Abstract
Pancreatic adenocarcinoma (PAAD) is the most malignant digestive tumor. The global incidence of pancreatic cancer has been rapidly trending upwards, necessitating an exploration of potential prognostic biomarkers and mechanisms of disease development. One of the most prevalent RNA modifications is 5-methylcytosine (m5C); however, its contribution to PAAD remains unclear. Data from The Cancer Genome Atlas (TCGA) database, including genes, copy number variations (CNVs), and simple nucleotide variations (SNVs), were obtained in the present study to identify gene signatures and prognostic values for m5C regulators in PAAD. Regulatory gene m5C changes were significantly correlated with TP53, BRCA1, CDKN2A, and ATM genes, which play important roles in PAAD pathogenesis. In particular, there was a significant relationship between m5C regulatory gene CNVs, especially in genes encoding epigenetic “writers”. According to m5C-regulated gene expression in clinically graded cases, one m5C-regulated genes, DNMT3A, showed both a strong effect on CNVs and a significant correlation between expression level and clinical grade (P < 0.05). Furthermore, low DNMT3A expression was not only associated with poor PAAD patient prognosis but also with the ribosomal processing. The relationship between low DNMT3A expression and poor prognosis was confirmed in an International Cancer Genome Consortium (ICGC) validation dataset.
Subject terms: Cancer, Computational biology and bioinformatics, Oncology
Introduction
Pancreatic adenocarcinoma (PAAD) is the most malignant tumor of the human digestive system. New cases of PAAD in the United States rank 10th among men, 9th among women, and 4th in cancer mortality according to data released by the American Cancer Society in 20191. For malignant tumors in China, PAAD ranks eighth for urban men and fifth for the mortality rate in large cities according to data from the National Cancer Center of China2, with a 5-year survival rate of less than 5%3,4. Globally, PAAD incidence and mortality increases with age and is slightly higher among men, although mortality has increased in both genders over the past few decades3,5–8. Despite the identification of some risk factors such as smoking, genetic diabetes, alcohol consumption, and lack of exercise, the causes of PAAD remain poorly understood9–14. Exploring potential prognostic biomarkers and mechanisms that drive pancreatic cancer are therefore urgently required.
RNA modifications play a pivotal role across many biological processes15–17. Among the 150 types of RNA modifications discovered, methylation has been identified as a common phenomenon and a key regulator of gene transcript expression, with 5-methylcytosine (m5C) being one of the most prevalent18,19. With the application of high-throughput sequencing techniques for detecting m5C RNA modifications (e.g., bisulfite sequencing), a whole-transcriptome map of m5C sites has become available in which m5C mainly appears in the anticodon loop and variable region of tRNAs and rRNAs and the coding sequences of mRNAs20–24. m5C deposition occurs through a methyltransferase complex involving three homologous factors, including methyltransferases (“writers”), demethylases (“erasers”), and m5C binding proteins (“readers”). m5C affects RNA structural stability and translational efficiency like other nucleobase modifications, and research has additionally revealed a potential role in promoting mRNA export and regulating tissue differentiation25,26. Dysregulation of m5C has been implicated in multiple abnormal cellular processes and human cancers27. For instance, tumor malignancy has been strongly correlated with levels of m5C in the DNA from breast cancer patient tissue. Measuring m5C levels in myeloid malignancies could also act as a valuable diagnostic and prognostic tool, not only for tailoring therapies but also assessing responses to anti-cancer drug activity and toxicity28–30.
Little is currently known about the relationship between m5C-related genes and PAAD. Based on The Cancer Genome Atlas (TCGA) database, researchers can easily access the gene expression, copy number variations (CNVs), and simple nucleotide variations (SNVs) data of human cancers. Both CNVs and SNVs can play crucial roles in cancer progression, but how the CNVs and SNVs of m5C-related genes contribute are poorly understood31. In the present study, we retrospectively analyzed the gene signatures and prognostic values of m5C regulators in PAAD using patient gene expression, CNV, and SNV data from the TCGA database. Changes to m5C regulatory genes were significantly associated with tumor stage, and a strong relationship between m5C regulatory gene CNVs and pancreatic cancer patient survival was established. These findings enabled the discovery of genetic alterations to m5C regulatory genes that contribute to PAAD patient outcomes and enhance our understanding of m5C epigenetic regulation in PAAD.
