Abstract
Asthma is a heterogeneous disease whose development is shaped by a variety of environmental and genetic factors. While several recent studies suggest that microbial dysbiosis in the gut may promote asthma, little is known about the relationship between the recently discovered lung microbiome and asthma. Innate lymphoid cells (ILCs) have also been shown recently to participate in asthma. To investigate the relationship between the lung microbiome, ILCs, and asthma, we recruited 23 healthy controls (HC), 42 patients with non-severe asthma, and 32 patients with severe asthma. Flow cytometry analysis showed severe asthma associated with fewer natural cytotoxicity receptor (NCR)+ILC3s in the lung. Similar changes in other ILC subsets, macrophages, and monocytes were not observed. The asthma patients did not differ from the HC in terms of the alpha and beta-diversity of the lung and gut microbiomes. However, lung function correlated positively with both NCR+ILC3 frequencies and microbial diversity in the lung. Sputum NCR+ILC3 frequencies correlated positively with lung microbiome diversity in the HC, but this relationship was inversed in severe asthma. Together, these data suggest that airway NCR+ILC3s may contribute to a healthy commensal diversity and normal lung function.
Keywords: Asthma, Innate lymphoid cells, Microbiota, Sputum, Host-microbial interaction
INTRODUCTION
Asthma is a chronic inflammatory disease that is characterized by airway hyperresponsiveness and chronic airway inflammation. It affects more than 300 million people worldwide each year (1,2). Asthma is a complex disease that is now known to comprise a variety of disease phenotypes and endotypes (3). Thus, while it was initially considered to be an allergic disease that is mediated by allergen-specific TH2 responses (4), 10%–33% of asthma patients suffer from non-allergic asthma, triggered by ozone exposure, obesity, and air pollutants (5,6). Since much of the research on asthma has focused on T cells, further research on the roles played by other immune cells is needed to improve our understanding of this heterogeneous disease.
Innate lymphoid cells (ILCs) have been found recently to participate in the pathogenesis of asthma (7,8,9). These cells produce distinct arrays of cytokines in response to different stimuli and have been divided into 3 types based on these cytokine profiles and their molecular/cell-surface signatures. Thus, the group 1, group 2, and group 3 ILCs (denoted ILC1s, ILC2s, and ILC3s, respectively) respectively produce TH1 (IFN-γ), TH2 (IL-4, IL-5, and IL-13), and TH17 (IL-17 and IL-22) cytokines and are therefore considered to be the innate counterparts of TH1, TH2, and TH17 cells, respectively (10). The ILC3s also display additional heterogeneity: they can be subdivided into 2 types (CCR6+ LTis and CCR6− ILC3) (11), with the CCR6− ILC3 cells, in turn, is divided on the basis of their expression of natural cytotoxicity receptor (NCR) into NCR+ILC3s and NCR−ILC3s (12).
Another immune cell type that may play an important role in asthma pathogenesis is the alveolar macrophage (AM). This innate immune cell is the most abundant immune cell in the lung (13,14) and contributes to chronic airway inflammation by producing proinflammatory cytokines such as TNF-α and IL-1β. Asthma associates with changes in macrophage polarization (15), which is driven by environmental stimuli such as bacterial components and cytokines and produces either the classically activated (M1) phenotype (which associates with TH1 inflammation), or the alternatively activated (M2) phenotype (which is linked to TH2 inflammation) (16,17). Our previous study with animal models of asthma and human asthma patients showed that 2 asthma phenotypes associate with interactions between lung ILCs and AMs: specifically, eosinophilic asthma associates with ILC2:M2 interactions while non-eosinophilic asthma is linked to ILC1/3:M1 interactions (18).
