SUMMARY
Background
Atopic dermatitis (AD) disease activity and severity is highly variable during childhood. Early attempts to identify subtypes based on disease trajectory have assessed AD presence over time without incorporating severity.
Objective
To identify childhood AD subtypes from symptom severity and trajectories, and determine associations with genetic risk factors, comorbidities and demographic and environmental variables.
Methods
We split data from children in the Avon Longitudinal Study of Parents and Children birth cohort into development and validation sets. To identify subtypes, we ran latent class analyses in the development set on AD symptom reports up to age 14. We regressed identified subtypes on non-genetic variables in mutually adjusted, multiply imputed (genetic: unadjusted, complete-case) multinomial regression analyses. We repeated analyses in the validation set and report confirmed results.
Results
11,866 children contributed to analyses. We identified one Unaffected/Rare class (66% of children) and four AD subtypes: Severe-Frequent (4%); Moderate-Frequent (7%); Moderate-Declining (11%); and Mild-Intermittent (12%). Symptom patterns within the first two subtypes appeared more homogeneous than the last two. Filaggrin null mutations (FLG), an AD polygenic risk score (PRS), being female, parental AD and comorbid asthma were associated with higher risk for some or all subtypes; FLG, AD-PRS and asthma associations were stronger along a subtype gradient arranged by increasing severity and frequency; FLG and AD-PRS further differentiated some phenotypes from each other.
Conclusions
Considering severity and AD trajectories leads to four well-defined and recognisable subtypes. The differential associations of risk factors among and between subtypes is novel and requires further research.
INTRODUCTION
Atopic dermatitis (AD) (eczema) is characterised by considerable heterogeneity including clinical morphology, disease severity, age at presentation, disease course, and comorbidities. Many attempts have been made to address this heterogeneity by identifying AD subtypes, which may improve diagnosis, more accurately estimate clinical prognoses, inform management or better predict treatment efficacy or effectiveness1. Older literature largely subtyped AD into allergic and non-allergic forms; recent evidence suggests that this dichotomisation is not clinically useful2 because IgE levels or sensitization status alone do not predict symptom resolution or treatment response3. Recent efforts using longitudinal cohorts have attempted to identify AD subtypes from trajectories of childhood disease activity4-6, or patterns of atopic disease overall7-10. However, none of these categorizations account for disease severity, which is the primary predictor of quality of life, sleep disruption, comorbidities including mental health outcomes, and need for systemic treatments11,12. Identifying factors early in the disease course that could differentiate between mild but persistent cases and severe but resolving cases is important both for understanding aetiology and outcomes and for informing patient prognoses and treatment options.
In this paper, we aim to identify AD subtypes based on trajectories of flexural dermatitis in children related to both presence and severity of disease, and quantify their relationships with known and suspected AD risk factors. We note that subtypes based on disease trajectories over time are not the same as directly observed phenotypes13. However, as the term ‘phenotypes’ is commonly used to describe AD subtypes, we use this nomenclature in this paper.
MATERIALS AND METHODS
Study population
We used data from the Avon Longitudinal Study of Parents and Children (ALSPAC)14,15. Pregnant women resident in Avon, UK with expected dates of delivery between 1st April 1991 to 31st December 1992 were invited to take part and 14,541 pregnancies were enrolled initially. Of these, there were 14,676 foetuses, resulting in 14,062 live births and 13,988 children who were alive at 1 year of age, 96% of whom are of white ethnicity. Please note that the study website contains details of all the data that is available through a fully searchable data dictionary and variable search tool16.
AD phenotype determination
AD presence was collected from questionnaires at eleven ages between 6 and 166 months (13.8 years). Mothers reported whether their children experienced flexural dermatitis (FD) via The International Study of Asthma and Allergies in Childhood (ISAAC)-validated question17: "Has your child had an itchy, dry skin rash in the joints and creases of his body (e.g., behind the knees, elbows, under the arms) in the past year?" Participants who responded ‘yes’ were followed up with an assessment of self-rated severity, “How bad was this?” with options: “no problem”, “mild”, “quite bad” and “very bad”. From these two responses we derived a five-category severity variable. Participants who did not complete the questionnaire or did not answer these questions were coded as ‘missing’.
Other variables
Family history of atopic diseases, environment exposures and social class were taken from parental questionnaires during pregnancy. Breastfeeding, comorbidities and siblings were taken from questionnaires at various ages (definitions in Supporting Information).
Genetic polymorphisms
Deoxyribonucleic acid (DNA) was obtained from blood samples using standard extraction techniques18. In children with genome-wide genotyping data, we created an AD polygenic risk score (PRS) using the 25 single nucleotide polymorphisms (SNPs) previously identified as associated with AD (P<5x10−08)19, where dosage genotypes were extracted and weighted by the (risk-increasing) genome-wide association study (GWAS) beta coefficient, summed and standardised to unit standard deviation (1-SD).
In children with filaggrin (FLG) loss-of-function genotyping data, null mutations in any of R2447X, S3247X, 2282del4 or R501X were coded as FLG-present, and FLG-absent was coded otherwise.
Statistical methods
We randomly split the ALSPAC cohort into two equally-sized groups, one (development cohort) for identifying AD phenotypes and associated characteristics, and the other (validation cohort) for internally validating the identified phenotypes. In both groups we excluded individuals with fewer than two of the 11 requested FD reports.
Phenotype identification
We identified phenotypes in the development group using latent class analysis (LCA). FD variables were modelled as joint categorical outcomes in children with and without FD symptoms to allow the LCA to identify a negative control phenotype without AD in addition to AD phenotypes.
We ran models with three to eight classes and compared various model fit statistics (Supporting Information), estimated class sizes, and clinical interpretation (by clinicians with AD management expertise) to determine the best classification. From the best fitting model, we estimated individuals’ posterior class probabilities and identified their most likely phenotype as the class with the highest probability. We drew heatmaps for each phenotype showing individuals’ symptom severity across ages, with a dendrogram to visualise common patterns.
Association of variables with phenotypes
We explored three methods20 to account for phenotype uncertainty (LCA class probabilities) in regression models (Figure S1). We chose the method that ignores uncertainty and treats phenotypes as known quantities, and report 99% confidence intervals to correct for this method’s potential for over-precision.