Results
m5C regulatory gene mutations in PAAD
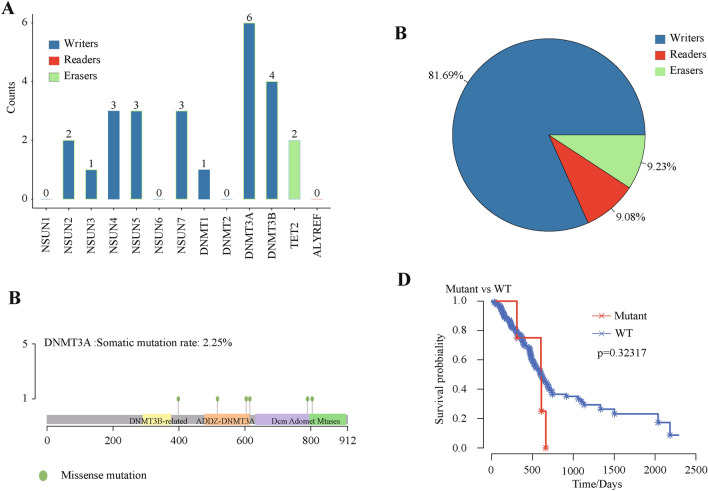
In the sequencing data set of 363 PAAD patients, m5C regulatory gene mutations appeared in 13 independent samples (Table 1). Further, these 13 PDAC samples were used to analyze the mutation characteristics of the m5C genes. The clinical information of these samples was shown in Supplementary Table 1. Compared with the eraser and reader genes, writer gene had a greater mutation frequency, as reader gene mutations were not detected (Fig. 1A). Six of the samples revealed a higher mutation frequency in the writer gene DNMT3A. In 183 PAAD samples with CNV data, m5C regulatory genes showed a high frequency of CNV (Fig. 1B), although CNVs within NSUN1 and DNMT2 genes were undetectable. For example, the reader gene ALYREF had a CNV event frequency of 18.48%, followed by the writer gene NSUN4 with a frequency of 15.22%. The eraser gene TET2 had the lowest frequency at 6.42% (Supplementary Table 2). Among them, the writer gene DNMT3A showed the highest mutation level (Fig. 1C).
Table 1.
Relationship between changes in m5C regulatory genes and high-frequency PAAD-related genes.
| Without SNV and CNV | With SNV and CNV | X2 | P | |||
|---|---|---|---|---|---|---|
| ATM | WT | 148 | 0 | 104.012609 | 2.0103E−24 | |
| n = 151 | Alternation | 0 | 3 | |||
| BRCA1 | WT | 149 | 0 | 84.1791333 | 4.5192E−20 | |
| n = 151 | Alternation | 0 | 2 | |||
| CDKN2A | WT | 150 | 1 | 18.3742163 | 1.815E−05 | |
| n = 151 | Alternation | 0 | 1 | |||
| TP53 | WT | 52 | 49 | 37.016707 | 1.1712E−09 | |
| n = 151 | Alternation | 0 | 50 | |||
| WT | 148 | 0 |
Figure 1.
CNVs, mutations, and SNVs of m5C regulatory genes in PAAD. A total of 363 PAAD patients from TCGA were analyzed. (A) Mutation frequencies in m5C regulatory genes with indicated functions. (B) Mutation sites in DNMT3A. (C) Seven functionally altered genes and patient survival. P value did not reach significance due to the small number of mutations. (D) CNV statistics of m5C regulatory genes in PAAD samples. Writers accounted for 81.69%, which was highest. Readers and erasers made up almost the same proportion at approximately 9%.
