compared to mouse. We assumed Ornstein–Uhlenbeck (OU) model to calculate expression distances among species and assessed relative evolutionary rates of expression divergence in NMR and woodchuck against that of mouse, taking human as an outgroup. We used the method given by Ruan et al.61. Locus 3p21.31 exhibited a significantly greater evolutionary rate of expression divergence in NMR and woodchuck from that of mouse (PWoodchuck = 0.0, PNMR = 1.8e–05), while adjacent regions showed insignificant or marginally significant rates (PWoodchuck = 0.02, 0.02, PNMR = 0.42, 0.08; Fig. 5a). The expression divergence was particularly higher towards the DNA breakpoints, suggesting the possibility of chromosomal position effect following genomic rearrangements (Fig. 5b). Indeed, the split 3p21.31 sub-clusters gained proximities to AT-rich regions, which exhibited lower overall ATAC-seq signal in NMR, implying a relatively heterochromatic environment in-between split sub-clusters of 3p21.31 (PGC = 1.5e–12, <2.2e–16, PATAC-seq = 1.2e–5, 7e-6; Fig. 5c). This was reinforced by the significant enrichment of lamina-associated domains (LADs) in the 3q orthologue in mouse (PLAD = 0.01, 0.0001; Fig. 5c). These observations suggested that the genomic interruption of 3p21.31 in hystricomorphs might have caused the expression divergence of genes through gained proximity to relatively impermissible chromatin neighbourhood. Indeed, we observed relative downregulation of genes located near the breakpoints in NMR, but not in mouse (Fig. 5d–f). In particular, the presence and the relative downregulation of hyaluronidases (HYALs) and chemokine receptors (CCRs), located near the terminal breakpoints of 3p21.31, was important given their association with cancer resistance and other hystricomorph-specific phenotypes like elastic skin, lack of inflammatory response, etc. (Fig. 5d). Among other downregulated genes were LARS2, a mitochondrial Leucyl t-RNA synthetase gene, known to have an association with hearing loss in human62, CDC25A associated with vision63,64, and UQCRC1 associated with oxidative phosphorylation and known to be downregulated in hypoxic conditions65,66 (Fig. 5d). Sciuromorph orthologue of 3p21.31 also exhibited expression divergence near the breakpoints. Unlike the 3q-insert in hystricomorphs, 19p13.11 insert in sciuromorph had relatively less contrasting GC content to that of 3p21.31, was depleted in LaminB1 enrichment, and had high expression levels of genes (Fig. 5e). The 3p21.31 genes near the breakpoint had higher expression levels in woodchuck when compared with the mouse. These genes include SCAP, DHX30, and PTPN23. The SCAP gene functions in cholesterol and lipid homoeostasis67, DHX30 regulates the ribosome assembly68 and PTPN23 is a protein tyrosine phosphatase69 (Fig. 5e, f). All three functions, namely lipid homoeostasis, ribosome assembly and protein (de)phosphorylation have diverged regulation in sciuromorpha due to periodic hibernations70,71. These observations together supported the implications of chromosomal position effects associated with 3p21.31 rearrangement on expression and phenotypic divergence of hystrico- and sciuromorphs.
Fig. 5. Hystrico- and sciuromorph-specific relaxation of functional constraints on 3p21.31.
a Rate of gene expression evolution in NMR and woodchuck, considering mouse as control and human as an outgroup. Expression rates are presented as Z score and the P values are from Z test. b Heatmap: quantile-normalised expression data of 3p21.31 genes in the skin, thymus, ovary, kidney, adrenal, heart, testes and liver (in that order) of mouse and of NMR. Line plot: mouse-to-NMR expression divergence of genes. c GC content, chromatin accessibility (ATAC-seq enrichment in NMR skin fibroblasts), and ChIP-Seq LaminB1 enrichment (mouse fibroblasts) along NMR scaffold JH602043.1. P values were calculated using two-sided Mann–Whitney U tests. Effect sizes: 0.54 and 0.73 for GC content; 0.17 and 0.17 for ATAC-seq; 0.073 and 0.18 for LaminB1 enrichment. d Quantile-normalised gene expression values of genes (for the same tissues as in b) located towards the breakpoint regions that are highlighted in the line plot of 3p21.31 expression divergence. The matching tissues of the mouse and NMR are marked by connecting lines between paired boxes. P values were calculated using two-sided Mann–Whitney U tests. Boxplot annotation: centre line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. Effect sizes: 0.19, 0.28, 0.70, 0.70, 0.06, 0.39, and 0.15 from left to right. A comparison of normalised expression values of mouse and woodchuck genes near the breakpoint region-4 is shown in the barplot. e Left panel: gene-wise expression divergence between mouse and woodchuck along 3p21.31 locus. Right panel: GC content (Alpine marmot), LaminB1 enrichment (in mouse orthologous regions), and gene expression (woodchuck liver) across the rearranged locus involving part of 3p21.31 and 19p13.11 loci. P values were calculated using two-sided Mann–Whitney U tests. Effect sizes: 0.54 for GC content, 7.9e-6 for LaminB1 enrichment. f Expression of 3p21.31 genes in mouse, NMR and woodchuck as a function of distance from the nearest breakpoint. Mean expression levels across tissues were plotted. Effect sizes: Pearson’s correlation coefficient (ρ) are mentioned in the plots. g Proportion of mouse-conserved mammalian CNEs present in NMR, woodchuck, guinea pig, rat and dog. P values were calculated using two-tailed t tests between rat and other species. Effect sizes: 1.91, 3.24, 5.12, 1.65 for NMR–rat, GP–rat, Woodchuck–rat and dog–rat comparisons respectively.
It, however, remains counterintuitive that the expression of TSGs also diverged in the cancer-resistant rodent clades. Particularly, the relative downregulation of 3p21.31 genes, including TSGs, in NMR was surprising. We propose the following explanations: (i) TSGs are growth suppressors, and their expression divergence may relate to body mass diversification in the cancer-resistant rodent clades, as explored in further sections; (ii) complementary cancer-resistance mechanisms like multiple paralogous copies of TSGs72, the longer half-life of TSGs73, low mutation rate74, stable epigenome25, efficient DNA repair26, accumulation of high-molecular-weight hyaluronan14, etc. may have relieved the constraints on expression levels of TSGs at 3p21.31 in hystricomorphs.
Further, it is recognised that conserved non-coding elements (CNEs) impose constraints on the linear clustering of genes75. We, therefore, analysed the relative loss of CNEs in 3p21.31 region. We observed that NMR, guinea pig and woodchuck exhibited significant loss of mouse-conserved mammalian CNEs in 3p21.31 when compared with that in rat and dog (an outgroup)(PNMR = 3.18e–3, PGP = 5.5e–5, PWoodchuck = 5.5e7, PDog = 0.016; Fig. 5g). This concomitantly highlighted that the constraints on 3p21.31 synteny were relieved in hystrico- and sciuromorphs.