We regressed sex, family history of AD, asthma, and hayfever, parental education, pet cats and dogs, maternal smoking, breastfeeding (ever and duration), comorbid asthma and older siblings in the household on phenotypes in unadjusted and mutually adjusted multinomial regressions. These factors have been previously associated with AD21. In the mutually adjusted model, we excluded ‘ever breastfed’ to avoid collinearity with breastfeeding duration and additionally adjusted for FLG null mutations to investigate independence of family history risk factors. We estimated crude genetic risk by regressing FLG, SNPs and the PRS on AD phenotypes in unadjusted multinomial regressions, treating SNP and PRS effects as linear.
Phenotype validation
To assess internal validity, we ran the final LCA model in the validation cohort and visually compared resulting phenotypes with those from the development cohort. We repeated the multinomial regressions and qualitatively compared results with development cohort results.
Missing data handling
We assumed missing FD reports were at random given observed reports and estimated the LCA using full information maximum likelihood (FIML). We checked the solution for sensitivity to departures from this assumption (Supporting Information). We assumed missing non-genetic regression variables were at random given observed variables, social class and comorbid hayfever, and used the multivariate imputation by chained equations22 procedure to impute them. We ran regression models on 10 imputed datasets and combined results using Rubin’s rules23. We performed complete-case genetic analyses, assuming genetic-data missingness was completely at random.
Software
LCAs were run in MPlus v8.2 (Muthén & Muthén, Los Angeles, CA, USA) and all other analyses in Stata v16.0 (StataCorp, College Station, TX, USA) and R v3.6.2 (R Foundation for Statistical Computing, Vienna, Austria).
Ethical approval
Ethical approval for this study was obtained from the ALSPAC Ethics and Law Committee (B2510) and the London School of Hygiene and Tropical Medicine Institutional Review Board (EPNCZK46).
RESULTS
Sample characteristics
13,972 children born as singletons or twins were eligible for inclusion. We randomly allocated half (n=6986) to each of the development and validation cohorts. Within each cohort, 85% had data on FD on two or more questionnaires (development: 5927; validation: 5939) and were included in the analyses. In the development and validation cohorts respectively, just under half were female (48%, 49%); 7.7% (7.4%) carried a FLG null mutation and 28% (29%), 19% (18%) and 43% (43%) had at least one parent with a history of AD, asthma and hayfever respectively. All characteristics were balanced between cohorts (Table 1). Reports of FD activity decreased over time. Generally, the activity peaked at 18 months while the lowest activity was observed at age 166 months; similar to the corresponding response rates (92%, 91% at 18 months and 57%, 56% at 166 months) (Table S1).
Table 1:
ALSPAC study characteristics
| Development cohort (n=5927) |
Validation cohort (n=5939) |
|
|---|---|---|
| Sex | ||
| Male | 3099 (52%) | 3031 (51%) |
| Female | 2828 (48%) | 2908 (49%) |
| Social class * | ||
| I | 693 (12%) | 677 (11%) |
| II | 2031 (34%) | 1999 (34%) |
| III(n) | 1203 (20%) | 1219 (21%) |
| III(m) | 387 (6.5%) | 414 (7%) |
| IV | 113 (1.9%) | 125 (2.1%) |
| V | 18 (0.3%) | 7 (0.1%) |
| Missing | 1482 (25%) | 1498 (25%) |
| At least one parent with university degree | ||
| No | 4393 (74%) | 4420 (74%) |
| Yes | 1288 (22%) | 1238 (21%) |
| Missing | 246 (4.1%) | 281 (4.7%) |
| Parental AD | ||
| No | 2563 (43%) | 2557 (43%) |
| Yes | 1678 (28%) | 1700 (29%) |
| Missing | 1686 (28%) | 1682 (28%) |
| Parental asthma | ||
| No | 2967 (50%) | 3041 (51%) |
| Yes | 1123 (19%) | 1059 (18%) |
| Missing | 1837 (31%) | 1839 (31%) |
| Parental hayfever | ||
| No | 1826 (31%) | 1812 (31%) |
| Yes | 2534 (43%) | 2549 (43%) |
| Missing | 1567 (26%) | 1578 (27%) |
| Any older siblings | ||
| No | 433 (7.3%) | 454 (7.6%) |
| Yes | 2105 (35%) | 1962 (33%) |
| Missing | 3389 (57%) | 3523 (59%) |
| Pet cat during pregnancy | ||
| No | 4011 (68%) | 3965 (67%) |
| Yes | 1723 (29%) | 1778 (30%) |
| Missing | 193 (3.2%) | 196 (3.3%) |
| Pet dog during pregnancy | ||
| No | 4353 (73%) | 4358 (73%) |
| Yes | 1381 (23%) | 1385 (23%) |
| Missing | 193 (3.2%) | 196 (3.3%) |
| Mum smoked during first 3m of pregnancy | ||
| No | 4458 (75%) | 4408 (74%) |
| Yes | 1307 (22%) | 1361 (23%) |
| Missing | 162 (2.7%) | 170 (2.9%) |
| Breastfeeding | ||
| Never | 1409 (24%) | 1349 (23%) |
| <3mths | 1214 (20%) | 1240 (21%) |
| 3-5mths | 855 (14%) | 908 (15%) |
| 6mths+ | 1857 (31%) | 1802 (30%) |
| missing | 592 (10%) | 640 (11%) |
| Asthma by 7.5 yrs | ||
| No | 3242 (55%) | 3188 (54%) |
| Yes | 816 (14%) | 829 (14%) |
| Missing | 1869 (31%) | 1922 (32%) |
| Hayfever by 10.5 years | ||
| No | 2877 (48%) | 2856 (48%) |
| Yes | 784 (13%) | 732 (12%) |
| Missing | 2266 (38%) | 2351 (40%) |
| FLG null mutation | ||
| No | 3985 (67%) | 3891 (66%) |
| Yes | 456 (7.7%) | 441 (7.4%) |
| Missing | 1486 (25%) | 1607 (27%) |
| Polygenic risk score (standardised) | ||
| Median [IQR] | −0.13 [−0.78, 0.63] | −0.10 [−0.74, 0.62] |
| Missing | 1971 (33%) | 2113 (36%) |
I Professional occupations, II Managerial and Technical occupations, III(n) Skilled occupations-non-manual, III(m) Skilled occupations-manual, IV Partly-skilled occupations, V Unskilled occupations.