A growing body of evidence suggests that the human microbiome is actively involved in the pathogenesis of asthma and other chronic respiratory diseases (19,20,21). The lungs were initially thought to be sterile until culture-independent techniques showed that the lower respiratory tract of healthy humans contains diverse commensal microbes (22). Of these, Bacteroidetes and Firmicutes are the most abundant phyla, followed by Proteobacteria and, to a lesser extent, Actinobacteria and Fusobacteria (23,24). Compared to healthy controls (HC), asthma patients exhibit airway dysbiosis characterized by elevated Proteobacteria or decreased Bacteroidetes prevalence (25,26,27). However, a lack of uniformity and standardization in the processing of respiratory samples used for microbiome characterization has resulted in inconsistencies between studies. Moreover, most of these studies are cross-sectional, which limits our understanding of the temporal relationship between airway microbiome changes and asthma, including any related immune changes. Longitudinal studies with standardized preparation/analysis protocols on this question are warranted.
To address these issues, we here characterized the interactions between the airway microbiome and immune cells in asthma by analyzing the airway microbiome in asthma patients and HC and assessing whether these variables correlated with asthma severity.
MATERIALS AND METHODS
Subjects
We recruited 23 healthy controls (HC) and 74 asthma patients from Seoul National University Hospital (Seoul, Korea) between December 2016 and June 2017. All patients with asthma met one of the following criteria: forced expiratory volume in 1 second (FEV1) changed over 12% by 200 ml after bronchodilator response, or significant airway hyperresponsiveness to methacholine or mannitol provocation. Patients with cancer, severe medical conditions, and other pulmonary diseases were excluded along with patients on immunotherapy or medications such as antibiotics, antifungal agents, antiviral drugs, probiotics, or any systemic steroids. The asthma patients were subdivided on the basis of their FEV1 or asthma control questionnaire (ACQ) score into those with severe (n=32) and non-severe (n=42) asthma: severe asthma was defined as ACQ score >1.50 or FEV1 40%–80% of the expected value despite daily use of an inhaled corticosteroid (ICS) dose equivalent to ≥1,000 μg fluticasone.
Study approval
All subjects enrolled in this study provided written informed consent. This study was approved by the Seoul National University Hospital Institutional Review Board (IRB approval number: 1607-148-778) and was conducted following the Declaration of Helsinki.
Preparation of airway and fecal samples for microbiome analysis
After measuring basal FEV1 and forced vital capacity (FVC), all participants were treated with 200 μg salbutamol (Ventolin®; GlaxoSmithKline, Brentford, England). To prompt induced sputum production, 3% hypertonic saline was inhaled repeatedly up to 4 times at 5-min intervals using an ultrasonic nebulizer with an output set at 4.5 ml/min (Omron Co., Tokyo, Japan). After each nebulization, the subjects rinsed their mouths and spat the induced sputum carefully into a Petri dish. Samples where squamous epithelial cells accounted for more than 20% of the total cell count were excluded from the analysis. To collect fecal samples, at least 5 g feces were collected with a sterile spoon and placed in a sterile container. All samples were stored at −80°C immediately after collection.
DNA extraction, PCR amplification, and 16S rRNA gene sequencing
Metagenomic DNA was extracted from 1 ml of whole sputum sample and 200 mg fecal sample by using the FastDNA® SPIN Kit for Soil DNA extraction (MP Biomedicals, Santa Ana, CA, USA). PCR amplification of the V3-V4 hypervariable region of the bacterial 16S rRNA gene was subsequently performed with 3 μl of extracted DNA template and 25 μl 2X KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Rome, Italy). The primers-318F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and 806R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATC C-3′) were used for 16S rRNA gene amplification. The second PCR was performed using the Nextera XT Index Kit (Illumina, San Diego, CA, USA) using a different adapter for each sample, and libraries were sequenced using Illumina Miseq.
Sequencing data processing
The raw sequencing data files were preprocessed before downstream data analysis by using the bioinformatics tools; PRINSEQ (28), Cutadapt (29), Paired-End reAd mergeR (30), USEARCH (31), and QIIME v1.9 (32). The Ezbiocloud database was used to identify operational taxonomic units (OTUs), which were defined as clusters of 16S sequences that had 97% or greater similarity (33). Alpha-diversity was quantified as OTU counts and Shannon and Simpson diversity indices. The weighted normalized UniFrac distance was plotted using data from principal component analysis (PCoA), and the significance of grouping in the ordination was evaluated by using permutational multivariate analysis of variance.