Accelerated rates of body mass evolution in hystrico- and sciuromorphs
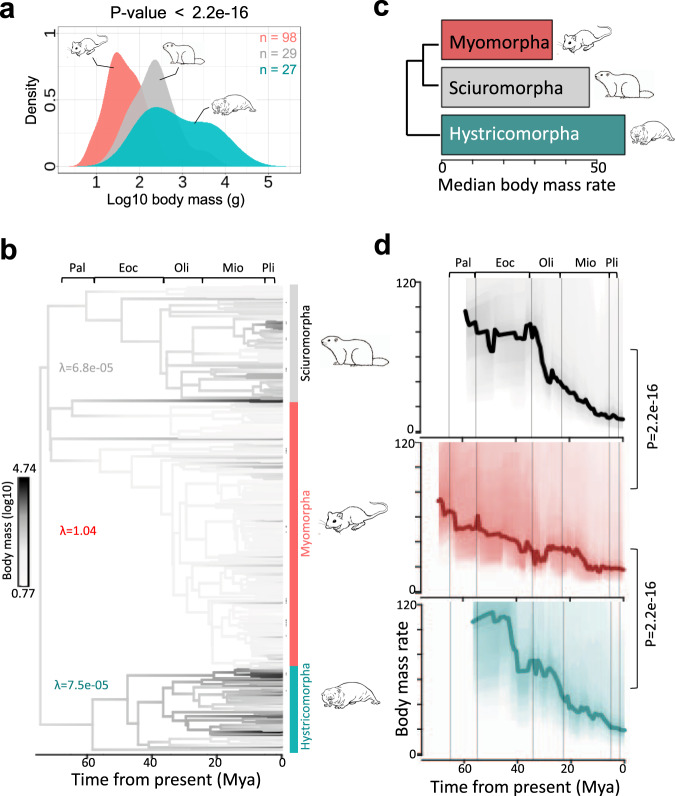
Tumour-suppressor genes are negative regulators of cell proliferation76–78. We indeed captured the ontology term ‘negative regulation of cell growth’ among 3p21.31 genes (FDR < 0.05, Fig. 7a). We, therefore, tested if the evolutionary rates of body mass variation differed in myomorphs, sciuromorphs and hystricomorphs. We first confirmed that their body mass differed significantly, and the latter two exhibited a greater mean and variance thereof (P = 2.2e–16; Fig. 6a, b). Indeed, the diversity of body mass in sciuro- and hystricomorphs was also apparent from the Pagel’s λ values of the three clades. Pagel’s λ captures the tendency of a trait distribution in a phylogenetic clade to match with the expected distribution under the assumption of the Brownian model of evolution79. A λ value closer to 1 signifies a higher degree of similarity of trait of a species to its phylogenetically closer, as compared to distant, species. Close to 1 value of Pagels’ λ is also sometimes considered as a proxy to higher evolutionary constraints on a trait. Pagels’ λ value significantly less than 1 might, but not necessarily, mark a lower evolutionary constraint on a trait. Accordingly, we observed that myomorpha clade had λ = 1.04, while sciuromopha and hystricmorpha clades had λ = 6.8e–05 and 7.5e = 05, respectively (Fig. 6b).
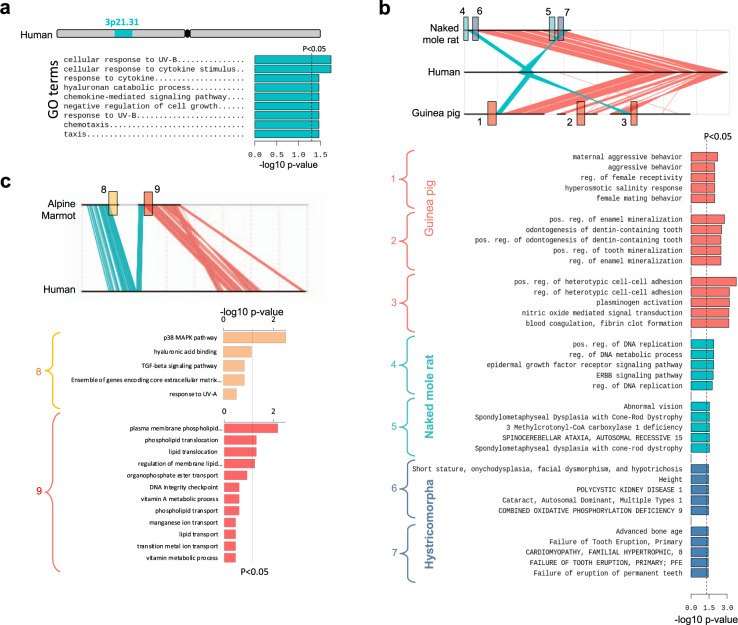
Fig. 7. Function of genes located within 3p21.31 and near the breakpoints in hystrico- and sciuromorphs.
a Enrichment of gene ontology terms in 3p21.31 locus. b, c Enrichment of gene ontology terms in regions near various breakpoints in NMR, guinea pig, and alpine marmot. The breakpoint regions 1–3 are specific to guinea pig. Regions 4–5 are specific to NMR, 6–7 are common to both the hystricomorphs, and 8–9 are specific to Aalpine marmot. All gene ontology analyses were performed using ToppGene suite with P-value correction through Benjamini–Hochberg method.
Fig. 6. Rate of body mass evolution in rodent clades.
a Distribution of log10 body mass of myo-, hystrico- and sciuromorphs. P value was calculated using Kruskal–Wallis test. b Ancestral reconstruction of body mass across phylogenetic tree of 312 rodent species. Pagel’s λ values were calculated using ‘phyloSig’ function of ‘phytools’. c Median rate of body mass evolution in myo-, hystrico- and sciuromorphs. d Rate of body mass evolution in myo-, hystrico- and sciruomorphs as a function time from the present. Body mass rates were calculated using the BAMM model. P values were calculated using two-sided Mann–Whitney U tests.
Through Bayesian Analysis of Macro-evolutionary Mixtures (BAMM)80, we calculated phylogenetic rates of body mass (log10) evolution across 312 rodents for 1 million generations. Fig. 6c, d showed significantly higher rates of body mass evolution in sciuro- and hystricomorphs, particularly during Eocene epoch, when compared with that of myomorphs (P = 2.2e–12). Indeed, Eocene marked the explosive radiations of mammals that produced the great diversity of mammals known today. The higher rate of body mass evolution in the ancestral sciuromorph and the ancestral hystricomorph also coincided with our observation that the 3p21.31 locus was split into all the analysed hystricomorphs and sciuromorphs. i.e., 3p21.31 locus likely split in the independent common ancestors of sciuromorphs and hystricomorphs. Based on these observations, we infer that the evolutionary constraint on the body mass was relaxed during the speciation of sciuromorpha and hystricomorpha rodents. This can be a consequence of Eocene climate change. The mid-Eocene is marked with emergence of more open habitats like savannah and scarce woodlands, as inferred from phytolith assemblage analyses. The constraints from cellulose fermentation due to dietary shift from browsing to grazing, the population dynamics of competitors and predatory carnivores, etc. might have impacted the rate of body mass evolution81,82.
Split 3p21.31 gained proximity to genes associated with hystrico- and sciuromorpha-specific traits
The genes within 3p21.31 were associated with negative regulation of growth, response to UV-B, response to cytokines/chemokines, hyaluronan catabolism, etc., hinting at the functions that are needed for terrestrial adaptations of most mammals, but are diverged in hystricopmorphs and sciuromorphs ('Discussion' section). To further understand the functional relevance of the observed genomic rearrangements, we tested the involved breakpoint regions, outside 3p21.31, in hystrico- and sciuromorphs for gene ontology enrichments. Genes near breakpoints in hystricomorphs were associated with hystricomorph-specific traits, like dental/enamel mineralisation, mating behaviour, blood coagulation, oxidative phosphorylation, DNA metabolic process, EGFR pathway, vision etc. (FDR < 0.05; Fig. 7b). Most of these terms are tightly linked to the ecological adaptations of hystricomorphs. For example, it is known that the teeth and the dentary organisations are adapted to assist in digging action in naked-mole rat, and grinding action in guinea pig83–85. Some hystricomorphs, like guinea pigs, exhibit diverse mating behaviour ranging from monogamy to promiscuity86. Naked-mole rats, in contrast, have an organised eusocial-type mating behaviour7. Blood coagulation has been proposed to be an adaptive trait of hystricomorphs to efficiently repair the wounds incurred due to digging, fighting, and being preyed63,87,88. Oxidative phosphorylation term is associated with hypoxic conditions in underground and semi-aquatic habitats of some of the hystricomorphs63,89–92. DNA metabolic process is associated with DNA repair mechanisms and it is well appreciated that some hystricomorphs have efficient DNA repair systems26,93. EGFR pathway is associated with wound healing and pain sensitivity or lack thereof94–96. The subterranean hystricomorphs are also known to have poor vision97, though whether or not loss of vision is adaptive or neutral is arguable. Similarly, 19p13.11 genes and 3q genes that gained proximities to 3p21.31 split sub-cluster in sciuromorphs were associated with MAP kinase pathway, hyaluronan binding, ECM, lipid transport, vitamin A metabolism, telomere maintenance, etc. (Fig. 7c and Supplementary Table 3). These functions have known associations with torpor and hibernation in sciuromorphs. Concerted changes have been observed in the MAP kinase pathway during hibernation, which may relate to stress tolerance in sciuromorphs during hibernation98. The cerebral cortex transcriptome of hibernating ground squirrels shows peaked expression of extracellular matrix components, including hyaluronan-binding proteins, laminins and collagens99. Dynamic regulation of ECM components may endow plasticity to the hibernating brain. Further, hibernating mammals primarily feed on stored lipids, which are accumulated in bulk during the activity period. Dramatic alterations in fuel utilisation are the hallmarks of hibernation, and explain the enrichment of lipid metabolism and transport-related terms70. Similarly, genes associated with vitamin A metabolism and transport are upregulated during hibernation, suggesting their potential role in transducing photoperiod100. Hibernating rodents also exhibit seasonal variation in telomere length101. In particular, their telomere lengths increase during hibernation, which may slow ageing.
