Abstract
Adenosine signaling plays a critical role in the maintenance of articular cartilage and may serve as a novel therapeutic for osteoarthritis (OA), a highly prevalent and morbid disease without effective therapeutics in the current market. Mice lacking adenosine A2A receptors (A2AR) develop spontaneous OA by 16 weeks of age, a finding relevant to human OA since loss of adenosine signaling due to diminished adenosine production (NT5E deficiency) also leads to development of OA in mice and humans. To better understand the mechanism by which A2AR and adenosine generation protect from OA development, we examined differential gene expression in neonatal chondrocytes from WT and A2AR null mice. Analysis of differentially expressed genes was analyzed by KEGG pathway analysis, and oPOSSUM and the flatiron database were used to identify transcription factor binding enrichment, and tissue-specific network analyses and patterns were compared to gene expression patterns in chondrocytes from patients with OA. There was a differential expression of 2211 genes (padj<0.05). Pathway enrichment analysis revealed that pro-inflammatory changes, increased metalloprotease, reduced matrix organization, and homeostasis are upregulated in A2AR null chondrocytes. Moreover, stress responses, including autophagy and HIF-1 signaling, seem to be important drivers of OA and bear marked resemblance to the human OA transcriptome. Although A2AR null mice are born with grossly intact articular cartilage, we identify here the molecular foundations for early-onset OA in these mice, further establishing their role as models for human disease and the potential use of adenosine as a treatment for human disease.
Keywords: Osteoarthritis, Chondrocyte transcriptome, Autophagy, Stress response, A2AR null mouse
Cristina M. Castro graduated with a BSc from the University of Puerto Rico at Mayagüez and began the Medical Scientist Training Program at NYU Grossman School of Medicine in 2014. She completed her PhD in Inflammation and Immunology in 2019. She focused on adenosine signaling as a promotor of mitochondrial metabolism and repair, primarily by mitigation of reactive oxygen species in vitro and in vivo. She will compete her MD training in May 2021 and will begin residency training in Internal Medicine at Harvard’s BIDMC this summer. She is interested in carrying out translational research during her career, focused on how inflammation accelerates aging and promotes autoimmunity.

Introduction
Over 30 million American adults suffer from osteoarthritis (OA) while current therapies fail to modulate the pathogenesis or progression of the disease [1]. The number of surgeries to treat OA is increasing rapidly, with nearly a million total hip and knee replacements carried out in 2010 and an expected 2-fold increase in total hip replacements and more than 5-fold increase in total knee replacements by 2030 due to both increasing obesity rates and aging of the population [2]. OA is a great financial burden on the healthcare system and restricts Americans’ livelihoods and productivity [2–4]. Costs—including loss of work, medications, hospitalization for joint replacement and/or repair surgery, and other types of therapy—are high and consume up to 0.5% of the gross domestic product of the USA [4].
Much work has been underway to develop a drug that is well tolerated and effective at halting and reversing OA pathology. Chondrocyte homeostasis is maintained by autocrine adenosine signaling, namely the sustained release of ATP which is converted extracellularly to adenosine which then binds to its A2A receptor (A2AR) to regulate chondrocyte function [5]. Our group has shown that ligation of the A2AR has translational value for human disease and could serve as the first disease-modifying drug for OA.
Application of liposomal preparations of either adenosine or the selective A2AR agonist (CGS21680) abrogates development of OA in models of post-traumatic OA (PTOA) in rats [5, 6] and obesity-induced OA in mice [6, 7]. Moreover, A2AR null mice develop spontaneous OA by 16 weeks [5, 8]. As previously reported, the absence of A2AR signaling in null mice yields increased the expression of metalloproteases (i.e., MMP-13) and collagen X (associated with chondrocyte hypertrophy) in tissue culture of neonatal chondrocytes and as early as 12 weeks in vivo, preceding onset of OA [5]. With aging or in the setting of OA, intracellular ATP levels drop in chondrocytes [9, 10] leading to a reduction in ATP release and extracellular adenosine resulting in diminished endogenous, autocrine A2AR stimulation and dysregulation of chondrocyte function [5, 8]. A2AR ligation also modulates aging and inflammation by regulating mitochondrial function [11] and stress responses, including autophagy [12]. Recent work on the human OA chondrocyte transcriptome by Fisch et al. suggests that there are disease-related changes in extracellular matrix organization and hypoxia-modulated and autophagy pathways [13]. Further evidence for the role of the A2AR and endogenous adenosine production is the observation that almost all patients with genetic absence of ecto-5′nucleotidase activity (5NTE), which hydrolyzes AMP to adenosine at the cell surface, develop premature osteoarthritis [14, 15]. Because we had already observed that neonatal A2AR null chondrocytes had increased expression of MMP-13 and collagen X [5], we wanted to test the hypothesis that the A2AR null chondrocyte transcriptome reveals major drivers of OA, recapitulating human OA [13], providing further evidence that the loss of autocrine signaling of the A2AR disrupts homeostasis, leading to the development of OA in humans. Moreover, these data further confirm that the A2AR is a novel target for new therapies for OA.