Isolation of immune cells from induced sputum and peripheral blood
To isolate sputum immune cells, induced sputum was mixed with the same volume of 0.1% dithiothreitol (Sigma, St. Louis, MO, USA) and incubated for 20 min at 37°C with shaking. The immune cells were then filtered with a 70-μm strainer and centrifuged at 1,400 rpm for 6 min. The pellet of sputum was resuspended with 2% FBS-containing PBS for flow cytometry analysis. To isolate PBMCs, whole blood was diluted with double the volume of PBS buffer (pH 7.2) and 2 mM EDTA, after which 35 ml of diluted sample was layered onto 15 ml Ficoll-Paque (GE Healthcare, Chicago, IL, USA). After centrifugation at 400×g for 30 min at 25°C without a brake, the PBMC layer was isolated and resuspended in RPMI1640 (Biowest, Riverside, MO, USA) with 10% FBS (Biowest) and antibiotics (penicillin-streptomycin; Sigma).
Flow cytometry analysis
The Fcγ receptors of immune cells were blocked with anti-CD16/CD32 Ab (Human BD Fc Block; BD Biosciences, Franklin Lakes, NJ, USA) before the cells were stained with fluorochrome-labeled mAbs. Cells from sputum and PBMCs were stained with the following Abs: anti-CD45 (HI30; purchased from BD Biosciences); anti-CD3ε (UCHT1), anti-CD11c (3.9), anti-CD11b (ICRF44), anti-CD14 (HCD14), anti-CD19 (HIB19), anti-CD49b (P1E6-C5), and anti-FcεRIα (AER-37), which were used as lineage markers and were purchased from BioLegend, San Diego, CA, USA; anti-CD15 (W6D3), anti-CD68 (Y1/82A), anti-CD117 (C-Kit, 104D2), anti-CD127 (A019D5), anti-CD206 (15-2), anti-CD45 (HI30), anti-CD16 (3G8), anti-NKp44 (P44-8), which were purchased from BioLegend; and anti-ST2L (B4E6; purchased from MD Bioproduct, Oakdale, MN, USA). Flow cytometry was performed using BD LSRFortessa™ and BD LSRFortessa™ X-20 (BD Biosciences) and analyzed by FlowJo (V10; Tree Star Inc., Ashland, OR, USA) software (BD Biosciences).
Statistics
Correlations between immune cell frequencies and microbial alpha-diversity in the sputum were investigated with the R programming language (https://www.r-project.org/; R Foundation, Vienna, Austria). The Shannon and Simpson alpha-diversity indices of the sputum microbial community of each subject were calculated with R package vegan (34). Pearson and Spearman correlation coefficients were calculated for each alpha-diversity and immune cell frequency pair. For Pearson correlations, the testing was conducted with an approximate t-distribution, with the degree of freedom being the number of tested samples minus 2. For Spearman correlations, the testing was performed by calculating the approximative null distribution with a conditional Monte Carlo procedure with 1,000,000 samples. This procedure was done with an R package coin (35,36). These steps were conducted with all available samples and separately for the HC, non-severe asthma patients, and severe asthma patients. The resulting p-values were adjusted with the Benjamini-Hochberg method (37).
To compare the patient/control groups in terms of immune cell frequencies or microbial alpha-diversity, the data were first subjected to the Shapiro-Wilk normality test to check whether they were normally distributed. Two groups were compared with either the Mann-Whitney U tests (non-parametric data) or unpaired t-tests (parametric data). Multiple independent groups were compared by using ordinary one-way ANOVA or Kruskal-Wallis test for parametric and non-parametric data, respectively. To analyze the correlation between the clinical index and ILC3 subset frequencies or microbial alpha-diversity, either Pearson or Spearman correlation test was performed depending on the normality of the data. The measured values are presented as mean±SD. GraphPad Prism 8 (GraphPad software, Inc., San Diego, CA, USA) was used for statistical analysis, and p-value <0.05 was considered statistically significant.