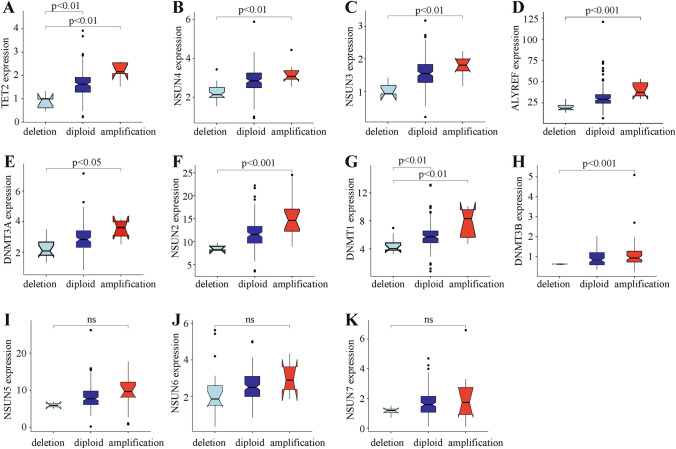
These results showed that in 177 PAAD samples, two out of 13 m5C regulatory genes were hardly detected. Excluding these two genes, mRNA expression levels of the remaining 11 m5C regulatory genes significantly correlated with their CNV patterns. Increased copy numbers for eight genes, which were distributed across all m5C regulation categories, were associated with higher mRNA expression. Deletion mutants also resulted in decreased mRNA expression. Additionally, three out of 11 m5C regulatory genes had no significant relationship with CNV expression and were concentrated only in writers genes (Fig. 2). The expression of all eraser and reader genes were significantly related to CNVs. We next used seven functionally altered genes to predict PAAD patient survival, but only uncovered a small number of mutations without predictive statistical significance (Fig. 1D).
Figure 2.
m5C regulatory gene correlation to clinicopathological molecular characteristics. (A–H) Relationship between CNVs and expression levels of m5C regulatory genes (significance P < 0.01). (I–K) Relationship between CNVs and expression level non-significant (ns) m5C regulatory genes.
m5C regulatory gene changes are related to molecular clinicopathological characteristics
Since TP53, BRCA1, CDKN2A, and ATM genes play crucial roles in the pathogenesis of PAAD, we investigated their relationship with m5C regulatory genes. Interestingly, variations in m5C regulatory genes associated significantly with alterations to TP53, BRCA1, CDKN2A, and ATM (Table 1).
Association between m5C regulatory genes and PAAD survival
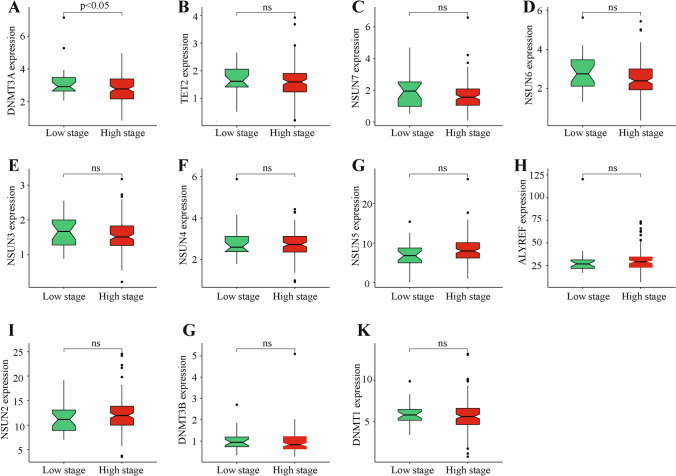
We then analyzed and clustered m5C regulatory gene expression in different clinically graded cases (Fig. 3). These results indicated that only one out of 11 m5C regulatory genes, DNMT3A, significantly correlated with the clinical grade of patients in that its high expression associated with high staging (Fig. 4). As previously described, DNMT3A also had the highest mutation rate and a positive correlation between CNV changes and expression level.
Figure 3.