Methods
Animals and Primary Chondrocyte Harvest
Mice employed in this study were kept under regular lighting conditions (12-h light/dark cycles) and given food and water ad libitum. Adenosine A2A receptor knockout (A2ARKO) mice, bred on a C57BL/6 background (referred to here as wild type, WT), were kindly provided by Dr. Jiang Fan Chen (Boston University School of Medicine, Boston, MA). Mice were bred and raised in the same facility (Science Building Animal Facility, NYU Langone Medical Center), receiving the same food and care. Phenotype had been confirmed by genotyping as previously reported [5]. Neonatal pups were sacrificed by decapitation and used for chondrocyte extraction from knee and hip joint as previously reported [5, 16]. Briefly, A2ARKO and WT pups from 1 to 5 days of age were sacrificed and their knees and femoral heads were collected and digested in two subsequent incubations of 3 mg/mL of collagenase D in serum-free DMEM for 45 min each at 37 °C. The cartilage chunks were agitated then incubated overnight with 0.5 mg/mL of collagenase D in serum-free DMEM at 37 °C. The following day, the digested cartilage was run through a 48-μm nylon mesh and centrifuged to pull down chondrocytes. Harvested chondrocytes were then cultured with full media and grown to confluence (without passage): DMEM supplemented with 4 mM L-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 10% fetal bovine serum (FBS) [16]. This protocol has been validated to yield chondrocyte predominant cell populations [5, 6, 16] since articular cartilage is composed of isolated chondrocytes within extracellular matrix of their own making. Indeed, we have previously used this technique in our prior work, where we were able to show (even after 1–2 passages) that primary chondrocytes retain the phenotype we observed in intact cartilage by immunohistochemistry [5]. This method, prior to RNA extraction, increases RNA yield compared to that achieved isolated directly from cartilage, which is only 2 cells thick in mice [17]. The New York University School of Medicine Institutional Animal Care and Use Committee approved all protocols for experimental procedures involving the use of animals according to US law.
RNA extraction and RNA sequencing
After harvesting, pooled chondrocytes from a single litter (males and females combined of either WT or A2ARKO) were allowed to grow to confluence without passage in a well of a 6-well plate (Thermo Scientific 140675). RNA was extracted from trypsinized primary cells using QUIAGEN’s RNeasy Mini Kit (cat. no. 74104) according to manufacturer’s instructions. Three litters of WT and A2ARKO were processed and submitted for RNA sequencing, namely WT1-3 and A2ARKO1-3. Isolated RNA was quantified for normalization and submitted to and processed by the Genome Technology Center (GTC) at NYU Langone Health using Illumina HiSeq 4000 to acquire chondrocytes’ transcriptome.
Sequencing data preprocessing
RNA library and sequencing files was generated by GTC at NYU Langone Health. Fastqc was run to assess the read quality and adapter contamination of resultant fastq files. Alignment was performed with STAR aligner using mouse reference genome (mm10). At least 48 million reads were uniquely aligned to the reference demonstrating high coverage of the transcriptome. Aligned BAM files were then used for gene feature count with HT-seq and normalized by library size and gene length to generate FPKM values.
Differential profiling and pathway analysis
Differential expression was performed with DEseq2 on raw count with simple linear formula (~ condition). We define genes with padj≤0.05 as differentially expressed (DE) where 2211 genes passed such threshold. Genes with padj≤0.05 and log2(Fold change)>1 were highlighted as yellow, and the rest of the genes were as blue in the volcano plot. Genes with padj less than which of Adora2 were text highlighted. Pathway enrichment was performed on gene sets in OA-related processes using Panther with fisher’s exact method and −log10(BH adjusted p value) was plotted as the x axis and the direction was determined by the mean fold change between WT and A2ARKO in each gene set. Gene-wise z-scored FPKM values of genes involved in autophagy are subsetted for heatmap visualization. Pearson correlation coefficient was used as clustering distance and complete linkage was used as clustering method.
Transcription factor binding enrichment analysis
Transcription factor (TF) binding enrichment analysis was performed to identify TFs where their binding sites are positively enriched in differentially expressed genes. R package biomaRt was utilized to annotate transcription factors with go term GO:0003700. As a result, we detected 631 transcription factors where 109 are DE. We further use all 2211 differentially expressed genes for transcription factor binding enrichment analysis with oPOSSUM mouse single site method. All genes in the oPOSSUM database were used Background gene set [18]. JASPAR CORE profiles were used as a pool of potential transcription factor binding sites. Conservation cutoff and matrix score threshold were set as 0.4 and 85% respectively for regions 1000 bp upstream of the differentially expressed genes. Returning positive enrichment z-score was used to indicate positive enrichment of transcription factor binding sites on the selected differentially expressed genes.
Tissue-specific network analysis
Differentially expressed transcription factors (TF) shown positive regulation (z-score>0) in our differentially expressed genes plus FOXO1 and FOXO3 are chosen for cartilage specific network construction. We utilized flatiron database as described previously [19]. Five types of data (transcription factor binding, protein-protein interaction, co-expression, microRNA targets, and perturbation) are chosen as reference for network edges, prioritized by uniqueness to the query genes with minimum interaction confidence as 0.08 and maximum number of genes equal to 50. Network visualization was performed with Cytoscape where edges with weight less than 0.2 were excluded [20]. Pathways were annotated based upon investigator’s curation.