The analyses of gene annotations, therefore, highlighted the adaptive significance of 3p21.31 rearrangement in the evolution of traits specific to hystrico- and sciuromorphs.
Discussion
Fronicke et al. had earlier proposed the eutherian ancestry of 3p21.31 cluster102. The lack of 3p21.31 clustering in marsupial and non-mammalian vertebrates suggested that the clustering was gained in the common ancestor of placental mammals. Remarkable regenerative capabilities of vertebrates like teleosts, reptiles, amphibians and marsupials likely conferred anti-tumorigenic properties to these vertebrates103,104. Mammals may have lost regenerative abilities during evolution, possibly as a trade-off between wound healing and tissue regeneration105. Being homoeothermic, mammals cannot tolerate excessive bleeding and are likely to die before any significant tissue regeneration105. Instead, efficient wound healing became essential in mammals, though at the cost of lost anti-tumorigenic ability105. Mammals, therefore, evolved with other mechanisms to select against tumorigenesis. Quality control of tumour-suppressor mechanisms can be one strategy to effectively avoid cancer. Early replication and consistently open chromatin state ensure genetic integrity of tumour-suppressor loci through minimising genetic and epigenetic lesions owing to spatially central localisation in the cell nucleus and ample availability of DNA proofreading and repair enzymes to the early replicating open chromatin106–111. An effective way of ensuring these properties is through positional clustering of genes33,112,113, as in the case of 3p21.31. The coordinated alteration in chromatin states and expression of genes in 3p21.31 had been observed by others57. It has been proposed that the clustering of TSGs evolved to suppress tumours in a coordinated manner57. Interestingly, Makino et al.60 captured 3p21.31 as one of the highly conserved ‘interacting gene clusters’, which are constrained by the protein–protein interactions of the linearly co-positioned genes60. The lower recombination rate of the 3p21.31 locus can be explained by linkage constraint to keep the combination of fitter alleles together. Yeaman114 had postulated that the genes important for adaptations to the local environment tend to cluster together in order to keep the combination of fitter alleles given that the adaptation is guided by the change in allelic frequency at multiple loci114. On similar lines, we propose that the clustering of genes associated with growth limitation, UV sensitivity and chemokine response at 3p21.31 together might represent the shared adaptation to the terrestrial habitat of mammals (Fig. 7a). The enrichment of the term ‘negative regulation of growth’ might relate to constraints on the body mass of terrestrial mammals by factors like (i) selection against cancer115, (ii) increased energetic cost of being endothermic116, (iii) energy investment in gestation117 and (iv) skeletal and muscular limitation of limbs to support a load of body mass under the influence of gravity118. Terrestrial adaptation to protect against UV radiation explains the enrichment of genes associated with the term ‘response to UV radiation’119,120 (Fig. 7a). Inflammation and deposition of matrix proteins are the essential steps in wound healing121,122. The enrichment of chemokine receptors and matrix proteins (collagen, laminin, etc.) in the 3p21.31 locus may explain the enrichment of ‘response to chemokine stimulus’ term (Fig. 7a). These proposals might largely explain the evolutionary constraints on the synteny of the 3p21.31 cluster in placental mammals.
Quantitative evidence of body mass variations in hystrico- and sciuromorphs is overwhelming1,123–125. We showed that the rate of body mass evolution was higher in the ancestral hystricomorph and sciuromorph, which coincided with the ancestral nature of the 3p21.31 split, and the higher expression rate of genes therein. Indeed, the expression divergence of most tumour-suppressor genes, including that of 3p21.31, is associated with growth-related phenotypes76–78. It is, therefore, tenable to reconcile that split of 3p21.31 locus and the expression divergence thereof might associate with the body mass divergence in hystrico- and sciuromorphs during Eocene, an epoch known to be associated with the emergence of open grasslands, widespread body mass divergence, and mammalian radiation. There are also evidence that many ancestral rodents, like those of South America, attained body mass in the open grassland due to the absence of competitors and the placental carnivores81. These associations imply the following possibilities:
Hystrico- and sciuromorphs were released from the evolutionary constraints on the body mass of ancestral rodent, due to the emergence of more open habitat, wider home range, and relative under-representation of competitors and predators. This might have relaxed the functional constraints on 3p21.31 synteny, i.e. the expression levels of growth suppressor genes were allowed to diverge. Figure 6a, b supported the relaxation of body mass constraint in hystrico- and sciuromorphs.
The complementary cancer-resistance mechanisms like efficient DNA repair, duplications of TSGs, the longer half-life of TSGs, stable epigenome, etc. might have allowed the relaxation of selection pressure to keep all TSGs of 3p21.31 together for coordinated tumour suppression. Consequently, the split of 3p21.31 was tolerated during the speciation of cancer-resistant rodent clades.
Hystrico- and sciuromorphs gained an adaptive advantage of the 3p21.31 split and expression divergence thereof. This was supported by the presence of genes associated with hystrico- and sciuromorpha-specific traits in the genomic regions that gained proximity to split 3p21.31 sub-clusters (Fig. 7 and Supplementary Table 3). The observed genomic rearrangements of the 3p21.31 locus may, therefore, be evolutionarily selected.
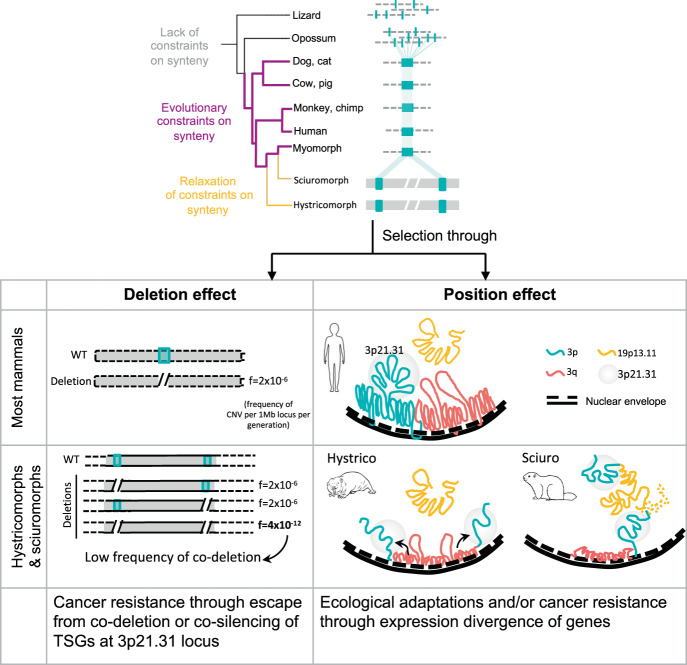
It was intriguing that 3p21.31 locus was split from different breakpoints, and had acquired different neighbourhoods in hystrico- and sciuromorphs. The convergent nature of 3p21.31 split in different phylogenetic clades argues strongly against the neutral drift, possibilities of genome assembly artefacts, etc., and instead represents independent ‘biological replicates’ confirming the outcome of an evolutionary experiment and natural selection thereof. Based on our observations, we propose a model for the evolution of cancer resistance in hystrico- and sciuromorph through genomic interruption of 3p21.31 TSG cluster (Fig. 8).
Fig. 8. The proposed model for the evolution of cancer resistance in hystrico- and sciuromorphs.
The frequency of CNVs per Mb per generation was obtained from Itsara et al.133.