AD phenotypes from infancy to adolescence
From data in the development cohort, we determined the optimal number of phenotypes from LCA models to be five, based on the Bayesian information criterion (Figure S2) and clinical interpretability (Figure S3). Children were generally clearly classified into phenotypes, with high probability of being in their most likely phenotype (mean 74-90%) and low probabilities of being in the other four (mean 0.1%-10%) (Figure S4, Table S2). There was no evidence the LCA solution was sensitive to missing data patterns and it was consistent across the development and validation cohorts (Figures S5-S6, Tables S2-S3).
Based on reported severity and duration of FD, we labelled the classes: Unaffected/Rare, to which 66% of children were assigned with highest probability; Severe-Frequent AD (4%); Moderate-Frequent AD (7%); Moderate-Declining AD (11%); and Mild-Intermittent AD (12%). Children in a particular phenotype did not always report the same symptoms as each other, but heatmaps in children with complete data showed similar patterns with more consistency among individuals in each Frequent phenotype than among individuals in the Declining and Intermittent phenotypes (Figure S7).
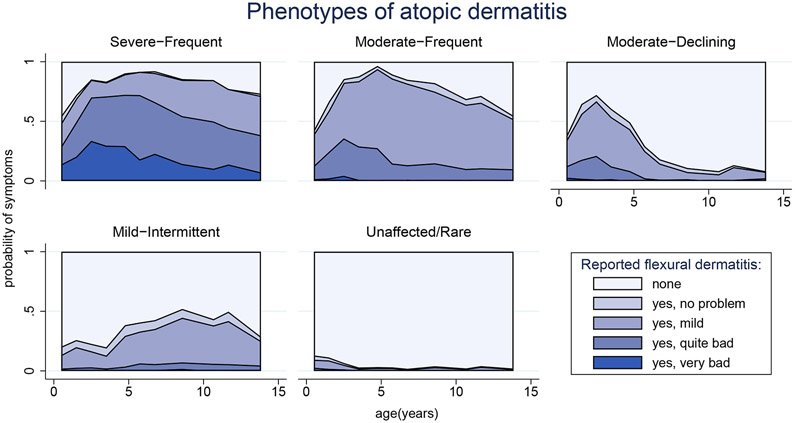
Figure 1 illustrates the distributions of FD severity at each age and how they changed over time for each phenotype. Children in the Severe-Frequent phenotype almost always reported FD and rarely characterised it as ‘no problem’. For example, at six months of age, FD was present and rated ‘very bad’ or ‘quite bad’ in 29% of the group, rising to 49% at 18 months; although heterogeneity was present within this and other subtypes. Children in the Moderate-Frequent phenotype also almost always reported FD, though they usually rated it less severely than children with Severe-Frequent AD and occasionally rated it no problem. Children with Moderate-Declining AD reported symptoms similar to those with Moderate-Frequent AD up to age 2.5, but after this age, their likelihood of FD declined. Children with Mild-Intermittent AD more often reported absence of FD than presence, and when present usually rated it mild or no problem. Up to 9% of the Unaffected/Rare group reported FD before age 2.5, mostly mild, but rarely reported it (<=3% of the time) thereafter. Proportions of individuals with missing symptoms are shown in Figure S8.
Figure 1: Atopic dermatitis phenotypes.
Five phenotypes of atopic dermatitis (including an Unaffected/Rare phenotype without atopic dermatitis) identified by latent class analysis on 5927 ALSPAC children whose mothers responded to at least two questionnaires between the ages of 6 months and 14 years. Probabilities of flexural dermatitis and its severity are shown by age for children assigned to each phenotype (missing reports not indicated): Severe-Frequent, n=230 (3.9%); Moderate-Frequent, n=408 (6.9%); Moderate-Declining, n=676 (11%); Mild-Intermittent, n=684 (12%); Unaffected/Rare, n=3929 (66%).
The distribution of children among phenotypes in the validation cohort was similar to the distribution in the development cohort, except that relatively fewer children were assigned to the Mild-Intermittent phenotype (8% vs 14%) and more to Unaffected/Rare (66% vs 62%).
Variables associated with phenotypes
Table 2 shows the distributions of clinical, demographic and genetic variables used in the regression analyses by most likely phenotype.
Table 2:
ALSPAC development cohort characteristics across each ‘most likely’ atopic dermatitis phenotype
| Severe- Frequent |
Moderate- Frequent |
Moderate- Declining |
Mild- Intermittent |
Unaffected /Rare |
|
|---|---|---|---|---|---|
| Phenotype N (%) | 230 (3.9%) | 408 (6.9%) | 676 (11%) | 684 (12%) | 3929 (66%) |
| Sex | |||||
| Male | 115 (3.7%) | 202 (6.5%) | 366 (12%) | 287 (9.3%) | 2129 (69%) |
| Female | 115 (4.1%) | 206 (7.3%) | 310 (11%) | 397 (14%) | 1800 (64%) |
| At least one parent with university degree | |||||
| No | 158 (3.6%) | 286 (6.5%) | 480 (11%) | 502 (11%) | 2967 (68%) |