Results
A2ARKO chondrocyte transcriptome reveals extracellular matrix catabolism and increased senescence, pro-inflammatory, and oxidative mediators
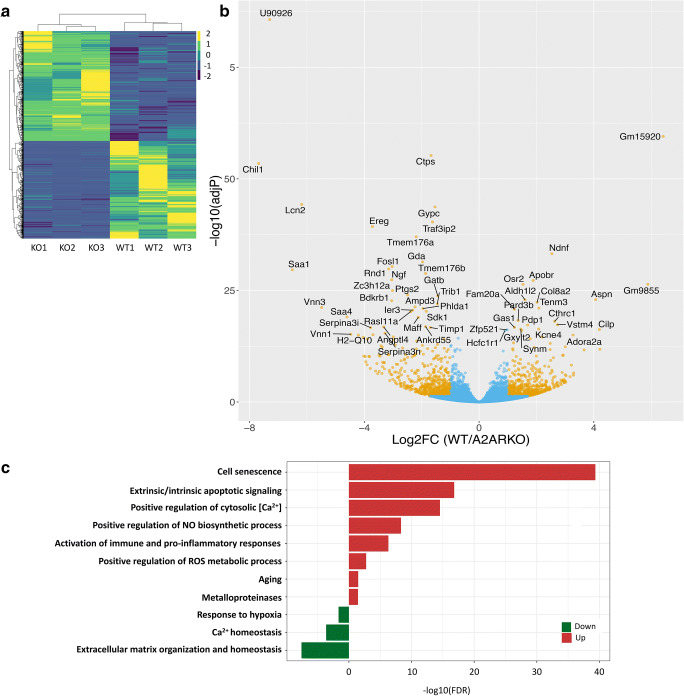
WT and A2AR null (A2ARKO) chondrocytes were isolated from neonatal mouse knees and submitted for RNA sequencing. Data were processed, and out of 16280 genes detected, 2211 were determined to be differentially expressed with a padj<0.05 (Fig. 1a). Differentially expressed (DE) genes are also illustrated as a volcano plot (Fig. 1b) where genes with padj≤0.05 and log2(Fold change)>1 were highlighted as yellow, and the rest of the genes were as blue. Raw data files can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146349. Genes with padj less than that of Adora2 were text highlighted. Pathway enrichment analysis was performed on differentially expressed genes revealing that pro-inflammatory changes and catabolic markers are upregulated in primary chondrocytes isolated in the neonate (Fig. 1c). We had already reported that A2ARKO primary chondrocytes showed signs of hypertrophy, including increased metalloprotease and collagen X expression, and A2ARKOs develop spontaneous OA as early as 12–16 weeks of age [5]. We have also previously shown that chondrocytes isolated from neonatal A2ARKO pups have unstable mitochondrial polarity and increased oxidative damage at baseline [11]. Along those findings, we here show that primary chondrocytes from neonates have increased activation of immune and pro-inflammatory responses, metalloproteinases, and reduced matrix organization and homeostasis (Fig. 1c). A2ARKO primary chondrocytes also show increased cell senescence and aging markers, increased extrinsic and intrinsic apoptotic signaling, and increased NO and ROS metabolism. These data indicate that these changes are present at birth leading to the compromise of cell viability and are major drivers of OA pathogenesis in our animal model.
Fig. 1.
RNA sequencing revealed over 600 differentially expressed (DE) genes and corresponding altered metabolic and stress response pathways. Primary chondrocytes from WT and A2ARKO animals were harvested and submitted for RNA sequencing. a Of 16280 genes detected, 2211 genes were found to be differentially expressed (DE). b Differentially expressed (DE) genes are represented as a volcano plot where log2Fold change is demonstrated in the x-axis and p value is shown in the y-axis. Genes colored in yellow are padj≤0.05 and abs|log2FoldChange|>1; genes with padj < Adora2a are text highlighted. c KEGG pathway analysis revealed increased pro-inflammatory and immune responses, metalloproteases with diminished extracellular matrix organization as previously published. ROS and RONS metabolism and biosynthesis were upregulated along with increased aging and senescence markers, extrinsic and intrinsic apoptotic signaling, and reduced responses to hypoxia and calcium homeostasis
Cartilage-specific analysis of transcription factors illustrate differentially expressed gene network for spontaneous OA development in A2ARKO chondrocytes
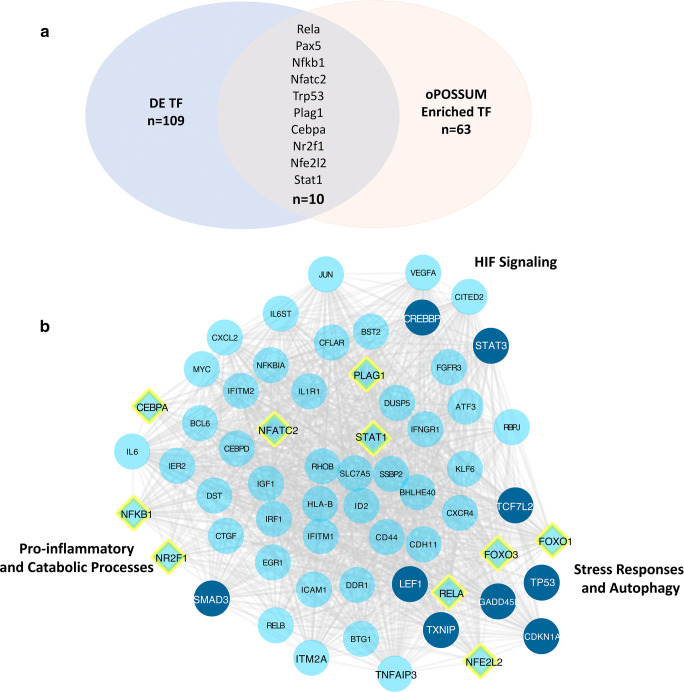
Transcription factor binding enrichment and tissue-specific network analysis were performed on the A2ARKO transcriptome to detect similarities between our animal model of OA and a recently published similar analysis of primary human OA chondrocytes [13]. We detected 631 transcription factors where 109 are differentially expressed (DE; Fig. 2a). We further used all 2211 differentially expressed genes for transcription factor (TF) binding enrichment analysis with oPOSSUM mouse single site method [18]. Differentially expressed transcription factors have shown positive regulation (z-score>0) in our differentially expressed genes. Diamonds represent enriched transcription factors or specific transcription factors of interest in our studies of the A2ARKO mouse. Genes depicted in dark blue circles represent genes of interest given their previously established role as drivers of OA: SMAD3, CREB-binding protein (CREBBP), LEF1, TXNIP, GADD45E, TCF7L2, TP53, and CDKN1A (Fig. 2b).