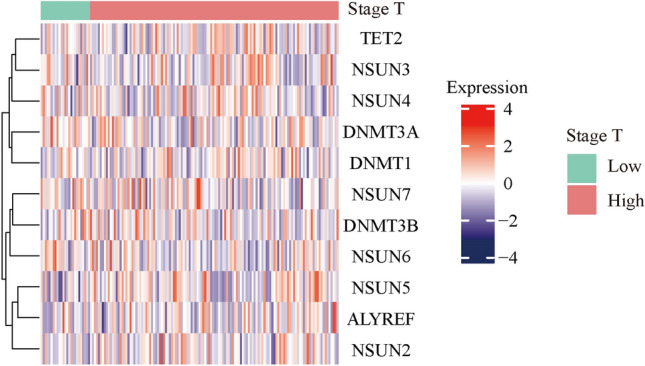
Heat map clustering between m5C regulatory gene expression and clinical tumor staging.
Figure 4.
m5C regulatory gene expression in different clinically graded PAAD cases. (A) Comparison of m5C regulatory gene expression in different clinically graded cases. DNMT3A expression showed a significant negative correlation with patient clinical grade (significance P < 0.05). (B–K) Comparison of m5C regulatory gene expression in different clinically graded cases that were non-significant (ns).
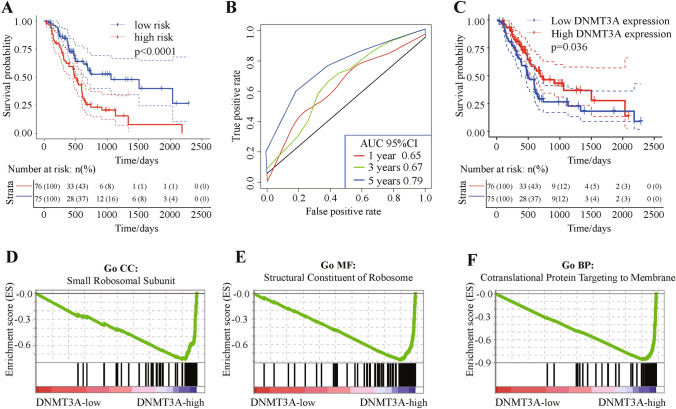
Given the relationship between clinical classification and patient prognosis, we believed that m5C regulatory gene expression would be related to patient prognosis. However, no significant correlation between m5C regulatory gene CNVs and patient prognosis was observed. In order to correlate the expression of some m5C regulatory genes with CNVs, we used single factor Cox regression to explore the relationship between m5C regulatory gene expression and patient prognosis. The expression of two genes showed a significant positive correlation with patient prognosis (P < 0.05), including DNMT3A (Table 2). Among these two genes, the expression level of DNMT3A was significantly related to CNV changes (Table 2). Using multivariate Cox regression analysis, we next found that m5C regulatory gene expression can significantly predict patient risk using AUC at 1, 3, and 5 years, with all AUC values greater than 0.65 (Fig. 5A,B). These results demonstrated that the expression of m5C regulatory genes can be used as a prognostic marker for pancreatic cancer.
Table 2.
Univariate Cox regression exploring the relationship between different m5C regulatory gene expression levels and patient prognosis.
| Beta | HR | (95% CI for HR) | Wald test | P-value | CNV sig | |
|---|---|---|---|---|---|---|
| DNMT3A | − 0.31 | 0.74 | (0.57–0.95) | 5.6 | 0.018 | Yes |
| NSUN6 | − 0.3 | 0.74 | (0.57–0.96) | 5.2 | 0.022 | No |
| NSUN7 | − 0.21 | 0.81 | (0.64–1) | 3.2 | 0.071 | No |
| NSUN2 | 0.044 | 1 | (0.98–1.1) | 2.1 | 0.15 | Yes |
| NSUN3 | 0.21 | 1.2 | (0.76–2) | 0.71 | 0.4 | Yes |
| NSUN5 | 0.02 | 1 | (0.97–1.1) | 0.53 | 0.47 | No |
| NSUN4 | − 0.04 | 0.96 | (0.72–1.3) | 0.07 | 0.79 | Yes |
| TET2 | − 0.045 | 0.96 | (0.68–1.4) | 0.06 | 0.8 | Yes |
| ALYREF | − 0.0013 | 1 | (0.98–1) | 0.02 | 0.88 | Yes |
| DNMT3B | 0.031 | 1 | (0.69–1.5) | 0.02 | 0.88 | Yes |
| DNMT1 | − 0.0011 | 1 | (0.89–1.1) | 0 | 0.99 | Yes |
Figure 5.