| Yes | 63 (4.9%) | 113 (8.8%) | 170 (13%) | 164 (13%) | 778 (60%) |
| Missing | 9 (3.7%) | 9 (3.7%) | 26 (11%) | 18 (7.3%) | 184 (75%) |
| Parental AD | |||||
| No | 73 (2.8%) | 138 (5.4%) | 278 (11%) | 296 (12%) | 1778 (69%) |
| Yes | 95 (5.7%) | 170 (10%) | 249 (15%) | 218 (13%) | 946 (56%) |
| Missing | 62 (3.7%) | 100 (5.9%) | 149 (8.8%) | 170 (10%) | 1205 (71%) |
| Parental asthma | |||||
| No | 99 (3.3%) | 192 (6.5%) | 344 (12%) | 364 (12%) | 1968 (66%) |
| Yes | 61 (5.4%) | 99 (8.8%) | 155 (14%) | 124 (11%) | 684 (61%) |
| Missing | 70 (3.8%) | 117 (6.4%) | 177 (9.6%) | 196 (11%) | 1277 (70%) |
| Parental hayfever | |||||
| No | 56 (3.1%) | 106 (5.8%) | 198 (11%) | 232 (13%) | 1234 (68%) |
| Yes | 122 (4.8%) | 207 (8.2%) | 325 (13%) | 287 (11%) | 1593 (63%) |
| Missing | 52 (3.3%) | 95 (6.1%) | 153 (9.8%) | 165 (11%) | 1102 (70%) |
| Any older siblings | |||||
| No | 17 (3.9%) | 38 (8.8%) | 62 (14%) | 49 (11%) | 267 (62%) |
| Yes | 86 (4.1%) | 168 (8%) | 248 (12%) | 285 (14%) | 1318 (63%) |
| Missing | 127 (3.7%) | 202 (6%) | 366 (11%) | 350 (10%) | 2344 (69%) |
| Pet cat during pregnancy | |||||
| No | 156 (3.9%) | 277 (6.9%) | 453 (11%) | 466 (12%) | 2659 (66%) |
| Yes | 67 (3.9%) | 123 (7.1%) | 205 (12%) | 205 (12%) | 1123 (65%) |
| Missing | 7 (3.6%) | 8 (4.1%) | 18 (9.3%) | 13 (6.7%) | 147 (76%) |
| Pet dog during pregnancy | |||||
| No | 168 (3.9%) | 327 (7.5%) | 505 (12%) | 524 (12%) | 2829 (65%) |
| Yes | 55 (4%) | 73 (5.3%) | 153 (11%) | 147 (11%) | 953 (69%) |
| Missing | 7 (3.6%) | 8 (4.1%) | 18 (9.3%) | 13 (6.7%) | 147 (76%) |
| Mum smoked during first 3 months of pregnancy | |||||
| No | 164 (3.7%) | 337 (7.6%) | 517 (12%) | 535 (12%) | 2905 (65%) |
| Yes | 59 (4.5%) | 63 (4.8%) | 146 (11%) | 131 (10%) | 908 (69%) |
| Missing | 7 (4.3%) | 8 (4.9%) | 13 (8%) | 18 (11%) | 116 (72%) |
| Breastfeeding | |||||
| Never | 49 (3.5%) | 81 (5.7%) | 139 (9.9%) | 160 (11%) | 980 (70%) |
| <3mths | 40 (3.3%) | 76 (6.3%) | 160 (13%) | 128 (11%) | 810 (67%) |
| 3-5mths | 27 (3.2%) | 64 (7.5%) | 98 (11%) | 109 (13%) | 557 (65%) |
| 6mths+ | 86 (4.6%) | 157 (8.5%) | 231 (12%) | 238 (13%) | 1145 (62%) |
| Missing | 28 (4.7%) | 30 (5.1%) | 48 (8.1%) | 49 (8.3%) | 437 (74%) |
| Asthma by 7.5 years | |||||
| No | 96 (3%) | 211 (6.5%) | 360 (11%) | 417 (13%) | 2158 (67%) |
| Yes | 62 (7.6%) | 90 (11%) | 106 (13%) | 119 (15%) | 439 (54%) |
| Missing | 72 (3.9%) | 107 (5.7%) | 210 (11%) | 148 (7.9%) | 1332 (71%) |
| FLG null mutation | |||||
| No | 121 (3%) | 261 (6.5%) | 461 (12%) | 481 (12%) | 2661 (67%) |
| Yes | 55 (12%) | 55 (12%) | 66 (14%) | 58 (13%) | 222 (49%) |
| Missing | 54 (3.6%) | 92 (6.2%) | 149 (10%) | 145 (9.8%) | 1046 (70%) |
| Polygenic risk score (standardised) | |||||
| Median [IQR] | 0.39 [−0.38, 1.47] | 0.18 [−0.73, 0.98] | −0.03 [−0.74, 0.76] | −0.02 [−0.73, 0.64] | −0.22 [−0.82, 0.54] |
| Missing* | 73 (32%) | 125 (31%) | 200 (30%) | 189 (28%) | 1384 (35%) |
missing polygenic risk scores are tabulated within phenotype, while missingness in other variables is tabulated across phenotypes.
We used the Unaffected/Rare phenotype as the reference group in multiple imputation analyses (Table S4). The unadjusted and adjusted estimates were similar, except for parental asthma, hayfever and education: each unadjusted estimate showed a higher risk for all but Mild-Intermittent AD, which attenuated after adjustment. The adjusted estimates in the development and validation cohorts were also similar (Figure 2).
Figure 2: Associations of demographic and clinical characteristics with atopic dermatitis phenotypes.
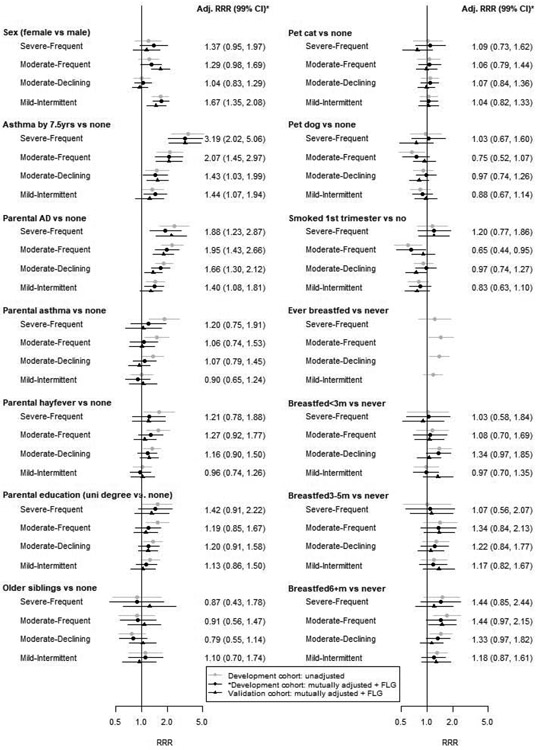
Unadjusted and mutually adjusted phenotype associations in the development cohort (grey and black circles), and adjusted associations in the validation cohort (black triangles). All adjusted associations include additional adjustment for FLG null mutations. Stated effects and confidence intervals are for adjusted associations in the development cohort. Adj=adjusted; RRR=relative risk ratio; CI=confidence interval.