Fig. 2.
Cartilage-specific analysis of transcription factors (TF) illustrates the differentially expressed (DE) gene network for spontaneous OA development in A2ARKO chondrocytes. a Transcription factor analysis yielded 10 transcription factors that were both differentially expressed and enriched from oPOSSUM: Rela, Pax5, Nfkb1, Nfatc2, Trp53, Plag1, Cebpa, Nr2f1, Nfe2l2, and Stat1. To this list, we also included FOXO1 and FOXO3, although they were not differentially expressed, since they regulate important processes implicated in OA. b Network diagram of differentially expressed genes and transcription factors. Diamonds represent enriched transcription factors or transcription factors of interest in our studies of the A2ARKO mouse while genes depicted in dark blue circles represent genes of interest: SMAD3, CREB-binding protein (CREBBP), LEF1, TXNIP, GADD45E, TCF7L2, TP53, and CDKN1A
Enriched transcription factors included CEBPA, NFKB1, NR2F1 NFATC2, PLAG1, STAT1, RELA, and NFE2L2. Although not differentially expressed in primary chondrocytes harvested from neonates, we chose to include FOXO1 and FOXO3 in our analysis given mouse knockouts of these genes have reduced expression of autophagy and homeostatic genes and led to earlier and more severe OA development [21]. Moreover, we have previously reported that A2AR stimulation promotes autophagy through these pathways [12] Along similar analysis from primary human OA chondrocytes [13], HIF-1 signaling, pro-inflammatory, catabolic, and deregulated stress responses including autophagy are promoted by differentially expressed transcription factors in primary A2AR null chondrocytes and illustrate their significance in the degenerative disease of OA (Fig. 2b). Of note, CREB-binding protein and STAT3 are implicated and associated to HIF-1 signaling and stress responses. Other genes in this network relate to inflammation, catabolism, cartilage homeostasis, stress response, and autophagy. SMAD3 is a downstream transcriptional mediator in the TGFβ signaling pathway. LEF1 and TCF7L2/TCF4 are Wnt pathway transcription factors that can inhibit cartilage homeostasis when complexed with beta-catenin partly by upregulation of MMP13 [22–25]. TXNIP is a pro-autophagy protein activated in the presence of increased oxidative stress [25]. GADD45B, TP53, and CDKN1A/p21 are involved in cellular senescence and chondrocyte apoptosis. In all, this transcription factor network illustrates disrupted cellular repair (i.e., autophagy) with increased stress response signaling as drivers of OA with remarkable similarities to data reported from the human OA transcriptome [13].
Discussion
On our previously reported animal model of spontaneous OA, the A2AR null mouse has chondrocyte pathology similar to that reported in human OA tissues [9, 13, 26–31], including increased pro-inflammatory, catabolic mediators [5], oxidative signaling [11], deregulated autophagy [12], and upregulated markers of aging and senescence. Moreover, these data suggest the transcriptome of primary A2AR null chondrocytes is similar to that of primary human chondrocytes harvested from OA patients [13], including deregulated cellular repair and activation of stress responses. Significantly, these changes occur in chondrocytes harvested from intact knees in neonatal null mice, indicating that these detrimental changes are present at birth, accelerating aging, compromising tissue homeostasis, and leading to early onset of spontaneous OA [32]. Moreover, our primary cell harvest was consistent with our prior work showing the molecular and histological effect of lack of A2AR signaling, further demonstrating this animal’s significance in the study of human disease.
Importantly, autophagy has been reported to be interrupted in OA [33, 34], and recently published work from our group suggests that A2AR ligation in human chondrocytic T/C28a2 cells promotes autophagy likely via nuclear localization and activation of upstream autophagy-promoting transcription factors FOXO1 and FOXO3 [12]. It was therefore pertinent to determine whether or not genes responsible for autophagic flux or regulation thereof were differentially expressed. Indeed, z-scored analysis of genes that were grouped by KEGG pathway analysis to autophagy regulation and signaling, autophagosome initiation and formation, autophagosome elongation, and autophagosome maturation and acidification was differentially expressed (data not shown), suggesting that autophagic flux is deregulated and likely impacted by pro-inflammatory, catabolic, and oxidative signaling in A2AR null chondrocytes. Importantly, we wanted to understand which transcription factors that regulate autophagy, senescence, and other survival and homeostasis pathways were differentially expressed in A2ARKO chondrocytes and therefore analyzed that in our data set following the methods reported by Fisch et al. in their study of human OA chondrocyte transcriptome [13]. It is unclear why we did not detect differential expression of FOXO1 and FOXO3 in the neonatal A2AR null chondrocytes, although post-translational changes in FOXO1 and FOXO3 may be more important for their role in maintaining chondrocyte homeostasis.