Relationship between m5C regulatory gene expression and patient prognosis, with DNMT3A gene functional enrichment analysis. The impact of 13 m5C regulatory genes on patient prognosis using multivariate Cox regression. (A, B) Multivariate Cox regression of survival (P < 0.001) and AUC curves (AUC curve areas > 0.65). (C) Relationship between DNMT3A expression and patient prognosis (P = 0.036). (D–F) GSEA enrichment analysis of the DNMT3A Gene showing low DNMT3A expression related to ribosomal processing.
Based on the above results, we performed a LASSO technique on the 13 m5C regulatory genes to more precisely identify prognostic markers of PAAD. Through 1,000 LASSO regressions, we observed that LASSO results repeatedly appeared more than 900 times. Only DNMT3A exhibited a significant result for expression level and CNV by single factor Cox analysis, and a significant correlation between expression level and clinical grade (P < 0.05) (Table 3).
Table 3.
Results of lasso analysis of m5C regulatory genes.
| Duplicates | Genes | CNV express sig | Functions | Survival sig | Stage sig |
|---|---|---|---|---|---|
| 985 | NSUN6 | No | Writer | No | No |
| 965 | DNMT3A | Yes | Writer | Yes | Yes |
| 939 | NSUN2 | Yes | Writer | No | No |
| 923 | NSUN3 | Yes | Writer | No | No |
| 234 | DNMT3B | Yes | Writer | No | No |
| 200 | TET2 | Yes | Eraser | No | No |
| 186 | DNMT1 | Yes | Writer | No | No |
| 105 | NSUN7 | No | Writer | No | No |
| 99 | NSUN4 | Yes | Writer | No | No |
| 88 | NSUN5 | No | Writer | No | No |
| 86 | ALYREF | Yes | Reader | No | No |
Given that the DNMT3A gene is a writer gene involved in vital m5C regulatory functions and to predict patient risk based on DNMT3A gene expression, we analyzed the relationship between DNMT3A expression and PAAD patient prognosis. Notably, the results illustrated that low DNMT3A expression significantly correlated with poor prognosis (Fig. 5C). These results suggested that DNMT3A gene expression could be a clinically significant biomarker for PAAD patients.
Functional enrichment analysis of DNMT3A gene in expression level
Because the DNMT3A gene is a methylation writer gene, we next investigated the role of m5C dysfunction in PAAD pathogenesis. We examined enriched gene sets in samples with varying mRNA expression of the DNMT3A gene. Gene enrichment analysis by gene set enrichment analysis (GSEA) showed that low DNMT3A expression was related to ribosomal processing (Fig. 5D–F). Ribosomes are essential for protein synthesis, and because of the elevated metabolic activity in tumor cells, protein synthesis requirements are much greater than normal cells. These findings also confirmed that the low expression of DNMT3A was related to the poor prognosis of patients.
Validation of DNMT3A act as an important pancreatic cancer target tiller invalidation data set
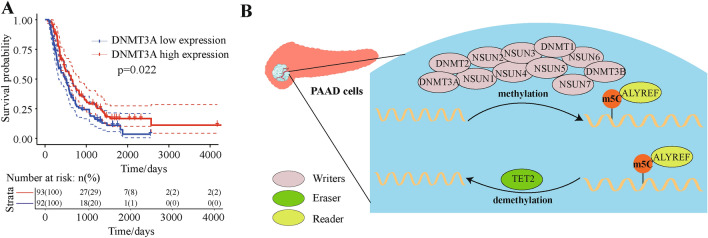
Finally, we used a validation data set containing 185 samples to analyze the relationship between DNMT3A gene expression and patient survival (Supplementary Table 3). Based on Cox regression analysis, low expression of DNMT3A was associated with poor prognosis (Fig. 6A). A schematic diagram showed the related mechanisms of m5C in PAAD cells (Fig. 6B), and these findings provided evidence of the role that m5C epigenetic regulation played in pancreatic cancer.