RESULTS
Patients with asthma have lower frequencies of NCR+ILC3s in their sputum than HC
To determine the changes of innate immune cells in asthmatics, we recruited 23 HC and 74 asthma patients (non-severe, n=42; severe, n=32) as described in Table 1. Lung functions represented by FVC (%), FEV1 (%), and FEV1/FVC ratio were decreased in the asthmatics.
Table 1. Clinical characteristics of the control and asthma subjects.
| Characteristics | HC | Non-severe asthmatics | Severe asthmatics | p-value |
|---|---|---|---|---|
| Number | 23 | 42 | 32 | |
| Age (yr) | 49.00 (39–62) | 58.00 (39–71) | 58.44 (38–85) | |
| Sex (M/F) | 8/15 | 12/30 | 13/19 | |
| BMI | 23.07 (18.36–32.87) | 24.09 (19.72–32.89) | 25.49 (17.95–37.53) | 0.0272 |
| AR | 0 | 29 (69.05) | 11 (47.83) | <0.0001 |
| AD | 0 | 3 (7.14) | 3 (13.04) | 0.2168 |
| FVC (%) | 103.7 (87–134) | 95.67 (62–127) | 79.79 (56–110) | <0.0001 |
| FEV1 (%) | 101.4 (85–124) | 85.95 (44–124) | 67.51 (23.7–126) | <0.0001 |
| FEV1/FVC | 81.77 (71–91) | 72.5 (56–88) | 65.52 (23.47–89) | <0.0001 |
| ACT | - | 18.07 (5–25) | 17.61 (5–24) | 0.2683 |
| ACQ | - | 6.10 (0–18) | 13.74 (2–26) | <0.0001 |
Values are expressed as number (%) or mean±SD. The p-values were generated by unpaired t-test.
BMI, body mass index; AR, atopic rhinitis; AD, atopic dermatitis; ACT, asthma control test.
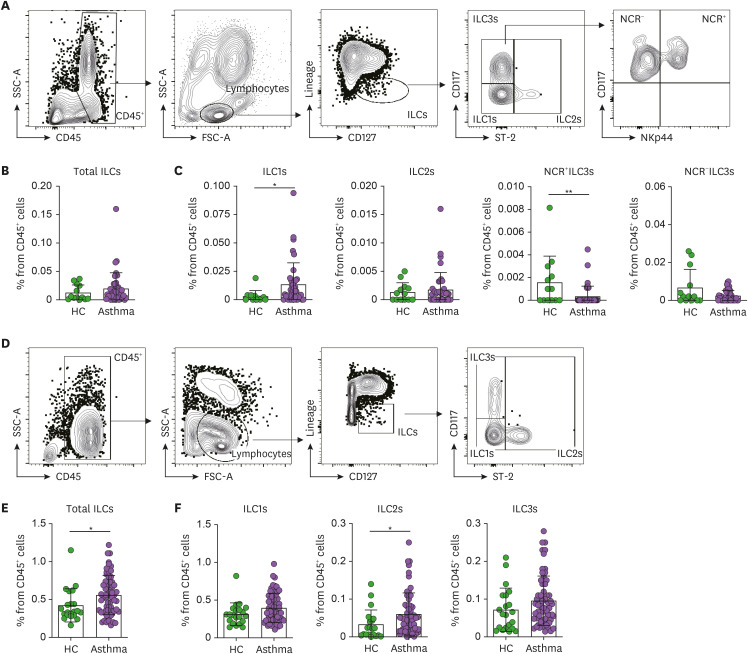
We first analyzed the frequencies of total ILCs, ILC1s, ILC2s, and ILC3 subsets in the induced sputum and PBMCs from the asthma patients and HC. The sputum ILCs were determined by first gating the total ILCs (Lineage−CD127+) in the CD45+lymphocytes and then subdivided into the ILC1s (ST-2−CD117−), ILC2s (ST-2+), NCR−ILC3s (NKp44−ST-2−CD117+), and NCR+ILC3s (NKp44+ST-2−CD117+) (Fig. 1A). With regard to the sputum, the asthma patients were similar to the HC in terms of total ILC, ILC2, and NCR−ILC3 frequencies (Fig. 1B and C), but had significantly more ILC1s and fewer NCR+ILC3s (Fig. 1C). With regard to the PBMCs (Fig. 1D), the asthma patients and HC were similar in terms of ILC1, and ILC3 frequencies but the asthma patients had significantly more total ILCs and ILC2s (Fig. 1E and F). The latter observations are consistent with previous reports (38,39).