As predicted by our KEGG pathway analysis and previously published work on the A2ARKO mouse, most of the differences in gene expression could be explained by activation of transcription factors such as NF-κB and its downstream signaling elements. Among affected genes associated with pro-inflammatory and catabolic processes, SMAD3 is of particular interest since our group has demonstrated that in vitro and in vivo ligation of A2AR promotes nuclear localization of phosphorylated SMAD2/3 which is associated with maintenance of cartilage [6, 7]. Increased SMAD3 expression in the null chondrocytes may, therefore, represent compensatory expression. HIF-1 signaling is associated with OA development in A2ARKO mice as previously reported in human OA chondrocytes [13]. Moreover, our group has previously described how diminished A2AR signaling in the null mouse or in primary chondrocytes harvested from neonatal knees promotes oxidative damage, mitochondrial dysfunction, and loss of viability in vivo and in vitro [11]. It is important to stress that despite a compromised state on the RNAseq level, A2ARKO chondrocytes were harvested from grossly intact cartilage in neonates more than 3 months before there is detectable evidence of osteoarthritis. Moreover, we saw compensatory increase in SOD2 expression in A2ARKO chondrocytes (data not shown). Increased hypoxia likely initially triggers enhanced compensatory strategies, but our data suggests that these inevitably burn out with increasing age. While hypoxia has been shown to modulate ecto-5′-nucleotidase/CD73 expression [35] and chondrogenesis differentiation [36, 37], over-active HIF-1 signaling may be responsible for the inevitable loss of chondrocyte viability [38] through overexpression of A2B receptors [39–41] and favor osteoblast differentiation with subsequent osteocyte formation [42]. Interval RNAseq and histologic experiments may help trace this timeline. In this regard, it should also be noted that cartilage is avascular and chondrocytes derive their nutrients and oxygen from synovial fluid which, itself, is relatively hypoxic [43, 44].
Of note, CREB-binding protein was highlighted in our network analysis, which has been reported to signal together with mitochondrial PKA (downstream of A2AR) to promote pro-survival responses [45]. CREB-binding protein also is a master regulator of mitochondrial mass [46]. Consistent with these findings, primary A2AR null chondrocytes have diminished mitochondrial volume and antagonism of the A2AR reduces mitochondrial network volumes in primary fully differentiated human chondrocytes [11]. Furthermore, this work also highlights the importance of STAT3, which has been shown to modulate oxidative phosphorylation complexes and thereby regulate mitochondrial function in various primary tissues [47].
We have carried out similar differential gene expression analyses on chondrocytes isolated from rats with post-traumatic OA (PTOA) which had received various intraarticular injections, including liposomal CGS21680, an A2AR-specific agonist, which reversed established PTOA in these animals [6]. When comparing the transcriptome of PTOA chondrocytes that received the A2AR agonist against those that had received the vehicle, we found transcription factor involvement and differentially expressed genes relating to stress responses (i.e., autophagy), HIF-1 signaling, inflammation, and anabolism (data not shown), suggesting that the mechanism of action of these liposomes directly targets the pathways gone awry in human OA [13] and that are found in our model of spontaneous OA, the A2AR null mouse. Consistent with our previous findings and the hypothesis that stimulation of the A2AR leads to OA reversal, we reported increased expression of STAT3, SMAD3, and TGIF1 in the treated rats [6].
In previous experiments, we have also shown that there is a crosstalk between the A2AR and Wnt pathways leading to osteoblast and fibroblast differentiation [48, 49]. LEF1 and TCF7L2 stimulate the expression of Wnt-stimulated genes in the presence of beta-catenin, leading to reduction in chondrocyte homeostasis and promotion of ossification and osteoblast differentiation [50]. Interestingly, we found that LEF1 was significantly downregulated in the rats with PTOA whether they had been treated with either empty liposomes or liposomes containing CGS21680, the A2AR agonist [6]. Moreover, LEF1 activates extracellular matrix catabolism by upregulating the protease MMP-13, and sirtuin 1 (SIRT1) deacetylates LEF1 to reduce its ability to activate MMP13 [22]. TXNIP is involved in autophagy activated by stress related to hypoxia and generation of ROS by complexing with mTORC1 inhibitor REDD1/DDIT4, leading to increased ATG4B activity and thus LC3 conjugation in autophagosomes [25]. GADD45B, CDKN1A/p21, and TP53 are all associated with cellular senescence. OA-related chondrocyte changes include chondrocyte apoptosis and senescence and increased p53 expression in chondrocytes is associated with typical OA changes in chondrocytes by activating chondrocyte apoptosis and/or cellular senescence (through CDKN2A/p16) [51, 52]. GADD45B causes G1 cell cycle arrest, and it may directly or indirectly activate p21 through CEBPβ [51]. Therefore, this A2ARKO transcription factor network includes genes associated with autophagy and homeostasis as well as those relevant to senescence and apoptosis, thus indicating that OA associated with reduced A2AR signaling probably involves an increase in senescence pathways with concomitant decrease in autophagy and homeostasis pathways, leading to cellular viability compromise, consistent with both previous findings in our animal models, but importantly, from patients [10, 13, 53–55].
In summary, the A2AR null mouse recapitulates inflammation and accelerated aging, specifically osteoarthritis, with close resemblance to the cellular pathology of primary human tissues. Future work should address mechanisms by which autophagy is increased through A2AR signaling and identify the link between nuclear/cytosolic signaling and gene expression and mitochondrial dysfunction in A2ARKO chondrocytes. The FOXO transcription factors are associated with autophagy but can induce apoptosis in the acetylated state; when deacetylated by SIRT1, the target genes are altered and promote a pro-survival state through autophagy and cellular homeostasis. These factors are also involved in cellular anti-aging and organismal longevity pathways, which would likely be therapeutic in OA. Unlike the FOXO transcription factors, p53 is deactivated by Sirt1 deacetylation. This suggests that A2AR signaling may require deacetylation to activate homeostatic stress response with FOXO1 and FOXO3 and reduce senescence by deacetylation-mediated p53 degradation. Furthermore, homeostatic antioxidant activity of SOD2 is also activated by a different sirtuin, SIRT3, which is a mitochondrial resident sirtuin. SOD2 gene expression can be increased in response to increased ROS and was upregulated A2ARKO chondrocytes; however, the function of the enzyme may still be reduced in the acetylated state. Studies with sirtuin inhibitors or siRNAs may be useful in determination of the pathway by which A2AR signaling enhances chondrocyte homeostasis and mitochondrial biogenesis.