Figure 6.
Relationship between DNMT3A expression and patient prognosis using validation data set. Analysis of patients from validation dataset ICGC-PACA-CA. (A) Relationship between DNMT3A expression and patient prognosis in the validation dataset (P = 0.022). (B) Mechanisms of m5C methylation regulation in PAAD.
Discussion
It is well known that m5C is an important chemical modification to mRNA involved in many pathological processes leading to cancer. The deposition of m5C occurs through a methyltransferase complex involving three homologous factors including methyltransferases known as writers (e.g., NSUN1, NSUN2, NSUN3, NSUN4, NSUN5, NSUN6, NSUN7, DNMT1, DNMT2, DNMT3A, and DNMT3B), demethylases known as erasers (e.g., TET2), and m5C binding proteins known as readers (e.g., ALYREF)32,33.
In the present study, we used 686 patients from the TCGA database with sequencing and CNV data to analyze the gene signatures and prognostic values of m5C regulators in PAAD patients. We identified genetic alterations to m5C regulatory genes and patient survival outcomes in pancreatic cancer, and an association between low expression of the writer gene DNMT3A and high tumor stage.
These findings provide clues to understanding m5C-mediated epigenetic regulation in pancreatic cancer and perhaps other types of cancer. For example, in patients treated with various chemotherapeutic agents, there was an unexpected and nonspecific amount of DNA methylation activity, as well as a global suppression of DNA repair genes and protein synthesis. Total m5C levels in DNA from patient breast cancer tissue correlates well with tumor malignancy28. In one study, cells expressing MYC-TET2 exhibited increased m5C staining, but a concomitant decrease in nuclear m5C staining30. In myeloid malignancies, m5C levels could provide valuable diagnostic and prognostic tools to tailor therapies and assess anti-cancer drug responses and toxicity, as DNMT3A was recently reported to be a key player in the progression of malignant glioma19. In another example, three patients with the highest m5C content in their normal colon appeared to have a germline predisposition to cancer (Lynch syndrome)29. Additionally, DNMT3A mutations were associated with adverse outcomes among patients with an intermediate-risk cytogenetic profile or FLT3 mutations, regardless of age, and were independently associated with a poor outcome by Cox proportional-hazards analyses34. The high mutation rate of DNMT3A may eventually lead to impaired biological functions, such as cyanosis, because its protein product affects the transmission of m5C signals in cells, which in turn causes tumor-related phenotype changes such as DNA repair activities and oncogenic tyrosine kinases (such as FLT3ITD, BCR-ABL1, JAK2V617F and MPLW515L) activating mutations.
Cancer-related pathways are dysregulated in the development of PAAD. Herein, the expression of TP53, BRCA1, CDKN2A, and ATM were associated with m5C regulatory gene CNVs and SNVs. Additionally, gene enrichment analysis showed that low DNMT3A expression was related to ribosomal processing, an essential part of tumor survival.
In conclusion, we have identified genetic alterations to m5C regulatory genes in pancreatic cancer patients that correlate with patient prognosis and survival, which provides evidence for the role that m5C epigenetic regulation plays in pancreatic cancer.
Methods
Data resource and processing
All PAAD clinical, copy number variations (CNVs), simple nucleotide variations (SNVs), and mRNA expression data were retrieved from TCGA-assembler from the TCGA website35,36 and downloaded in September 2019. The validation dataset used to validate findings was from the International Cancer Genome Consortium (ICGC) (https://dcc.icgc.org/) using the Pancreatic Cancer Data Set. For transcriptome data, we obtained 177 samples and downloaded data for reading counts. Data were normalized by R package DESeq. For SNV data, we obtained a total of 175 samples, and the downloaded data was level 3 data processed by Mu Tect37. For CNV data, there were 183 samples processed by R package RTCGA. Here, the deletion referred to as the Segment Mean was less than 0.2, and the application that was considered as the Segment Mean was greater than 0.2. For clinical data, there were 185 samples after integrating the data and excluding samples with a survival time less than 90 days, leaving a total of 151 pancreatic cancer samples for further analysis.