After adjusting for the other variables and FLG, we found that comorbid asthma, parental AD and sex were associated with AD phenotype (all p<0.0001). Children with asthma were three times more likely to be in the Severe-Frequent group (relative risk ratio (RRR) 3.06 99% confidence interval (CI) [1.80, 5.20]), twice as likely to be Moderate-Frequent (RRR 1.98 [1.41, 2.79]); and 1.41 (1.02, 1.94) and 1.44 (1.07, 1.95) times as likely to be Moderate-Declining and Mild-Intermittent, respectively. There was strong evidence (p=0.0001) for this differential association with each phenotype. Children with a family history of AD had twice the risk of Severe-Frequent (RRR 1.97, 1.32, 2.94) or Moderate-Frequent (RRR 1.91 [1.40, 2.62]) disease, 1.61 (1.24, 2.09) times the risk of Moderate-Declining disease and 1.39 (1.07, 1.81) times the risk of Mild-Intermittent disease, but there was no evidence that these associations were truly differential (p=0.0987). Girls were more likely than boys to have Mild-Intermittent AD (RRR 1.67 (1.34, 2.08)). There was no evidence that the other Table 2 variables were independently associated with AD phenotypes.
The patterns of associations above were replicated in the validation cohort with similar estimates and confidence intervals, except that differential parental AD RRRs were detectable across phenotypes (p=0.0039). Additionally, in the validation cohort there was borderline evidence (p=0.0063) that maternal smoking was associated with phenotypes after adjusting for other variables; there was similar evidence (p=0.0135) for the same association in the development cohort. Children whose mothers smoked were less likely to have Moderate-Frequent AD in the development cohort and less likely to have Moderate-Declining AD in the validation cohort; the other associations were inconclusive.
Genetic phenotype associates
Of children included in this study, 8773 (74%) had FLG genotype data and 7782 (66%) had available genome-wide genotype data.
FLG loss-of-function mutations
FLG was strongly associated with AD phenotypes (p=4.26x10−32), with strong evidence of higher risks for more severe and frequently-active phenotypes (p=1.23x10−10). Children with FLG mutations were more than four times more likely to have Severe-Frequent AD (RRR 4.32 [3.07, 6.07]), more than twice as likely to have Moderate-Frequent AD (RRR 2.41 [1.81, 3.22]), 1.80 (1.39, 2.34) times more likely to have Moderate-Declining AD and 1.52 (1.13, 2.04) times more likely to have Mild-Intermittent AD than children without FLG mutations (Table 3).
Table 3:
Associations between genetic profiles and atopic dermatitis phenotypes in ALSPAC children
| Severe-Frequent* N=305 |
Moderate- Frequent* N=634 |
Moderate- Declining* N=1008 |
Mild-Intermittent* N=845 |
|||||
|---|---|---|---|---|---|---|---|---|
| Chr. | Nearest gene |
Global p- value† |
RRR (99% CI) | RRR (99% CI) | RRR (99% CI) | RRR (99% CI) | Equality p- value‡ |
|
| FLG null mutation | 4.26E-32 | 4.32 (3.07, 6.07) | 2.41 (1.81, 3.22) | 1.80 (1.39, 2.34) | 1.52 (1.13, 2.04) | 1.23E-10 | ||
| Polygenic risk score | 1.22E-28 | 1.67 (1.45, 1.93) | 1.37 (1.23, 1.52) | 1.17 (1.07, 1.27) | 1.13 (1.02, 1.24) | 8.60E-11 | ||
| Individual SNP associations | ||||||||
| rs11205006_a | 1 | LCE5A, FLG-AS1 | 9.94E-07 | 1.55 (1.24, 1.93) | 1.22 (1.03, 1.44) | 1.11 (0.97, 1.28) | 1.10 (0.95, 1.28) | 3.20E-03 |
| rs2228145_c | 1 | IL6R | 3.69E-04 | 1.31 (1.06, 1.63) | 1.15 (0.99, 1.34) | 0.95 (0.83, 1.08) | 0.93 (0.81, 1.07) | 1.49E-04 |
| rs61813875_g | 1 | LCE3E, CRCT1 | 8.63E-10 | 2.99 (1.79, 4.98) | 2.07 (1.34, 3.19) | 1.96 (1.36, 2.84) | 1.33 (0.84, 2.09) | 9.82E-03 |
| rs1057258_c | 2 | INPP5D | 5.81E-02 | 1.21 (0.90, 1.63) | 0.99 (0.81, 1.21) | 0.97 (0.82, 1.14) | 0.86 (0.72, 1.01) | 4.81E-02 |
| rs112111458_a | 2 | CD207 | 6.68E-02 | 1.18 (0.85, 1.65) | 1.00 (0.80, 1.25) | 1.17 (0.96, 1.42) | 0.91 (0.75, 1.10) | 4.42E-02 |
| rs13015714_g | 2 | IL18R1 | 5.40E-02 | 1.07 (0.83, 1.38) | 1.23 (1.03, 1.47) | 1.06 (0.92, 1.24) | 1.02 (0.87, 1.20) | 1.74E-01 |
| rs17389644_a | 4 | IL21 | 9.94E-02 | 1.17 (0.91, 1.51) | 1.07 (0.89, 1.29) | 1.15 (0.99, 1.33) | 1.02 (0.87, 1.21) | 4.36E-01 |
| rs10214237_t | 5 | IL7R | 3.01E-01 | 0.96 (0.75, 1.21) | 1.06 (0.89, 1.26) | 1.01 (0.88, 1.17) | 1.13 (0.96, 1.32) | 3.66E-01 |
| rs2897442_c | 5 | KIF3A | 4.78E-02 | 1.29 (1.02, 1.63) | 1.11 (0.93, 1.32) | 0.99 (0.86, 1.15) | 0.99 (0.85, 1.16) | 4.02E-02 |
| rs12153855_t | 6 | TNXB | 8.75E-03 | 1.04 (0.72, 1.50) | 1.42 (1.06, 1.91) | 1.07 (0.86, 1.33) | 1.22 (0.96, 1.56) | 1.32E-01 |
| rs6473227_c | 8 | ZBTB10 | 2.49E-01 | 1.12 (0.90, 1.40) | 1.09 (0.93, 1.28) | 1.07 (0.94, 1.22) | 1.06 (0.93, 1.22) | 9.47E-01 |