It is also notable that, unlike myomorphs, most hystrico- and sciuromorphs exhibit di-urnality, which strongly associates with high speciation rates in mammals. The large structural variations in genome promotes speciation by reducing the fertility of heterozygotes due to disruptive chromosomal segregations and the meiotic silencing of unpaired chromosomes126. Coherently, hystricomorpha exhibits a high rate of karyotype evolution127,128 as well as one of the largest shifts towards high speciation rate across mammals. These observations together support the possibility that: (i) the high rate of speciation may itself have innovated unique traits like cancer resistance among others, or conversely, (ii) the relative ‘evolvability’129 of lineages, marked by a distinctly higher rate of trait diversification, in this case, may have shaped the dynamics of speciation, further widening the scope our findings.
In summary, we showed that the linear clustering of genes, while having functional and evolutionary significance, might have predisposed mammals to pro-tumorigenic copy number variations. Genomic interruptions of proto-oncogene and tumour-suppressor gene clusters might, therefore, endow cancer resistance to some mammals, like hystricomorphs and sciuromorphs. Convergent evolution of 3p21.31 split further negated the possibility of neutral evolution or technical artefacts and strengthened the underlying evolutionary significance. Altogether, our observations highlight that linear and spatial alterations in genome organisation might guide the evolution of lineage-specific phenotypes.
Methods
We did not use any statistical method to predetermine sample size. We did not randomise any experiments. We were not blinded to allocations during experiments and outcome assessment. We mentioned the sample sizes and statistical tests wherever applicable. The Source of each dataset is listed in Supplementary Table 1.
Analysis of genomic rearrangements between NMR and human
We obtained coordinates of orthologous genes of human (hg38) and NMR (hetGla2) from Ensembl biomart (https://www.ensembl.org/biomart/martview). Only one-to-one orthologues were considered to map synteny and genomic rearrangements. We calculated distances between transcription start sites of consecutive gene-pairs of human genome, and scatter-plotted against those of corresponding NMR orthologues using the ‘geom_point’ function of ‘ggplot2’ R-package (https://ggplot2.tidyverse.org/).
Dot plot
To obtain the dot-plot view of human chr3 and NMR scaffold JH602043.1, we scatter-plotted the gene-TSS coordinates of human chr3 and that of corresponding orthologues in NMR using the ‘geom_point’ function of ‘ggplot2’ in R-package.
Analysis of cancer-associated properties of human chr3
We obtained the lists of tumour-suppressor genes (TSGs, n = 2070) and proto-oncogenes (POGs, n = 2511) from CancerMine database (https://bionlp.bcgsc.ca/cancermine/). Genes were binned in 5-mb bins, and the proportions of TSGs and POGs were calculated by: (i) dividing TSG and POG counts by total number of TSG and POGs, respectively, in the genome, and (b) dividing TSG and POG counts by the total number of genes in each bin. For copy number variation analysis, we obtained log2 ratio of tumour-to-normal values of genomic copy numbers across human chr3 from TCGA Pan-Cancer (TCGA-PANCAN) data available at UCSC Xena browser (https://xenabrowser.net/). Similarly, the gene expression data of normal and cancer samples were obtained from ‘IlluminaHiSeq pancan-normalised’ track of Xena browser for various different TCGA cancer types. The heatmaps of log2-transformed values were obtained directly from the Xena browser and colours were transformed to green (#339999) and red (#FF6666) colours using gimp tool (https://www.gimp.org). Somatic point mutation data in TCGA-pancan cohort was also directly obtained from Xena browser.
Mapping of 3p21.31 locus across other genomes
We obtained the chromosomal coordinates of orthologous genes of human (hg38) in the following species: Alpine marmot (marMar2.1), woodchuck (GCA_901343595.1), 13-lined ground squirrel (SpeTri2.0), arctic ground squirrel (ASM342692v1), daurian ground squirrel (ASM240643v1), mongolian gerbil (MunDraft-v1.0), golden hamster (MesAur1.0), chinese hamster (CriGri_1.0), blind mole rat (S.galili_v1.0), damara mole rat (DMR_v1.0), brazilian guinea pig (CavAp1.0), degu (OctDeg1.0), chinchilla (ChiLan1.0), NMR (hetGla2, GCA_014060925.1), mouse (GRCm38.p6), rat (Rnor_6.0), domestic guinea pig (Cavpor3.0), bonobo (panpan1.1), chimpanzee (Pan_tro_3.0), gorilla (gorGor4), orangutan (PPYG2), gibbon (Nleu_3.0), macaque (Mmul_8.0.1), olive baboon (Panu_3.0), rabbit (OryCun2.0), cow (ARS-UCD1.2), sheep (Oar_v3.1), pig (Sscrofa11.1), cat (Felis_catus_9.0), dog (CanFam3.1), horse (EquCab3.0), opossum (monDom5), lizard (AnoCar2.0), and crocodile (CroPor_comp1) from Ensembl biomart (https://www.ensembl.org/biomart/martview/). One-to-many orthologues, and scaffolds with <5 genes were removed. To plot the synteny maps, we paired together the TSS coordinates of orthologous genes of two species in a single data column. An empty line separated each pair. The second data column contained a binary vector; ‘0’ for the coordinates of the first species and ‘1’ for that of other species. The two columns were scatter-plotted using ‘plot’ function with the type = ”l” parameter on R-package. Wherever the split 3p21.31 clusters could not be mapped on a single scaffold, locations of genes flanking the breakpoint were assessed. The split occurred between Plxnb1 and Nme6 genes in hystricomorphs, and between Nme6 and Camp genes in sciuromorphs. If the genes flanking the breakpoint were on different scaffolds and their immediate neighbourhood had genes from elsewhere (from 3q and 19p13 in hystrico- and sciuromorphs, respectively), the 3p21.31 is considered as split.
Hi-C analysis
SRA files of liver Hi-C datasets of mouse, dog, rabbit, and rhesus macaque were downloaded from Gene Expression Omnibus (GEO) with accession ID GSE65126 (Supplementary Table 1). Human liver Hi-C was downloaded from GSE58752 (Supplementary Table 1). NMR Hi-C data of embryonic fibroblasts was obtained from SRR8204318. SRA files were converted into fastq files using ‘fastq-dump’ of NCBI SRA Toolkit (https://github.com/ncbi/sra-tools) and processed using HiCUP (https://www.bioinformatics.babraham.ac.uk/projects/hicup/) with the parameter for restriction enzyme as ‘HindIII’ for human, mouse, dog, rabbit and macaque Hi-C data, and as ‘DpnII’ for NMR. The valid Hi-C pairs from HiCUP were built into contact maps, which were further normalised at 200 kb resolution using ‘simpleNorm’ method available in ‘Homer’ package (http://homer.ucsd.edu/homer/). Normalised contact maps of 3p21.31 and + /− 15MB neighbouring regions in human and other species were drawn using ‘heatmap’ function with scale ‘none’ on R. Snapshots (chr3:30–70 mb at 40 kb resolution) of Hi-C contact maps of other human tissues were directly obtained from PennState Hi-C browser (http://promoter.bx.psu.edu/hi-c/). The insulation domainograms were obtained using ‘insulation_domainogram’ function of GENOVA R-package (https://github.com/robinweide/GENOVA). The leading singular vectors of normalised Hi-C matrices were obtained using ‘svd’ function in R (https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/svd). For the Hi-C matrices of various human tissues, the compartment scores (leading principal component) were directly obtained from Schmitt et al.130.
Analysis of linkage disequilibrium (LD) and recombination rates
We used LD Score to analyse the linkage of alleles on human chr3. LD Score is the property of an SNP defined as the summation of Pearson’s correlation values r2, with all other SNPs in a 1 cM (centi Morgan) window. Human LD Score data for European-ancestry was obtained from download-server of Broad Institute (https://data.broadinstitute.org/mpg/snpsnap/database_download.html). LD scores were plotted using ‘geom_polygon’ function of ‘ggplot2’ R-package. P values for comparison between 3p21.31 (chr3:44–51 mb) and its neighbouring regions (upstream: 30–37 mb and downstream: 63–70 mb) were calculated using two-sided Mann–Whitney U tests on R. The recombination rates (at 5 mb resolution) of human and mouse genomes were obtained from Jensen-Seaman et al.131. The coordinates were lifted over to hg19 (human) and mm9 (mouse) assemblies using UCSC’s ‘liftover’ utility (https://genome.ucsc.edu/cgi-bin/hgLiftOver). The recombination rates were plotted without any further processing.