The least absolute shrinkage and selection operator (LASSO) analysis
The LASSO method was introduced to identify important predictors of m5C regulatory genes using the R package glmnet as previously described38,39. It is a commonly used high-dimensional index regression method that sub-selects the m5C regulatory genes involved in PAAD patient prognosis by applying a penalty proportional to the contraction of the regression coefficient. Based on this method, we constructed m5C regulatory gene signatures with prognostic values, and calculated a risk score for each PAAD patient. All patients were divided into high-risk and low-risk groups with the median risk score as the cut-off score and used Kaplan–Meier curves to estimate the survival of PAAD patients in different groups.
Gene set enrichment analysis (GSEA)
GSEA is a computational method determining whether an a priori defined set of genes shows statistically significant, concordant differences between two biological phenotypes40. GSEA was provided by the JAVA program and downloaded from the website (http://software.broadinstitute.org). We divided all samples into two groups according to the median expression value of the DNMT3A gene. Gene sets with a normalized P < 0.05 and false discovery rate (FDR) < 0.25 were considered to be significantly enriched.
Statistical analysis
All statistical data were analyzed using SPSS 23 (IBM, Chicago, USA) and R language. The association between CNV and SNV of m5C regulatory genes and clinicopathological characteristics were analyzed by Chi-square test. The association between three pancreatic cancer Kaplan–Meier curve and we used log-rank test to evaluate the prognosis value of alteration of m5C regulatory gene. All statistical results with a P-value less than 0.05 were considered significant.
Supplementary Information
Acknowledgements
We thank the patients and investigators who participated in TCGA and ICGC for providing data.
Abbreviations
- PAAD
Pancreatic adenocarcinoma
- m5C
5-Methylcytosine
- TCGA
The Cancer Genome Atlas
- CNVs
Copy number variations
- SNVs
Simple nucleotide variations
- ICGC
International Cancer Genome Consortium
- GSEA
Gene set enrichment analysis
- FDR
False discovery rate
- LASSO
The least absolute shrinkage and selection operator
- HR
Hazard ratio
- CI
Confidence interval
Author contributions
All of the authors worked collaboratively on the work presented here. GWZ and HYT defined the research theme and discussed analyses, interpretation, and presentation. YX, Zhang QY and Zhang MG drafted the manuscript and analyzed the data. Zhang QY and FG helped with references collection. Zheng QY collected data and helped draft the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by funds from Science and Technology Innovation Talents in Henan Universities (19HASTIT003); and Key Scientific Research Project of Henan Higher Education Institutions of China (21A320026).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiao Yu, Qiyao Zhang and Fang Gao.
Contributor Information
Yuting He, Email: fccheyt1@zzu.edu.cn.
Wenzhi Guo, Email: fccguowz@zzu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96470-w.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen W. Cancer statistics: updated cancer burden in China. Chin. J. Cancer Res. 2015;27:1. doi: 10.3978/j.issn.1000-9604.2015.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M, Cascinu S, Kleeff J, Labianca R, Löhr J-M, Neoptolemos J, Real FX, Van Laethem J-L, Heinemann V. Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology. 2015;15:8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Xue J, Jaffee EM, Habtezion A. Role of immune cells and immune-based therapies in pancreatitis and pancreatic ductal adenocarcinoma. Gastroenterology. 2013;144(6):1230–1240. doi: 10.1053/j.gastro.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Megibow AJ. Pancreatic adenocarcinoma: designing the examination to evaluate the clinical questions. Radiology. 1992;183(2):297–303. doi: 10.1148/radiology.183.2.1561324. [DOI] [PubMed] [Google Scholar]