| rs10995251_c | 10 | ZNF365 | 5.82E-01 | 0.99 (0.79, 1.23) | 0.99 (0.84, 1.16) | 1.08 (0.95, 1.24) | 1.03 (0.89, 1.18) | 5.87E-01 |
| rs12295535_t | 11 | PRR5L | 8.22E-03 | 1.55 (0.92, 2.61) | 1.21 (0.80, 1.84) | 1.37 (0.99, 1.91) | 0.78 (0.50, 1.22) | 1.83E-02 |
| rs479844_g | 11 | OVOL1 | 1.17E-05 | 1.51 (1.21, 1.90) | 1.17 (1.00, 1.37) | 1.06 (0.93, 1.21) | 1.06 (0.92, 1.22) | 1.34E-03 |
| rs7127307_t | 11 | ETS1 | 5.28E-01 | 1.05 (0.84, 1.30) | 1.07 (0.92, 1.25) | 1.00 (0.88, 1.13) | 1.07 (0.94, 1.23) | 6.39E-01 |
| rs7927894_t | 11 | C11orf30 | 2.48E-02 | 1.21 (0.98, 1.50) | 1.09 (0.94, 1.28) | 0.96 (0.84, 1.09) | 1.10 (0.96, 1.26) | 3.74E-02 |
| rs2227483_t | 12 | IL22 | 9.80E-01 | 1.03 (0.83, 1.29) | 1.01 (0.86, 1.18) | 1.00 (0.88, 1.14) | 1.03 (0.90, 1.18) | 9.75E-01 |
| rs2143950_t | 14 | PPP2R3C | 1.19E-02 | 1.24 (0.95, 1.62) | 1.26 (1.04, 1.53) | 1.01 (0.86, 1.20) | 1.06 (0.89, 1.27) | 5.74E-02 |
| rs7146581_c | 14 | TRAF3 | 1.83E-01 | 1.09 (0.84, 1.42) | 1.16 (0.96, 1.40) | 1.09 (0.94, 1.27) | 1.04 (0.88, 1.22) | 6.78E-01 |
| rs2041733_t | 16 | CLEC16A | 4.41E-01 | 0.98 (0.79, 1.22) | 1.12 (0.96, 1.30) | 1.00 (0.88, 1.14) | 1.03 (0.90, 1.18) | 4.20E-01 |
| rs11657987_t | 17 | PGS1 | 9.36E-02 | 1.22 (0.99, 1.51) | 1.09 (0.94, 1.27) | 1.02 (0.90, 1.15) | 1.06 (0.92, 1.21) | 2.44E-01 |
| rs16948048_g | 17 | ZNF652 | 7.69E-02 | 1.18 (0.95, 1.46) | 0.92 (0.79, 1.08) | 0.98 (0.86, 1.11) | 0.92 (0.80, 1.06) | 5.50E-02 |
| rs17881320_t | 17 | STAT3 | 2.25E-01 | 1.30 (0.91, 1.86) | 1.12 (0.85, 1.47) | 1.13 (0.90, 1.41) | 1.09 (0.86, 1.39) | 7.37E-01 |
| rs2164983_a | 19 | ACTL9 | 1.26E-05 | 1.34 (1.02, 1.77) | 1.40 (1.15, 1.70) | 1.16 (0.98, 1.37) | 1.18 (0.99, 1.42) | 1.49E-01 |
| rs6010620_g | 20 | RTEL1 | 9.39E-02 | 1.14 (0.88, 1.47) | 1.17 (0.97, 1.41) | 1.05 (0.90, 1.21) | 1.11 (0.94, 1.30) | 6.05E-01 |
compared with the Unaffected/Rare population, n=4990
null hypothesis: RRR1=RRR2=RRR3=RRR4=0
null hypothesis: RRR1=RRR2=RRR3=RRR4. FLG=filaggrin gene; SNP=single nucleotide polymorphism; Chr=chromosome; RRR=relative risk ratio; CI=confidence interval.
Standardised Genetic Score
The PRS was strongly associated with AD phenotypes (p=1.22x10−28), with strong evidence of higher risks for more severe and frequently-active phenotypes (p=8.60x10−11). A score increase of 1-SD was associated with a relative risk increase of: 1.67 (1.45, 1.93) for Severe-Frequent; 1.37 (1.23, 1.52) for Moderate-Frequent; 1.17 (1.07, 1.27) for Moderate-Declining; and 1.13 (1.02, 1.24) for Mild-Intermittent AD. Removing rs11205006 from the score, which tags the FLG locus, and the remainder of the Chromosome 1 SNPs made little difference to these associations (Table S5).
SNPs
Eight SNPs on Chromosomes 1, 6, 11, 14 and 19 were associated with AD phenotypes (p=8.63x10−10 - 1.19x10−02; Table 3). Of these, five were associated with Severe-Frequent AD (RRR range 1.31-2.99); six with Moderate-Frequent (RRR range 1.17-2.07); and one with Moderate-Declining (RRR 1.96). There was no evidence that any single SNP was associated with Mild-Intermittent AD.
Association differences between AD phenotypes
We explored whether any of the variables we investigated could distinguish between the four AD phenotypes. In six post-hoc Bonferroni-corrected pairwise comparisons from the multinomial regression models, we found evidence that the relative risk associated with asthma, sex, FLG null mutations and the AD-PRS differed among some phenotype pairs (Table S6). In particular, the AD-PRS and FLG were able to discriminate between four of six pairs showing a significantly higher relative risk in the more severe and/or frequently affected phenotypes. The remaining comparisons were inconclusive, generally producing confidence intervals consistent with important effects in both directions.
DISCUSSION
We identified four phenotypes of AD based on disease severity and trajectories during childhood: Severe-Frequent, Moderate-Frequent, Moderate-Declining and Mild-Intermittent disease. Providers and patients might be reassured that in all subgroups, the probability of reporting ‘quite bad’ or ‘very bad’ symptoms declined with age (with evidence of within group heterogeneity). Associations with some established risk factors (FLG null mutations, AD-PRS, family history of AD) and comorbidities (asthma) were present in all phenotypes with evidence of a gradient of association from more severe to less severe phenotypes. AD-PRS and FLG mutations were further able to differentiate many of our phenotypes from each other, highlighting the need for increased understanding of drivers of disease severity/activity throughout life in people with AD, including the role of gene-environment interactions.