Figure 1. Frequencies of ILC subsets in the induced sputum and peripheral blood. (A) Gating strategy used to identify the ILCs in the sputum. The total ILCs in the CD45+ lymphocytes were gated as Lineage (CD3ε, CD11c, CD11b, CD14, CD19, CD49b, and FcεRIα)-negative and CD127 (IL-7R)-positive cells. The ILC1s, ILC2s, and ILC3s were then determined by assessing ST-2 and CD117 expression. (B, C) Frequencies of total ILCs (B) and each ILC subset (C) in the HC (n=15) and asthma patients (n=41). The 2 ILC3 subtypes were identified based on their NCR (NKp44) expression. (D) Gating strategy used to identify the ILCs in the peripheral blood. The total ILCs in the CD45+ lymphocytes were gated as Lineage−CD127+ cells. The ILC1, ILC2, and ILC3 subsets were then determined by assessing ST-2 and CD117 expression. (E, F) Frequencies of total ILCs (E) and each ILC subset (F) in the HC (n=21) and asthma patients (n=62). The healthy control and asthma groups were compared in terms of ILC frequencies using the Mann-Whitney U test or unpaired t-tests. Each dot represents one human sample. The data are presented as mean±SD.
*p<0.05; **p<0.01.
NCR+ILC3 frequencies in the sputum correlate negatively with asthma severity
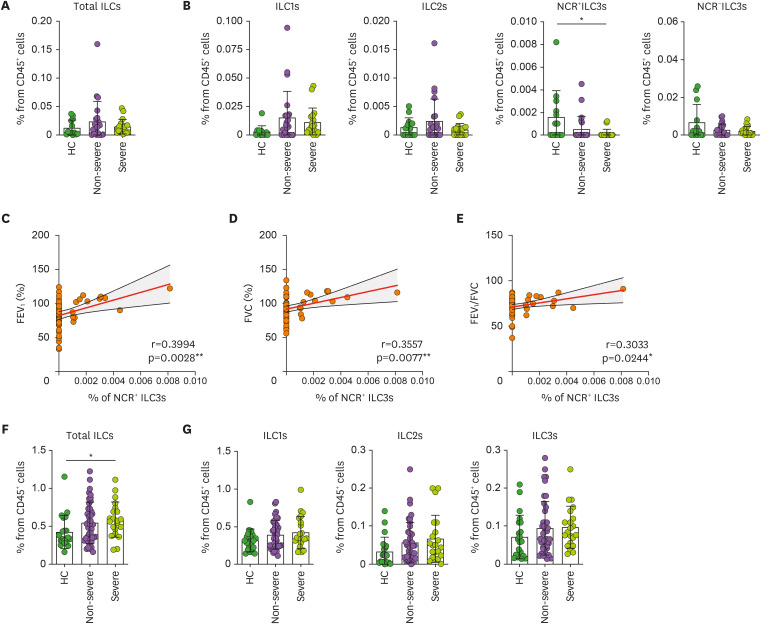
As the ILC subsets in sputum and blood showed significant changes in asthma status, we further classified the asthma patients into non-severe and severe asthma according to asthma severity; the severe asthma group was defined as those who had higher than 1.50 in ACQ score or FEV1 in the range between 40%–80% despite using a daily ICS.