- 7.Malvezzi M, Carioli G, Bertuccio P, Rosso T, Boffetta P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2016 with focus on leukaemias. Ann. Oncol. 2016;27:725–731. doi: 10.1093/annonc/mdw022. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Yang G-H, Lu X-H, Huang Z-J, Li H. Pancreatic cancer mortality in China (1991–2000) World J. Gastroenterol WJG. 2003;9:1819. doi: 10.3748/wjg.v9.i8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezzati M, Henley SJ, Lopez AD, Thun MJ. Role of smoking in global and regional cancer epidemiology: current patterns and data needs. Int. J. Cancer. 2005;116:963–971. doi: 10.1002/ijc.21100. [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo M. Pancreatic cancer. N. Engl. J. Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 11.Parkin DM, Boyd L, Walker L. 16 The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br. J. Cancer. 2011;105:77. doi: 10.1038/bjc.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willett WC, MacMahon B. Diet and cancer—an overview. N. Engl. J. Med. 1984;310:697–703. doi: 10.1056/NEJM198403083101006. [DOI] [PubMed] [Google Scholar]
- 13.Hua Y-Q, Zhu Y-D, Xie G-Q, Zhang K, Sheng J, Zhu Z-F, Ning Z-Y, Chen H, Chen Z, Meng Z-Q. Long non-coding SBF2-AS1 acting as a competing endogenous RNA to sponge microRNA-142–3p to participate in gemcitabine resistance in pancreatic cancer via upregulating TWF1. Aging (Albany NY) 2019;11:5579. doi: 10.18632/aging.102307. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Kong Y, Yao Q, Zhang X, Fu Y, Li J, Liu C, Wang Z. Three hypomethylated genes were associated with poor overall survival in pancreatic cancer patients. Aging (Albany NY) 2019;11:885. doi: 10.18632/aging.101785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Pan T. RNA epigenetics. Transl. Res. 2015;165:28–35. doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marbaniang CN, Vogel J. Emerging roles of RNA modifications in bacteria. Curr. Opin. Microbiol. 2016;30:50–57. doi: 10.1016/j.mib.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Omer AD, Ziesche S, Decatur WA, Fournier MJ. Dennis pp. RNA-modifying machines in archaea. Mol. Microbiol. 2003;48:617–629. doi: 10.1046/j.1365-2958.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- 18.Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2017;2017(46):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m6A methylation in cancer. Can. Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 20.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013;31:458. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2008;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA–methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7:e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, Li A, Wang X, Bhattarai DP, Xiao W. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m 5 C reader. Cell Res. 2017;27:606. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barciszewska AM, Murawa D, Gawronska I, Murawa P, Nowak S, Barciszewska MZ. Analysis of 5-methylcytosine in DNA of breast and colon cancer tissues. IUBMB Life. 2007;59:765–770. doi: 10.1080/15216540701697412. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Can. Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 30.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gigek CO, Calcagno DQ, Rasmussen LT, Santos LC, Leal MF, Wisnieski F, Burbano RR, Lourenco LG, Lopes-Filho GJ, Smith MAC. Genetic variants in gastric cancer: risks and clinical implications. Exp. Mol. Pathol. 2017;103:101–111. doi: 10.1016/j.yexmp.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Huang L-H, Wang R, Gama-Sosa MA, Shenoy S, Ehrlich M. A protein from human placental nuclei binds preferentially to 5-methylcytosine-rich DNA. Nature. 1984;308:293. doi: 10.1038/308293a0. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Perrera V, Saplaoura E, Apelt F, Bahin M, Kramdi A, Olas J, Mueller-Roeber B, Sokolowska E, Zhang W. m5C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 2019;29(2465–76):e5. doi: 10.1016/j.cub.2019.06.042. [DOI] [PubMed] [Google Scholar]
- 34.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag. Res. 2018;10:3857. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Network CGAR. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113. doi: 10.1186/s13072-019-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang D, Li R, Li Y, Zhu X. A 7-lncRNA signature predict prognosis of Uterine corpus endometrial carcinoma. J. Cell. Biochem. 2019 doi: 10.1002/jcb.29164. [DOI] [PubMed] [Google Scholar]
- 40.Xue C, He Y, Zhu W, Chen X, Yu Y, Hu Q, Chen J, Liu L, Ren F, Ren Z. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am. J. Transl. Res. 2018;10:4152. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.