Acknowledgements
We would like to thank the Science Building Animal Facility and Genome Technology Center (GTC) at NYU Langone Health for care of our animals, RNA sequencing, and assistance.
Author contribution
Conception and design, C.M. Castro, C. Corciulo, B.N. Cronstein; collection and assembly of data, C.M. Castro, C. Corciulo; analysis and interpretation of the data, C.M. Castro, C. Corciulo, B. Friedman, Z. Li, S. Jacob; drafting of the article, C.M. Castro; critical revision of the article for important intellectual content, C.M. Castro, C. Corciulo, B. Friedman, Z. Li, B.N. Cronstein; statistical expertise, Z. Li, S. Jacob, D. Fenyo; obtaining of funding, B.N. Cronstein, D. Fenyo; administrative, technical, or logistic support, B.N. Cronstein; D. Fenyo.
Funding
This work was supported by grants from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (RO1-AR054897 and RO1-AR056672), the NYU-HHC Clinical and Translational Science Institute (UL1-TR000038 from the National Center for Advancing Translational Sciences), and the Arthritis Foundation. CMC would like to thank the Medical Scientist Training Program at NYU School of Medicine (T32GM136573) and the National Institutes of Health (3RO1AR056672-07S1) for her training and funding.
Data availability
GEO Accession for raw data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146349
Declarations
Conflicts of interest
Drs. Corciulo and Cronstein have received a patent for the use of intra-articular injection of liposomal preparations of adenosine and A2AR agonists for the treatment of osteoarthritis. In addition, Drs. Corciulo and Cronstein are among the founders of Regenosine, a company incorporated to develop adenosine receptor–related therapies for osteoarthritis.
Ethical approval
The New York University School of Medicine Institutional Animal Care and Use Committee approved all protocols for experimental procedures involving the use of animals according to US law.
Consent to participate
No human subjects were recruited in this study. This section therefore does not apply to this work.
Consent for publication
All authors stated above have reviewed and consented to the content of this publication.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cristina M. Castro, Email: cristina.castro@nyumc.org
Carmen Corciulo, Email: carmen.corciulo@gu.se.
Benjamin Friedman, Email: Benjamin.friedman@nyumc.org.
Zhi Li, Email: zhi.li@nyulangone.org.
Samson Jacob, Email: samson.jacob@nyumc.org.
David Fenyo, Email: David.fenyo@nyulangone.org.
Bruce N. Cronstein, Email: bruce.cronstein@nyumc.org
References
- 1.Gohal C, Shanmugaraj A, Tate P, Horner NS, Bedi A, Adili A, Khan M (2018) Effectiveness of valgus offloading knee braces in the treatment of medial compartment knee osteoarthritis: a systematic review. Sports Health 1941738118763913 [DOI] [PMC free article] [PubMed]
- 2.Bitton R (2009) The economic burden of osteoarthritis. Am J Manag Care 15:S230–S235 [PubMed]
- 3.Nho SJ, Kymes SM, Callaghan JJ, Felson DT (2013) The burden of hip osteoarthritis in the United States: epidemiologic and economic considerations. J Am Acad Orthop Surg 21(Suppl 1):S1–S6 [DOI] [PubMed]
- 4.Puig-Junoy J, Ruiz Zamora A (2015) Socio-economic costs of osteoarthritis: a systematic review of cost-of-illness studies. Semin Arthritis Rheum 44:531–541 [DOI] [PubMed]
- 5.Corciulo C, Lendhey M, Wilder T, Schoen H, Cornelissen AS, Chang G, Kennedy OD, Cronstein BN (2017) Endogenous adenosine maintains cartilage homeostasis and exogenous adenosine inhibits osteoarthritis progression. Nat Commun 8:15019 [DOI] [PMC free article] [PubMed]
- 6.Corciulo C, Castro CM, Coughlin T, Jacob S, Li Z, Fenyo D, Rifkin DB, Kennedy OD, Cronstein BN (2020) Intraarticular injection of liposomal adenosine reduces cartilage damage in established murine and rat models of osteoarthritis. Sci Rep 10:13477 [DOI] [PMC free article] [PubMed]
- 7.Corciulo C, Castro C, Coughlin T, Jacob S, Fenyo D, Rifkin D, Kennedy OD, Angle S, Cronstein BN (2019) A2A adenosine receptor stimulation regenerates cartilage in osteoarthritis animal model. Osteoarthr Cartil 27:S174
- 8.Bekisz JM, Lopez CD, Corciulo C, Mediero A, Coelho PG, Witek L, Flores RL, Cronstein BN (2018) The role of adenosine receptor activation in attenuating cartilaginous inflammation. Inflammation 41:1135–1141 [DOI] [PubMed]
- 9.Terkeltaub R, Johnson K, Murphy A, Ghosh S (2002) Invited review: the mitochondrion in osteoarthritis. Mitochondrion 1:301–319 [DOI] [PubMed]
- 10.Loeser RF, Collins JA, Diekman BO (2016) Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol 12:412–420 [DOI] [PMC free article] [PubMed]