The non-severe and severe asthma patients did not differ in terms of total ILC, ILC1, ILC2, and NCR−ILC3 frequencies in their sputum, but the severe asthma patients had significantly fewer NCR+ILC3s than the HC (Fig. 2A and B). The sputum NCR+ILC3s of the HC and asthma patients also correlated positively and significantly with 3 lung function indices, namely, FEV1 (Fig. 2C), FVC (Fig. 2D), and FEV1/FVC ratio (Fig. 2E). Thus, worse lung function is associated with lower NCR+ILC3 frequencies in the lung. With regard to the blood, only the total blood ILC frequencies were higher in severe asthma patients than in HC (Fig. 2F), but any ILC subset frequencies did not differ among groups (Fig. 2G).
Figure 2. Correlation between sputum NCR+ILC3 frequencies and asthma severity. (A, B) Frequencies of the total ILCs (A) and ILC subsets (B) in the sputum of HC (n=15), non-severe asthma patients (n=23), and severe asthma patients (n=18). (C-E) Correlation between NCR+ILC3 frequencies in the sputum and pulmonary function indicators, namely, FEV1 (C), FVC (D), and FEV1/FVC (E). (F, G) Frequencies of the total ILCs (F) and ILC subsets (G) in the blood of HC (n=21), non-severe asthma patients (n=41), and severe asthma patients (n=21). The healthy control, non-severe asthma, and severe asthma groups were compared by ordinary one-way ANOVA (parametric data) or Kruskal-Wallis test (non-parametric data). The correlations between sputum NCR+ILC3 frequencies and pulmonary function indicators were determined by Pearson or Spearman r correlation tests depending on data normality. Each dot represents one human sample. The data are presented as mean±SD.
*p<0.05.
Since lung-resident immune cell profiles may reflect the asthmogenic/asthma-promoting immune conditions better than circulating immune cells, these data suggest that sputum NCR+ILC3s relate more closely to asthma severity than blood ILC subsets. It is also interesting that low NCR+ILC3 frequencies in the sputum correlate with greater disease severity since it has been shown that the IL-22 produced by NCR+ILC3s maintains mucosal barrier integrity and host-commensal homeostasis (40). These observations together led us to speculate that NCR+ILC3s in the lung may prevent or limit poor lung function by shaping the airway microbiome.
Asthma does not associate with AM and circulating monocyte frequencies
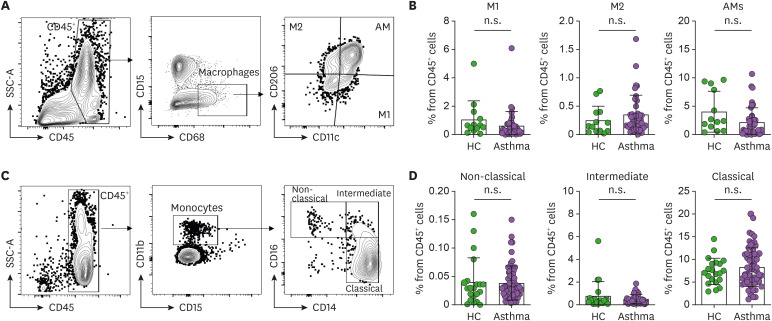
Since our previous study showed that asthma phenotypes are associated closely with macrophage polarization in the airways (18), we next investigated the macrophage populations in the induced sputum and peripheral blood of the asthma patients and HC. Thus, the macrophages in the sputum were defined as CD45+CD68+CD15− cells and then classified as M1 (CD206−CD11c+), M2 (CD206+CD11c−), and AMs (CD206+CD11c+) (Fig. 3A). Similarly, the monocytes in the blood (the circulating counterpart of lung-resident macrophages) were defined as CD45+CD11b+CD15− cells and then classified according to CD14 and CD16 expression as non-classical monocytes (CD14lowCD16hi), intermediate monocytes (CD14hiCD16hi), and classical monocytes (CD14hiCD16low) (Fig. 3C). These analyses showed that the asthma patients did not differ from the HC in terms of the frequencies of any of these populations (Fig. 3B and D).