Our findings add novel information about patterns of disease severity over time, and are consistent with prior cross-sectional studies on disease severity and longitudinal studies focused on disease activity alone. While disease activity (i.e. the presence of any symptoms at a particular age) and disease severity (i.e. the impact of those symptoms) are sometimes correlated, they are separate concepts, and it is important to differentiate them24. For example, a small patch of mild eczema that persists for years may be quite different from extensive severe disease that quickly resolves.
Paternoster et al5 conducted a previous study in ALSPAC with external validation25, Hu et al6 examined a multi-ethnic population up to age 10, and Roduit et al4 examined an European population up to age 6. All reported phenotypes described onset timing and persistence during childhood, and between them there were some commonly described phenotypes (early-persistent, early-transient, late-onset). Hu et al concluded that established risk factors have little capacity to differentiate eczema phenotypes. By additionally incorporating severity, the most clinically relevant aspect of AD, we have been able to show that genetic factors, in particular, have potential to differentiate between phenotypes.
Our study has several strengths. We used a well-characterised, population-based cohort, hence, our results are generalisable to similar settings and populations. Our large sample (n=11,866) offered precision; and our phenotypes were internally reproducible, lending confidence to our findings. We applied robust statistical methods to minimise imprecision and bias in our regression estimates.
Our study also has limitations. We used a measure of self-reported AD, which has not been directly validated in ALSPAC, but has been shown to be a reasonable predictor in other contexts, including validation by physical exam in the UK26 and physician assessment in a multi-centre US study27. Missing data could have introduced bias into our estimates, although we tried to minimise this risk with FIML estimation and multiple imputation techniques. There is uncertainty in phenotype assignment that could affect regression results, however, we compared several methods of accounting for it and found little difference in our results, suggesting insensitivity to this issue. Finally, there are few data sources recording AD severity, and we were unable to find a large cohort with similarly characterised children to externally validate our findings, though we did attempt this using a small cohort (Supporting Information).
The LCA method involves probabilistic classification into subgroups and individual disease trajectories can vary within subgroups. For example, on average, active disease becomes less likely over time in the Moderate-Declining group, but an individual classified into this phenotype may still have active FD in late childhood. Future work may consider how early aggressive treatment for severe AD might modify the natural history28.
Our study adds important information about patterns of AD severity over time. The identified phenotypes were stable across sensitivity analyses and had strong genetic associations, providing support for using them in future work toward a better aetiological understanding of AD, including the role of environmental risk factors throughout the life course, and toward better clinical trial design. Additional research is underway to determine the utility of our phenotypes for prognosis and to inform priorities for early intervention.
Supplementary Material
What’s already known about this topic?
Atopic dermatitis (AD) is a heterogeneous condition in terms of both disease activity and severity.
Childhood AD phenotypes in previous studies have focused only on disease activity.
What does this study add?
We incorporate disease severity over time to derive four clinically recognisable AD phenotypes using data-driven methods.
Disease severity improved over time in all phenotypes (even in those with high probability of activity in late childhood and adolescence).
Several established risk factors, including genetics associates, were associated with our proposed phenotypes, with most factors more strongly associated with phenotypes reporting the worst symptoms. Fewer factors differentiated between more frequent and declining/intermittent phenotypes.
ACKNOWLEDGEMENTS
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We also thank Drs Sadia Haider and Toshinori Nakamura at the National Heart & Lung Institute, Imperial College London, for their valuable input into the design of this research.
Funding
This article presents independent research supported by a Wellcome Trust Senior Research Fellowship in Clinical Science to SML (205039/Z/16/Z) and a National Institute of Arthritis and Musculoskeletal and Skin Diseases Career Development Award to KA (K23AR073915). This work was also supported by British Skin Foundation Innovative grant (8066) and Health Data Research UK (LOND1), which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome Trust. It was also supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement no 668954. The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and AM will serve as guarantor for the contents of this paper.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funders.
Footnotes
Conflicts of Interest
AC reports personal fees from Novartis, personal fees from Thermo Fisher Scientific, personal fees from Philips, personal fees from Sanofi, personal fees from Stallergenes Greer, outside the submitted work. KA reports consultancy for Target RWE and institutional grant funding from Pfizer, outside the submitted work. ADI has received consultancy fees from AbbVie, Arena Pharmaceuticals, Benevolent AI, Chugai, Dermavant, Genentech, LEO Pharma, Eli Lilly, Menlo Therapeutics, Novartis, Pfizer, Regeneron, Sanofi, and UCB.