Chromatin accessibility analysis
DNase-Seq data for 42 human and 27 mouse tissues were downloaded from ENCODE as bigWig files (Supplementary Table 1). These files were converted to ‘bedgraph’ files using the UCSC’s ‘bigWigToBedGraph’ utility. We binned DNase-seq signals at 1 Mb resolution and plotted mean (+/− 95% confidence intervals) values along 3p21.31 and adjacent loci. P values for comparison of DNase-seq enrichment at 3p21.31 region and neighbouring regions (25–35 mb upstream and 65–75 mb downstream) were calculated using two-sided Mann–Whitney U tests on R. The ATAC-seq data of NMR skin-fibroblast (Supplementary Table 1) was mapped along JH602043.1 scaffold (hetGla2) at 500 kb bins and smoothened using ‘running.mean’ function of ‘igraph’ R-package. P values for the comparisons of ATAC-seq signals at 0–4.6 mb and 37–42 mb regions, and at 37–42 mb and 73.4–78.8 mb regions of NMR scaffold were calculated using two-sided Mann–Whitney U tests on R.
Replication timing analysis
The ‘bigwig’ files of Repli-chip data for four human cell-lines (GM06990, H9ES, HELA, and IMR90) and six mouse cell-lines (CH12, EPISC, ES, L1210F, MEFM, and MEL) were obtained from ENCODE (Supplementary Table 1) and converted to ‘bedgraph’ files using UCSC’s ‘bigWigToBedGraph’. We binned the Repli-chip signals at 1 Mb resolution and plotted mean (+/− 95% confidence intervals) values of Repli-chip enrichment along human chr3 and mouse chr9. P values were calculated using the same approach as in preceeding section.
Gene expression, co-expression and divergence analyses
We calculated RPKM values from RNA-seq reads of the skin, heart, ovary, testis, liver, kidney and thymus tissues of adult NMR, guinea pig, human and mouse (Supplementary Table 1). For woodchuck, we obtained the data for the kidney, spleen and liver of adults (Supplementary Table 1). The datasets were quantile-normalised across tissues and species using ‘normalise.quantiles’ function of ‘preprocessCore’ R-package (https://rdrr.io/bioc/preprocessCore/) for the pair-wise comparisons of the expression levels. Median expression values across tissues were plotted for human chr3, and for corresponding orthologues in mouse. For individual genes, the quantile-normalised values were plotted as boxplots using ‘geom_boxplot’ function of ‘ggplot2’ R-package. P values for differential gene expression were calculated using two-sided Mann–Whitney U tests on R. For co-expression analysis, we calculated Pearson’s correlation coefficients among all genes ordered along human chromosome 3 using ‘cor’ function in R. The gene order of human chr3 was considered as reference for all co-expression matrices. The correlation matrices were further smoothened using the equation ni,j = (2mij + mi-1,j + mi,j-1 + mi+1,j+1 + mi,j+1)/6 in three sequential iterations, where n is the smoothened matrix, m is the original matrix and i and j are the row and column indexes of matrices. This equation was implemented using ‘matrixSmooth’ function of ‘oce’ R-package(https://rdrr.io/cran/oce/man/oce.html). We plotted co-expression heatmaps using ‘pheatmap’ package using ‘spectral’ colour brewing scheme. The leading singular vector (SV) of co-expression matrix was calculated using ‘svd’ function in R. Expression divergence (d) of a gene between two different species was calculated using equation di = 1- ρ(xi, yi) where xi and yi are vectors of quantile-normalised expression values of gene i across multiple tissues of species x and y. The divergence values were smoothed using ‘running.mean’ function of ‘igraph’ R-package with binwidth=10.
Analysis of conserved non-coding elements (CNEs)
Hg19 chromosomal cooridnates of mammalian CNEs present in 3p21.31 region were obtained from132 and lifted over to mouse (mm10) using ‘liftover’ (https://genome-store.ucsc.edu/) chains at 0.95 mapping coverage. The mouse-conserved CNEs were further lifted over to NMR (HetGla2), guinea pig (CavPor3), Woodchuck (GCA_901343595.1), rat (Rn6), and dog (CanFam3) using the same parameters. The CNEs that were partially or fully deleted in respective species were considered as lost cases in that species. The proportions of lost cases were calculated for each species.
Gene ontology (GO)
All gene ontology analyses were performed using ToppGene Suite (https://toppgene.cchmc.org/) with Benjamini–Hochberg correction of P values.
GC content along NMR and alpine marmot scaffolds
The genome sequences (fasta) of NMR and alpine marmot were downloaded from the UCSC genome browser, and NCBI respectively. The DNA sequences of NMR scaffold JH602043.1 and alpine marmot scaffold CZRN01000025.1 were fetched and the percentage of ‘G + C’ nucleotides in 100-kb bins were calculated. The values were smoothened using ‘running.mean’ function of ‘igraph’ R-package with binwidth=10. P values for the difference in GC contents between 0–4.6 mb and 37–42 mb, and between 37–42 mb and 73.4–78.8 mb regions were calculated using two-sided Mann–Whitney U tests on R. For alpine marmot, the size of split 3p21.31 sub-cluster and the 19p13.11 locus were comparable, and therefore GC% values in the whole regions were used to calculate p values using two-sided Mann–Whitney U tests.
LaminB1 enrichment analysis
LaminB1 ChIP-Seq (MEF) data of mouse orthologous regions of NMR scaffold JH602043.1 and alpine marmot scaffold CZRN01000025.1 were taken from GSE17051 (Supplementary Table 1). Smoothened values were plotted along scaffolds. P values for comparison of enrichment at 0–4.6 mb and 37–42 mb, and at 37–42 mb and 73.4–78.8 mb NMR orthologous regions in mouse were calculated using two-sided Mann–Whitney U tests. Like in the case of GC content, the LaminB1 enrichment values in the whole of split 3p21.31 and 19p13.11 regions were used to calculate P values using two-sided Mann–Whitney U tests.
Evolutionary rate calculations
Evolutionary rates of lineage-specific expression divergence for the genes within 3p21.31 locus and upstream/downstream loci (50 genes each side) were calculated and tested for significance using ‘TreeExp’ R-package (https://github.com/hr1912/TreeExp). NMR and woodchuck were taken as ‘test’, mouse was considered as ‘reference’, and human served as ‘out-group’ in the analysis. Body mass rates for 312 rodent species were calculated using ‘BAMM’ package (http://bamm-project.org/). We plotted body mass rate as a function of evolutionary time using ‘BAMMtools’ R-package (https://cran.r-project.org/web/packages/BAMMtools/index.html). Phylogenetic tree (Newick format) was obtained from TimeTree (http://www.timetree.org/) (Supplementary Table 1). Body mass and activity data of extant rodents and their inferred ancestors were superimposed onto phylogenetic trees using ‘contMap’ function of ‘phytools’ R-package (http://www.phytools.org/). Pagel’s λ was calculated using ‘phyloSig’ function of ‘phytools’. Density plots of body mass for the rodent suborders were made using ‘geom_density’ function of ‘ggplot2’ R-package.
Statistical tests
We mostly used two-sided Mann–Whitney U test and two-tailed t test to assess the statistical significance. Effect sizes were calculated using Cohen’s d and η2 for t tests and Mann–Whitney U tests using Eqs. (1) and (2), respectively.
| 1 |
Where, m1 and m2 are mean of sample 1 and 2, respectively, and Spooled is the pooled standard deviation of two samples.
| 2 |
where, and
n1 and n2 are sizes of samples 1 and 2. U is the Mann–Whitney U value. mU is the mean U value. z is the Z score of U value calculated as above.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Fellowship support to Y.J., K.R.C. and J.B. from DBT, UGC, and DST, respectively, are duly acknowledged.
Author contributions
Y.J. and K.R.C. performed most of the analyses. A.A.V. conceived the idea and produced the initial results. J.B., M.L., M.B. and H.S. helped in the data analyses. K.S.S. improvised and supervised the project, contributed to the data analyses and figure preparation and wrote the manuscript.
Data availability
The mapped datasets are available as online Supplementary Data.
Code availability
The custom scripts used for data analyses are available on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yachna Jain, Keerthivasan Raanin Chandradoss, Anjoom A. V.