- 11.Castro CM, Corciulo C, Solesio ME, Liang F, Pavlov EV, Cronstein BN (2020) Adenosine A2A receptor (A2AR) stimulation enhances mitochondrial metabolism and mitigates reactive oxygen species-mediated mitochondrial injury. FASEB J 34(4):5027–5045. 10.1096/fj.201902459R. [DOI] [PMC free article] [PubMed]
- 12.Friedman B, Corciulo C, Castro CM, Cronstein BN (2021) Adenosine A2A receptor signaling promotes FoxO associated autophagy in chondrocytes. Sci Rep 11:968. 10.1038/s41598-020-80244-x. [DOI] [PMC free article] [PubMed]
- 13.Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK (2018) Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis and cartilage / OARS, Osteoarthr Res Society 26:1531–1538 [DOI] [PMC free article] [PubMed]
- 14.Ichikawa N, Taniguchi A, Kaneko H, Kawamoto M, Sekita C, Nakajima A, Yamanaka H (2015) Arterial calcification due to deficiency of CD73 (ACDC) as one of rheumatic diseases associated with periarticular calcification. J Clin Rheumatol 21:216–220 [DOI] [PubMed]
- 15.St. Hilaire C, Ziegler SG, Markello TC, Brusco A, Groden C, Gill F, Carlson-Donohoe H, Lederman RJ, Chen MY, Yang D, Siegenthaler MP, Arduino C, Mancini C, Freudenthal B, Stanescu HC, Zdebik AA, Chaganti RK, Nussbaum RL, Kleta R, Gahl WA, Boehm M (2011) NT5E mutations and arterial calcifications. N Engl J Med 364:432–442 [DOI] [PMC free article] [PubMed]
- 16.Thirion S, Berenbaum F (2004) Culture and phenotyping of chondrocytes in primary culture. Methods Mol Med 100:1–14 [DOI] [PubMed]
- 17.Chu CR, Szczodry M, Bruno S (2010) Animal models for cartilage regeneration and repair. Tiss Eng Part B Rev 16:105–115 [DOI] [PMC free article] [PubMed]
- 18.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW (2012) oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2:987–1002 [DOI] [PMC free article] [PubMed]
- 19.Barzi A, Miksad R, Surinach A, Corvino FA, Wang S, Torres AZ, Mamlouk K, Pulgar S, Valderrama A, Bekaii-Saab T, Ahn D (2020) Real-world dosing patterns and outcomes of patients with metastatic pancreatic cancer treated with a liposomal irinotecan regimen in the United States. Pancreas 49:193–200 [DOI] [PMC free article] [PubMed]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504 [DOI] [PMC free article] [PubMed]
- 21.Matsuzaki T, Alvarez-Garcia O, Mokuda S, Nagira K, Olmer M, Gamini R, Miyata K, Akasaki Y, Su AI, Asahara H, Lotz MK (2018) FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med 10:eaan0746 [DOI] [PMC free article] [PubMed]
- 22.Elayyan J, Lee EJ, Gabay O, Smith CA, Qiq O, Reich E, Mobasheri A, Henrotin Y, Kimber SJ, Dvir-Ginzberg M (2017) LEF1-mediated MMP13 gene expression is repressed by SIRT1 in human chondrocytes. FASEB J 31:3116–3125 [DOI] [PubMed]
- 23.Arce L, Yokoyama NN, Waterman ML (2006) Diversity of LEF/TCF action in development and disease. Oncogene 25:7492–7504 [DOI] [PubMed]
- 24.Hoppler S, Kavanagh CL (2007) Wnt signalling: variety at the core. J Cell Sci 120:385–393 [DOI] [PubMed]
- 25.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, Nash Z, Yang Z, Liesa M, Yoshioka J, Matsuzawa S, Shirihai OS, Lee RT, Reed JC, Ellisen LW (2015) A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun 6:7014 [DOI] [PMC free article] [PubMed]
- 26.Blanco FJ, Rego I, Ruiz-Romero C (2011) The role of mitochondria in osteoarthritis. Nat Rev Rheumatol 7:161–169 [DOI] [PubMed]
- 27.Maneiro E, Martin MA, de Andres MC, Lopez-Armada MJ, Fernandez-Sueiro JL, del Hoyo P, Galdo F, Arenas J, Blanco FJ (2003) Mitochondrial respiratory activity is altered in osteoarthritic human articular chondrocytes. Arthritis Rheum 48:700–708 [DOI] [PubMed]
- 28.Loeser RF (2017) The role of aging in the development of osteoarthritis. Trans Am Clin Climatol Assoc 128:44–54 [PMC free article] [PubMed]
- 29.Pagano G, Talamanca AA, Castello G, Cordero MD, d’Ischia M, Gadaleta MN, Pallardo FV, Petrovic S, Tiano L, Zatterale A (2014) Oxidative stress and mitochondrial dysfunction across broad-ranging pathologies: toward mitochondria-targeted clinical strategies. Oxidative Med Cell Longev 2014:541230 [DOI] [PMC free article] [PubMed]
- 30.Soto-Hermida A, Fernandez-Moreno M, Oreiro N, Fernandez-Lopez C, Pertega S, Cortes-Pereira E, Rego-Perez I, Blanco FJ (2014) Mitochondrial DNA (mtDNA) haplogroups influence the progression of knee osteoarthritis. Data from the Osteoarthritis Initiative (OAI). PLoS One 9:e112735 [DOI] [PMC free article] [PubMed]
- 31.Blanco FJ, Lopez-Armada MJ, Maneiro E (2004) Mitochondrial dysfunction in osteoarthritis. Mitochondrion 4:715–728 [DOI] [PubMed]
- 32.Higuchi-Sanabria R, Frankino PA, Paul JW 3rd, Tronnes SU, Dillin A (2018) A futile battle? Protein quality control and the stress of aging. Dev Cell 44:139–163 [DOI] [PMC free article] [PubMed]
- 33.Loeser RF (2009) Aging and osteoarthritis: the role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthritis and cartilage / OARS, Osteoarthr Res Society 17:971–979 [DOI] [PMC free article] [PubMed]