All others report no conflicts of interest.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/BJD.19885
REFERENCES
- 1.Custovic A, Henderson J, Simpson A. Does understanding endotypes translate to better asthma management options for all? J Allergy Clin Immunol [Internet]. 2019July1 [cited 2020 May 13]; 144(1):25–33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31145940 [DOI] [PubMed] [Google Scholar]
- 2.Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol [Internet]. 2006/July/04. 2006;118(1):209–13. Available from: https://www.ncbi.nlm.nih.gov/pubmed/16815157 [DOI] [PubMed] [Google Scholar]
- 3.Flohr C, Johansson SG, Wahlgren CF, Williams H. How atopic is atopic dermatitis? J Allergy Clin Immunol [Internet]. 2004;114(1):150–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15241359 [DOI] [PubMed] [Google Scholar]
- 4.Roduit C, Frei R, Depner M, Karvonen AM, Renz H, Braun-Fahrlander C, et al. Phenotypes of Atopic Dermatitis Depending on the Timing of Onset and Progression in Childhood. JAMA Pediatr [Internet]. 2017/05/23. 2017;171(7):655–62. Available from: https://www.ncbi.nlm.nih.gov/pubmed/28531273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paternoster L, Savenije OEM, Heron J, Evans DM, Vonk JM, Brunekreef B, et al. Identification of atopic dermatitis subgroups in children from 2 longitudinal birth cohorts. J Allergy Clin Immunol [Internet]. 2017/November/14. 2017; Available from: https://www.ncbi.nlm.nih.gov/pubmed/29129583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Duijts L, Erler NS, Elbert NJ, Piketty C, Bourdès V, et al. Most associations of early-life environmental exposures and genetic risk factors poorly differentiate between eczema phenotypes: the Generation R Study. Br J Dermatol [Internet]. 2019December30 [cited 2020 Nov 10]; 181(6):1190–7. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/bjd.17879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herr M, Just J, Nikasinovic L, Foucault C, Le Marec AM, Giordanella JP, et al. Risk factors and characteristics of respiratory and allergic phenotypes in early childhood. J Allergy Clin Immunol. 2012;130(2). [DOI] [PubMed] [Google Scholar]
- 8.Belgrave DCM, Granell R, Simpson A, Guiver J, Bishop C, Buchan I, et al. Developmental Profiles of Eczema, Wheeze, and Rhinitis: Two Population-Based Birth Cohort Studies. PLoS Med. 2014;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bousquet J, Anto JM, Wickman M, Keil T, Valenta R, Haahtela T, et al. Are allergic multimorbidities and IgE polysensitization associated with the persistence or reoccurrence of foetal type 2 signalling? the MeDALL hypothesis. Allergy Eur J Allergy Clin Immunol. 2015;70(9):1062–78. [DOI] [PubMed] [Google Scholar]
- 10.Hose AJ, Depner M, Illi S, Lau S, Keil T, Wahn U, et al. Latent class analysis reveals clinically relevant atopy phenotypes in 2 birth cohorts. J Allergy Clin Immunol. 2017June1;139(6):1935–1945.e12. [DOI] [PubMed] [Google Scholar]
- 11.Langan SM, Irvine AD, Weidinger S. Atopic dermatitis. Vol. 396, The Lancet. Lancet Publishing Group; 2020. p. 345–60. [DOI] [PubMed] [Google Scholar]
- 12.Simpson EL, Bruin-Weller M, Flohr C, Ardern-Jones MR, Barbarot S, Deleuran M, et al. When does atopic dermatitis warrant systemic therapy? Recommendations from an expert panel of the International Eczema Council. J Am Acad Dermatol. 2017October;77(4):623–33. [DOI] [PubMed] [Google Scholar]
- 13.Oksel C, Haider S, Fontanella S, Frainay C, Custovic A. Classification of pediatric asthma: From phenotype discovery to clinical practice [Internet]. Vol. 6, Frontiers in Pediatrics. Frontiers Media S.A.; 2018. [cited 2020 May 13]. p. 258. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30298124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort Profile: The ‘Children of the 90s’-the index offspring of the Avon Longitudinal Study of Parents and Children. [cited 2020 Feb 10]; Available from: http://ije.oxfordjournals.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser A, Macdonald-wallis C, Tilling K, Boyd A, Golding J, Davey smith G, et al. Cohort profile: The avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol [Internet]. 2013February [cited 2020 Nov 10];42(1):97–110. Available from: https://pubmed.ncbi.nlm.nih.gov/22507742/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Explore data and samples ∣ Avon Longitudinal Study of Parents and Children ∣ University of Bristol [Internet]. [cited 2020 Nov 10]. Available from: http://www.bristol.ac.uk/alspac/researchers/our-data/ [Google Scholar]
- 17.Asher M, Keil U, Anderson H, Beasley R, Crane J, Martinez F, et al. International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur Respir J. 1995;8(3). [DOI] [PubMed] [Google Scholar]
- 18.Paternoster L, Standl M, Chen CM, Ramasamy A, Bøpnnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet. 2012;44(2):187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multiancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015December1;47(12):1449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark SL, Muthén B. Relating Latent Class Analysis Results to Variables not Included in the Analysis. 2009. [Google Scholar]
- 21.Blakeway H, Van-de-Velde V, Allen VB, Kravvas G, Palla L, Page MJ, et al. What is the evidence for interactions between filaggrin null mutations and environmental exposures in the aetiology of atopic dermatitis? A systematic review. British Journal of Dermatology. Blackwell Publishing Ltd; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White IR, Royston P, Wood AM. Multiple imputation using chained equations: Issues and guidance for practice. Stat Med [Internet]. 2011February20 [cited 2020 Feb 10];30(4):377–99. Available from: http://doi.wiley.com/10.1002/sim.4067 [DOI] [PubMed] [Google Scholar]
- 23.Multiple Imputation for Nonresponse in Surveys - Rubin Donald B. - Google Books [Internet]. [cited 2020 Apr 20]. Available from: https://books.google.co.uk/books?hl=en&lr=&id=bQBtw6rx_mUC&oi=fnd&pg=PR24&dq=Rubin,+D.B.+(1987).+Multiple+Imputation+for+Nonresponse+in+Surveys.+New+York:+John+Wiley+and+Sons.&ots=8OvM9L6-bQ&sig=jNcnlbxsEl8VOYswI-KMcw912m8#v=onepage&q=Rubin%2C D.B. (1987). Multiple Imputation for Nonresponse in Surveys. New York%3A John Wiley and Sons.&f=false [Google Scholar]
- 24.Abuabara K, Margolis DJ, Langan SM. The Long-term Course of Atopic Dermatitis. Dermatol Clin. 2017;35(3):291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wijga AH, Kerkhof M, Gehring U, De jongste JC, Postma DS, Aalberse RC, et al. Cohort profile: The prevention and incidence of asthma and mite allergy (PIAMA) birth cohort. Int J Epidemiol. 2014;43(2):527–35. [DOI] [PubMed] [Google Scholar]
- 26.McNally NJ, Williams HC, Phillips DR, Strachan DP. Is there a geographical variation in eczema prevalence in the U.K.? Evidence from the 1958 British birth cohort study. Br J Dermatol. 2000;142(4):712–20. [DOI] [PubMed] [Google Scholar]
- 27.Silverberg JI, Patel N, Immaneni S, Rusniak B, Silverberg NB, Debashis R, et al. Assessment of atopic dermatitis using self-report and caregiver report: A multicentre validation study. Br J Dermatol. 2015;173(6):1400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boguniewicz M, Fonacier L, Guttman-Yassky E, Ong PY, Silverberg J, Farrar JR. Atopic dermatitis yardstick: Practical recommendations for an evolving therapeutic landscape. Ann Allergy, Asthma Immunol [Internet]. 2018January1 [cited 2020 Jul 20];120(1): 10–22.e2. Available from: 10.1016/j.anai.2017.10.039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.