Supplementary information
The online version contains supplementary material available at 10.1038/s41514-021-00072-9.
References
- 1.Álvarez, A., Arévalo, R. L. M. & Verzi, D. H. Diversification patterns and size evolution in caviomorph rodents. Biol. J. Linn. Soc.121, 907–922 (2017).
- 2.Long, J. L. Introduced Mammals of the World: Their History, Distribution and Abundance, Vol. 1 (CSIRO Publishing, 2003).
- 3.Matthews LH. Hibernation in mammals and birds. Br. Med. Bull. 1961;17:9–13. doi: 10.1093/oxfordjournals.bmb.a069880. [DOI] [PubMed] [Google Scholar]
- 4.Nedergaard J, Cannon B. Mammalian hibernation. Philos. Trans. R. Soc. L. B Biol. Sci. 1990;326:669–685. doi: 10.1098/rstb.1990.0038. [DOI] [PubMed] [Google Scholar]
- 5.Ronald, M. Nowak, E. P. W. Walker’s Mammals of the World. Vol. 1 (Johns Hopkins University Press, 1999).
- 6.Michael, A. Mares, H. H. G. Mammalian Biology in South America, Vol. 1 (Pymatuning Laboratory of Ecology, University of Pittsburgh, 1981).
- 7.Jarvis JU. Eusociality in a mammal: cooperative breeding in naked mole-rat colonies. Science. 1981;212:571–573. doi: 10.1126/science.7209555. [DOI] [PubMed] [Google Scholar]
- 8.Park TJ, et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356:307–311. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- 9.Park TJ, et al. Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber) PLoS Biol. 2008;6:e13. doi: 10.1371/journal.pbio.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiser F. Evolution of daily torpor and hibernation in birds and mammals: importance of body size. Clin. Exp. Pharm. Physiol. 1998;25:736–739. doi: 10.1111/j.1440-1681.1998.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiao B, et al. HIF-1alpha contributes to hypoxia adaptation of the naked mole rat. Oncotarget. 2017;8:109941–109951. doi: 10.18632/oncotarget.22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buffenstein R. The naked mole-rat: a new long-living model for human aging research. J. Gerontol. A Biol. Sci. Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 13.Seluanov A, Gladyshev VN, Vijg J, Gorbunova V. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer. 2018;18:433–441. doi: 10.1038/s41568-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–349. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wainberg MA, Minden P, Weiss DW. Vertical transmission of tumour resistance in guinea pigs. Nature. 1976;259:213–215. doi: 10.1038/259213a0. [DOI] [PubMed] [Google Scholar]
- 16.Gorbunova V, Seluanov A. Coevolution of telomerase activity and body mass in mammals: from mice to beavers. Mech. Ageing Dev. 2009;130:3–9. doi: 10.1016/j.mad.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caulin AF, Maley CC. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol. Evol. 2011;26:175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lui JC, Baron J. Mechanisms limiting body growth in mammals. Endocr. Rev. 2011;32:422–440. doi: 10.1210/er.2011-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moeller ME, et al. Warts signaling controls organ and body growth through regulation of ecdysone. Curr. Biol. 2017;27:1652–1659. doi: 10.1016/j.cub.2017.04.048. [DOI] [PubMed] [Google Scholar]
- 20.Galis F, Metz JA. Anti-cancer selection as a source of developmental and evolutionary constraints. Bioessays. 2003;25:1035–1039. doi: 10.1002/bies.10366. [DOI] [PubMed] [Google Scholar]
- 21.Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, Vijg J. Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat. Rev. Genet. 2014;15:531–540. doi: 10.1038/nrg3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wensink, M. J. Size, longevity and cancer: age structure. Proc. Biol. Sci. 283, 20161510 (2016). [DOI] [PMC free article] [PubMed]
- 23.Seton-Rogers S. Metastasis: multitasking hyaluronic acid. Nat. Rev. Cancer. 2012;12:228. doi: 10.1038/nrc3254. [DOI] [PubMed] [Google Scholar]
- 24.Lan T, et al. Cross-linked hyaluronic acid gel inhibits metastasis and growth of gastric and hepatic cancer cells: in vitro and in vivo studies. Oncotarget. 2016;7:65418–65428. doi: 10.18632/oncotarget.11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan L, et al. Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Rep. 2017;9:1721–1734. doi: 10.1016/j.stemcr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evdokimov A, et al. Naked mole rat cells display more efficient excision repair than mouse cells. Aging. 2018;10:1454–1473. doi: 10.18632/aging.101482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrera-Álvarez, S., Karlsson, E., Ryder, O. A., Lindblad-Toh, K. & Crawford, A. J. How to make a rodent giant: genomic basis and tradeoffs of gigantism in the capybara, the world’s largest rodent. Mol. Biol. Evol. 10.1093/molbev/msaa285 (2020). [DOI] [PMC free article] [PubMed]
- 28.Debout C, Taouji S, Izard J. Increase of a guinea pig natural killer cell (Kurloff cell) during leukemogenesis. Cancer Lett. 1995;97:117–122. doi: 10.1016/0304-3835(95)03960-5. [DOI] [PubMed] [Google Scholar]
- 29.Sulak, M. et al. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife5, e11994 (2016). [DOI] [PMC free article] [PubMed]
- 30.Tollis M, et al. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the Humpback Whale (Megaptera novaeangliae) Mol. Biol. Evol. 2019;36:1746–1763. doi: 10.1093/molbev/msz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Z, et al. Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat. Ecol. Evol. 2019;3:1110–1120. doi: 10.1038/s41559-019-0913-3. [DOI] [PubMed] [Google Scholar]
- 32.Knudson, A. G. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer1, 157–162 (2001). [DOI] [PubMed]
- 33.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 34.Andrews T, et al. The clustering of functionally related genes contributes to CNV-mediated disease. Genome Res. 2015;25:802–813. doi: 10.1101/gr.184325.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl Acad. Sci. USA. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmood SF, et al. PPAPDC1B and WHSC1L1 are common drivers of the 8p11-12 amplicon, not only in breast tumors but also in pancreatic adenocarcinomas and lung tumors. Am. J. Pathol. 2013;183:1634–1644. doi: 10.1016/j.ajpath.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 37.Zheng J, Li C, Wang C, Ai Z. Common genetic variant on 14q13.3 contributes to thyroid cancer susceptibility: evidence based on 12 studies. Mol. Genet. Genomics. 2015;290:1125–1133. doi: 10.1007/s00438-014-0981-7. [DOI] [PubMed] [Google Scholar]
- 38.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl Acad. Sci. USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho N, et al. Appraising the relevance of DNA copy number loss and gain in prostate cancer using whole genome DNA sequence data. PLoS Genet. 2017;13:e1007001. doi: 10.1371/journal.pgen.1007001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asnaghi L, et al. Identification of multiple DNA copy number alterations including frequent 8p11.22 amplification in conjunctival squamous cell carcinoma. Investig. Ophthalmol. Vis. Sci. 2014;55:8604–8613. doi: 10.1167/iovs.14-14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishii H, et al. FEZ1/LZTS1 gene at 8p22 suppresses cancer cell growth and regulates mitosis. Proc. Natl Acad. Sci. USA. 2001;98:10374–10379. doi: 10.1073/pnas.181222898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Br. Funct. Genom. Proteomic. 2007;6:19–39. doi: 10.1093/bfgp/elm007. [DOI] [PubMed] [Google Scholar]
- 43.Ji L, Minna JD, Roth JA. 3p21.3 tumor suppressor cluster: prospects for translational applications. Futur. Oncol. 2005;1:79–92. doi: 10.1517/14796694.1.1.79. [DOI] [PubMed] [Google Scholar]
- 44.Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26:7283–7301. doi: 10.1038/sj.onc.1210547. [DOI] [PubMed] [Google Scholar]
- 45.Wu WK, et al. Oncogenes without a neighboring tumor-suppressor gene are more prone to amplification. Mol. Biol. Evol. 2017;34:903–907. doi: 10.1093/molbev/msx170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mérot, C., Oomen, R. A., Tigano, A. & Wellenreuther, M. A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol. Evol.35, 561–572 (2020). [DOI] [PubMed]
- 47.Wellenreuther, M. & Bernatchez, L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol.33, 427–440 (2018). [DOI] [PubMed]
- 48.Castiglia, R. Sympatric sister species in rodents are more chromosomally differentiated than allopatric ones: implications for the role of chromosomal rearrangements in speciation. Mamm. Rev. 44, 1–4 (2014).