- 34.Goutas A, Syrrou C, Papathanasiou I, Tsezou A, Trachana V (2018) The autophagic response to oxidative stress in osteoarthritic chondrocytes is deregulated. Free Radic Biol Med 126:122–132 [DOI] [PubMed]
- 35.Thompson LF, Eltzschig HK, Ibla JC, Van De Wiele CJ, Resta R, Morote-Garcia JC, Colgan SP (2004) Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J Exp Med 200:1395–1405 [DOI] [PMC free article] [PubMed]
- 36.Kanichai M, Ferguson D, Prendergast PJ, Campbell VA (2008) Hypoxia promotes chondrogenesis in rat mesenchymal stem cells: a role for AKT and hypoxia-inducible factor (HIF)-1alpha. J Cell Physiol 216:708–715 [DOI] [PubMed]
- 37.Lee HH, Chang CC, Shieh MJ, Wang JP, Chen YT, Young TH, Hung SC (2013) Hypoxia enhances chondrogenesis and prevents terminal differentiation through PI3K/Akt/FoxO dependent anti-apoptotic effect. Sci Rep 3:2683 [DOI] [PMC free article] [PubMed]
- 38.Pinto-Cardoso R, Pereira-Costa F, Pedro Faria J, Bandarrinha P, Bessa-Andres C, Correia-de-Sa P, Bernardo Noronha-Matos J (2020) Adenosinergic signalling in chondrogenesis and cartilage homeostasis: friend or foe? Biochem Pharmacol 174:113784 [DOI] [PubMed]
- 39.Lu N, Malemud CJ (2019) Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated gene expression. Int J Mol Sci 20 [DOI] [PMC free article] [PubMed]
- 40.Kwon JH, Lee J, Kim J, Jo YH, Kirchner VA, Kim N, Kwak BJ, Hwang S, Song GW, Lee SG, Yoon YI, Park GC, Tak E (2019) HIF-1alpha regulates A2B adenosine receptor expression in liver cancer cells. Exp Ther Med 18:4231–4240 [DOI] [PMC free article] [PubMed]
- 41.Feoktistov I, Ryzhov S, Zhong H, Goldstein AE, Matafonov A, Zeng D, Biaggioni I (2004) Hypoxia modulates adenosine receptors in human endothelial and smooth muscle cells toward an A2B angiogenic phenotype. Hypertension 44:649–654 [DOI] [PubMed]
- 42.Corciulo C, Wilder T, Cronstein BN (2016) Adenosine A2B receptors play an important role in bone homeostasis. Purinergic Signal 12:537–547 [DOI] [PMC free article] [PubMed]
- 43.Gibson JS, Milner PI, White R, Fairfax TP, Wilkins RJ (2008) Oxygen and reactive oxygen species in articular cartilage: modulators of ionic homeostasis. Pflugers Arch 455:563–573 [DOI] [PubMed]
- 44.Zhou S, Cui Z, Urban JP (2004) Factors influencing the oxygen concentration gradient from the synovial surface of articular cartilage to the cartilage-bone interface: a modeling study. Arthritis Rheum 50:3915–3924 [DOI] [PubMed]
- 45.Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ (2005) Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA 102:13915–13920 [DOI] [PMC free article] [PubMed]
- 46.Uittenbogaard M, Brantner CA, Chiaramello A (2018) Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells. Cell Death Dis 9:360 [DOI] [PMC free article] [PubMed]
- 47.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC (2009) Function of mitochondrial Stat3 in cellular respiration. Science (New York, NY) 323:793–797 [DOI] [PMC free article] [PubMed]
- 48.Zhang J, Corciulo C, Liu H, Wilder T, Ito M, Cronstein B (2017) Adenosine A2a receptor blockade diminishes Wnt/beta-catenin signaling in a murine model of bleomycin-induced dermal fibrosis. Am J Pathol 187:1935–1944 [DOI] [PMC free article] [PubMed]
- 49.Shaikh G, Zhang J, Perez-Aso M, Mediero A, Cronstein B (2016) Adenosine A2A receptor promotes collagen type III synthesis via beta-catenin activation in human dermal fibroblasts. Br J Pharmacol 173:3279–3291 [DOI] [PMC free article] [PubMed]
- 50.Borhani S, Corciulo C, Larranaga-Vera A, Cronstein BN (2019) Adenosine A2A receptor (A2AR) activation triggers Akt signaling and enhances nuclear localization of beta-catenin in osteoblasts. FASEB J:fj201900014R [DOI] [PMC free article] [PubMed]
- 51.Ashraf S, Cha BH, Kim JS, Ahn J, Han I, Park H, Lee SH (2016) Regulation of senescence associated signaling mechanisms in chondrocytes for cartilage tissue regeneration. Osteoarthr Cartil 24:196–205 [DOI] [PubMed]
- 52.Hashimoto S, Nishiyama T, Hayashi S, Fujishiro T, Takebe K, Kanzaki N, Kuroda R, Kurosaka M (2009) Role of p53 in human chondrocyte apoptosis in response to shear strain. Arthritis Rheum 60:2340–2349 [DOI] [PubMed]
- 53.Lotz MK, Carames B (2011) Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat Rev Rheumatol 7:579–587 [DOI] [PMC free article] [PubMed]
- 54.Loeser RF (2011) Aging and osteoarthritis. Curr Opin Rheumatol 23:492–496 [DOI] [PMC free article] [PubMed]
- 55.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M (2010) Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum 62:791–801 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GEO Accession for raw data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146349