- 49.Faria, R., Johannesson, K., Butlin, R. K. & Westram, A. M. Evolving inversions. Trends Ecol. Evol.34, 239–248 (2019). [DOI] [PubMed]
- 50.Faria, R. & Navarro, A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol.25, 660–669 (2010). [DOI] [PubMed]
- 51.Marquès-Bonet, T. et al. Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet.20, 524–529 (2004). [DOI] [PubMed]
- 52.Bagadia M, et al. Evolutionary loss of genomic proximity to conserved noncoding elements impacted the gene expression dynamics during mammalian brain development. Genetics. 2019;211:1239–1254. doi: 10.1534/genetics.119.301973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, X. et al. Beaver and naked mole rat genomes reveal common paths to longevity. Cell Rep. 32, 107949 (2020). [DOI] [PMC free article] [PubMed]
- 54.Fields AP, Justilien V, Murray NR. The chromosome 3q26 OncCassette: a multigenic driver of human cancer. Adv. Biol. Regul. 2016;60:47–63. doi: 10.1016/j.jbior.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J. Thorac. Oncol. 2008;3:212–215. doi: 10.1097/JTO.0b013e3181663544. [DOI] [PubMed] [Google Scholar]
- 56.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senchenko VN, et al. Simultaneous down-regulation of tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung cancer. BMC Cancer. 2010;10:75. doi: 10.1186/1471-2407-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagadia M, Singh A, Singh Sandhu K. Three dimensional organization of genome might have guided the dynamics of gene order evolution in eukaryotes. Genome Biol. Evol. 2016;8:946–954. doi: 10.1093/gbe/evw050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singer, G. A. C., Lloyd, A. T., Huminiecki, L. B. & Wolfe, K. H. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol. Biol. Evol. 22, 767–775 (2005). [DOI] [PubMed]
- 60.Makino T, McLysaght A. Interacting gene clusters and the evolution of the vertebrate immune system. Mol. Biol. Evol. 2008;25:1855–1862. doi: 10.1093/molbev/msn137. [DOI] [PubMed] [Google Scholar]
- 61.Ruan H, Su Z, Gu X. TreeExp1.0: R package for analyzing expression evolution based on RNA-Seq data. J. Exp. Zool. B Mol. Dev. Evol. 2016;326:394–402. doi: 10.1002/jez.b.22707. [DOI] [PubMed] [Google Scholar]
- 62.Pierce SB, et al. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet. 2013;92:614–620. doi: 10.1016/j.ajhg.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies KT, Bennett NC, Tsagkogeorga G, Rossiter SJ, Faulkes CG. Family wide molecular adaptations to underground life in African mole-rats revealed by phylogenomic analysis. Mol. Biol. Evol. 2015;32:3089–3107. doi: 10.1093/molbev/msv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh L, et al. Expression of CDC25A and CDC25B phosphatase proteins in human retinoblastoma and its correlation with clinicopathological parameters. Br. J. Ophthalmol. 2015;99:457–463. doi: 10.1136/bjophthalmol-2014-305830. [DOI] [PubMed] [Google Scholar]
- 65.Shan W, Li J, Xu W, Li H, Zuo Z. Critical role of UQCRC1 in embryo survival, brain ischemic tolerance and normal cognition in mice. Cell Mol. Life Sci. 2019;76:1381–1396. doi: 10.1007/s00018-019-03007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martos-Sitcha JA, Bermejo-Nogales A, Calduch-Giner JA, Perez-Sanchez J. Gene expression profiling of whole blood cells supports a more efficient mitochondrial respiration in hypoxia-challenged gilthead sea bream (Sparus aurata) Front. Zool. 2017;14:34. doi: 10.1186/s12983-017-0220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng X, Li J, Guo D. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy. Curr. Top. Med. Chem. 2018;18:484–493. doi: 10.2174/1568026618666180523104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015;10:920–932. doi: 10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 69.Zhang S, et al. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev. 2017;31:1939–1957. doi: 10.1101/gad.304261.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Otis JP, Sahoo D, Drover VA, Yen CL, Carey HV. Cholesterol and lipoprotein dynamics in a hibernating mammal. PLoS ONE. 2011;6:e29111. doi: 10.1371/journal.pone.0029111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frerichs KU, et al. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc. Natl Acad. Sci. USA. 1998;95:14511–14516. doi: 10.1073/pnas.95.24.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tollis, M., Schneider-Utaka, A. K. & Maley, C. C. The evolution of human cancer gene duplications across mammals. Mol. Biol. Evol. 37, 2875–2886 (2020). [DOI] [PMC free article] [PubMed]
- 73.Deuker, M. M. et al. Unprovoked stabilization and nuclear accumulation of the naked mole-rat p53 protein. Sci. Rep. 10, 1–11 (2020). [DOI] [PMC free article] [PubMed]
- 74.Kim, E. B. et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature479, 223–227 (2011). [DOI] [PMC free article] [PubMed]
- 75.Kikuta H, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson KL, Justice RW, Bryant PJ. Drosophila in cancer research: the first fifty tumor suppressor genes. J. Cell Sci. Suppl. 1994;18:19–33. doi: 10.1242/jcs.1994.Supplement_18.4. [DOI] [PubMed] [Google Scholar]
- 77.Lovrecic L, Bertok S, Zerjav Tansek M. A new case of an extremely rare 3p21.31 interstitial deletion. Mol. Syndr. 2016;7:93–98. doi: 10.1159/000445227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jagannathan R, et al. AJUBA LIM proteins limit hippo activity in proliferating cells by sequestering the Hippo core kinase complex in the cytosol. Mol. Cell Biol. 2016;36:2526–2542. doi: 10.1128/MCB.00136-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 80.Rabosky, D. L. et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4, 1–8 (2013). [DOI] [PubMed]
- 81.Lovegrove BG, Haines L. The evolution of placental mammal body sizes: evolutionary history, form, and function. Oecologia. 2004;138:13–27. doi: 10.1007/s00442-003-1376-3. [DOI] [PubMed] [Google Scholar]
- 82.Lovegrove BG, Mowoe MO. The evolution of mammal body sizes: responses to Cenozoic climate change in North American mammals. J. Evol. Biol. 2013;26:1317–1329. doi: 10.1111/jeb.12138. [DOI] [PubMed] [Google Scholar]
- 83.Cox PG, et al. Functional evolution of the feeding system in rodents. PLoS ONE. 2012;7:e36299. doi: 10.1371/journal.pone.0036299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgan CC, Verzi DH, Olivares AI, Vieytes EC. Craniodental and forelimb specializations for digging in the South American subterranean rodent Ctenomys (Hystricomorpha, Ctenomyidae) Mamm. Biol. 2017;87:118–124. doi: 10.1016/j.mambio.2017.07.005. [DOI] [Google Scholar]
- 85.Alvarez A, Perez I, Verzi DH. Ecological and phylogenetic influence on mandible shape variation of South American caviomorph rodents (Rodentia: Hystricomorpha) Biol. J. Linn. Soc. 2011;102:828–837. doi: 10.1111/j.1095-8312.2011.01622.x. [DOI] [Google Scholar]
- 86.Oliver Adrian NS. Diversity of social and mating systems in cavies: a review. J. Mammal. 2011;92:39–53. doi: 10.1644/09-MAMM-S-405.1. [DOI] [Google Scholar]
- 87.Danial‐Farran N, et al. Adaptive evolution of coagulation and blood properties in hypoxia tolerant Spalax in Israel. J. Zool. 2017;303:226–235. doi: 10.1111/jzo.12480. [DOI] [Google Scholar]
- 88.Kimani, J. M. Comparative Skin Morphology and Wound Healing in Kenyan African Mole Rat Naked Mole Rat (Heterocephalus Glaber).Department of Veterinary Anatomy and Physiology. PhD, University of Nairobi (2013).
- 89.Labinskyy N, et al. Comparison of endothelial function, O2-* and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. Am. J. Physiol. Hear. Circ. Physiol. 2006;291:H2698–H2704. doi: 10.1152/ajpheart.00534.2006. [DOI] [PubMed] [Google Scholar]
- 90.Lambert AJ, et al. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 91.Larson J, Park TJ. Extreme hypoxia tolerance of naked mole-rat brain. Neuroreport. 2009;20:1634–1637. doi: 10.1097/WNR.0b013e32833370cf. [DOI] [PubMed] [Google Scholar]
- 92.Lewis KN, Andziak B, Yang T, Buffenstein R. The naked mole-rat response to oxidative stress: just deal with it. Antioxid. Redox Signal. 2013;19:1388–1399. doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tian X, et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell. 2019;177:622–638. doi: 10.1016/j.cell.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burgel PR, Nadel JA. Epidermal growth factor receptor-mediated innate immune responses and their roles in airway diseases. Eur. Respir. J. 2008;32:1068–1081. doi: 10.1183/09031936.00172007. [DOI] [PubMed] [Google Scholar]
- 95.Repertinger SK, et al. EGFR enhances early healing after cutaneous incisional wounding. J. Investig. Dermatol. 2004;123:982–989. doi: 10.1111/j.0022-202X.2004.23478.x. [DOI] [PubMed] [Google Scholar]
- 96.Omerbasic D, et al. Hypofunctional TrkA accounts for the absence of pain sensitization in the african naked mole-rat. Cell Rep. 2016;17:748–758. doi: 10.1016/j.celrep.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crish SD, Dengler-Crish CM, Catania KC. Central visual system of the naked mole-rat (Heterocephalus glaber) Anat. Rec. A Disco. Mol. Cell Evol. Biol. 2006;288:205–212. doi: 10.1002/ar.a.20288. [DOI] [PubMed] [Google Scholar]
- 98.Rouble AN, Tessier SN, Storey KB. Characterization of adipocyte stress response pathways during hibernation in thirteen-lined ground squirrels. Mol. Cell Biochem. 2014;393:271–282. doi: 10.1007/s11010-014-2070-y. [DOI] [PubMed] [Google Scholar]
- 99.Schwartz C, Hampton M, Andrews MT. Seasonal and regional differences in gene expression in the brain of a hibernating mammal. PLoS ONE. 2013;8:e58427. doi: 10.1371/journal.pone.0058427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwartz C, Andrews MT. Circannual transitions in gene expression: lessons from seasonal adaptations. Curr. Top. Dev. Biol. 2013;105:247–273. doi: 10.1016/B978-0-12-396968-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Turbill C, Ruf T, Smith S, Bieber C. Seasonal variation in telomere length of a hibernating rodent. Biol. Lett. 2013;9:20121095. doi: 10.1098/rsbl.2012.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fronicke L, Wienberg J, Stone G, Adams L, Stanyon R. Towards the delineation of the ancestral eutherian genome organization: comparative genome maps of human and the African elephant (Loxodonta africana) generated by chromosome painting. Proc. Biol. Sci. 2003;270:1331–1340. doi: 10.1098/rspb.2003.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oviedo NJ, Beane WS. Regeneration: the origin of cancer or a possible cure? Semin Cell Dev. Biol. 2009;20:557–564. doi: 10.1016/j.semcdb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waddington CH. Cancer and the theory of organisers. Nature. 1935;135:606–608. doi: 10.1038/135606a0. [DOI] [Google Scholar]
- 105.Liozner LD. Evolution of the regeneration capabilities of animals. Usp. Sovrem. Biol. 1975;79:459–467. [PubMed] [Google Scholar]
- 106.Muller CA, Nieduszynski CA. DNA replication timing influences gene expression level. J. Cell Biol. 2017;216:1907–1914. doi: 10.1083/jcb.201701061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith KS, Liu LL, Ganesan S, Michor F, De S. Nuclear topology modulates the mutational landscapes of cancer genomes. Nat. Struct. Mol. Biol. 2017;24:1000–1006. doi: 10.1038/nsmb.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sima J, Gilbert DM. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr. Opin. Genet. Dev. 2014;25:93–100. doi: 10.1016/j.gde.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun L, et al. Preferential protection of genetic fidelity within open chromatin by the mismatch repair machinery. J. Biol. Chem. 2016;291:17692–17705. doi: 10.1074/jbc.M116.719971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 112.Pal C, Hurst LD. Evidence for co-evolution of gene order and recombination rate. Nat. Genet. 2003;33:392–395. doi: 10.1038/ng1111. [DOI] [PubMed] [Google Scholar]
- 113.Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat. Genet. 2007;39:945–949. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- 114.Yeaman S. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc. Natl Acad. Sci. USA. 2013;110:E1743–E1751. doi: 10.1073/pnas.1219381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tollis M, Boddy AM, Maley CC. Peto’s Paradox: how has evolution solved the problem of cancer prevention? BMC Biol. 2017;15:60. doi: 10.1186/s12915-017-0401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith FA, et al. The evolution of maximum body size of terrestrial mammals. Science. 2010;330:1216–1219. doi: 10.1126/science.1194830. [DOI] [PubMed] [Google Scholar]
- 117.Webster AJ, Gittleman JL, Purvis A. The life history legacy of evolutionary body size change in carnivores. J. Evol. Biol. 2004;17:396–407. doi: 10.1046/j.1420-9101.2003.00664.x. [DOI] [PubMed] [Google Scholar]
- 118.Biewener AA. Scaling body support in mammals: limb posture and muscle mechanics. Science. 1989;245:45–48. doi: 10.1126/science.2740914. [DOI] [PubMed] [Google Scholar]
- 119.Rauhala L, et al. Low dose ultraviolet B irradiation increases hyaluronan synthesis in epidermal keratinocytes via sequential induction of hyaluronan synthases Has1-3 mediated by p38 and Ca2+/calmodulin-dependent protein kinase II (CaMKII) signaling. J. Biol. Chem. 2013;288:17999–18012. doi: 10.1074/jbc.M113.472530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ding Q, Hu Y, Xu S, Wang J, Jin L. Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol. Biol. Evol. 2014;31:683–695. doi: 10.1093/molbev/mst260. [DOI] [PubMed] [Google Scholar]
- 121.Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev. Mol. Med. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boniakowski AE, et al. Murine macrophage chemokine receptor CCR2 plays a crucial role in macrophage recruitment and regulated inflammation in wound healing. Eur. J. Immunol. 2018;48:1445–1455. doi: 10.1002/eji.201747400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilson LA, Sanchez-Villagra MR. Heterochrony and patterns of cranial suture closure in hystricognath rodents. J. Anat. 2009;214:339–354. doi: 10.1111/j.1469-7580.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Upham, N. S. & Patterson, B. D. Ecological Diversification and Biogeography in the Neogene: Evolution of a Major Lineage of American and Caribbean Rodents (Caviomorpha, Octodontoidea).Faculty of the Division of the Biological Sciences. PhD, University of Chicago (2014).
- 125.Upham NS, Patterson BD. Diversification and biogeography of the Neotropical caviomorph lineage Octodontoidea (Rodentia: Hystricognathi) Mol. Phylogenet Evol. 2012;63:417–429. doi: 10.1016/j.ympev.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 126.Robinson TJ, King M. Species evolution: the role of chromosome change. Syst. Biol. 1995;44:578–580. [Google Scholar]
- 127.Romanenko, S. A., Perelman, P. L., Trifonov, V. A. & Graphodatsky, A. S. Chromosomal evolution in Rodentia. Heredity108, 4–16 (2012). [DOI] [PMC free article] [PubMed]
- 128.Romanenko, S. A. et al. A first generation comparative chromosome map between guinea pig (Cavia porcellus) and humans. PLoS ONE10, e0127937 (2015). [DOI] [PMC free article] [PubMed]
- 129.Pigliucci, M. Is evolvability evolvable? Nat. Rev. Genet. 9, 75–82 (2008). [DOI] [PubMed]
- 130.Schmitt AD, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marcovitz A, Jia R, Bejerano G. "Reverse Genomics" Predicts Function of Human Conserved Noncoding Elements. Molecular Biology and Evolution. 2016;33(5):1358–1369. doi: 10.1093/molbev/msw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Itsara A, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mapped datasets are available as online Supplementary Data.
The custom scripts used for data analyses are available on request.