Figure 3. Frequencies of macrophage and monocyte subsets in the induced sputum and peripheral blood (A) Gating strategy used to identify polarized phenotypes of sputum macrophages. The total macrophages in CD45+ cells were gated as CD15−CD68+ cells, after which M1 macrophages, M2 macrophages, and AMs were identified on the basis of CD11c and CD206 expression. (B) Frequencies of the 3 macrophage populations in the sputum of the HC (n=15) and asthma patients (n=41). (C) Gating strategy used to identify peripheral blood monocyte subsets. The total monocytes in CD45+ cells were gated as CD15−CD11b+ cells, after which classical (CD14hiCD16low), intermediate (CD14hiCD16hi), and non-classical monocyte (CD14lowCD16hi) subsets were identified on the basis of their CD14 and CD16 expression. (D) Frequencies of the 3 monocyte subsets in the blood of HC (n=21) and asthma patients (n=62). The healthy control and asthmatic groups were compared by either the Mann-Whitney U test or unpaired t-tests. Each dot represents one human sample. The data are presented as mean±SD.
n.s., not significant.
Asthma does not associate with altered lung or gut microbiome composition and diversity
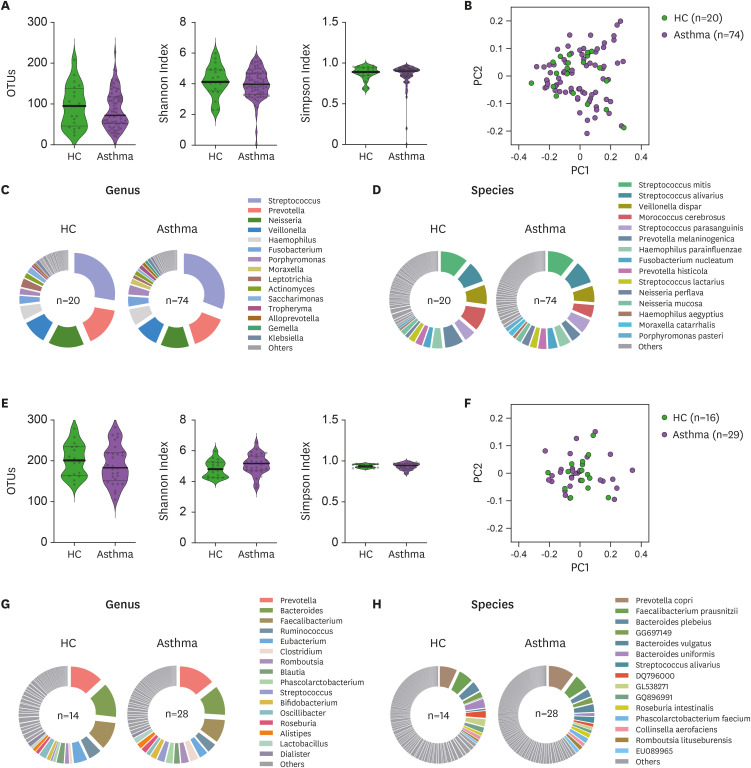
Given that (i) we observed severe asthma associated with reduced NCR+ILC3s in the lung and (ii) these cells are known to regulate host-commensal homeostasis (40), we asked whether asthma associates with changes in the airway commensal bacteria. Thus, we first assessed the alpha-diversity of the microbiomes in the sputum of HC and asthma patients by counting OTUs and using the Shannon and Simpson indices. The 2 groups did not differ in terms of airway microbiome diversity (Fig. 4A). Compositional differences in the airway microbiome were then assessed by using PCoA, but the HC and asthma patients did not form distinct clusters (Fig. 4B). Similarly, the 2 groups did not differ in terms of the relative abundance of microbiome genera (Fig. 4C) or species (Fig. 4D) in the sputum.
Figure 4. Microbiome diversity and abundance in the airway and gut of HC and asthma patients. (A) Sputum microbiome alpha-diversity in the HC (n=20) and asthma patients (n=74) was measured by counting the OTUs or using the Shannon and Simpson indices. (B) Sputum microbiome beta-diversity in the HC (n=20) and asthma patients (n=74) was measured by using the weighted normalized UniFrac distance matrix. (C, D) Phi-charts showing the relative microbial abundances in the sputum from HC (n=20) and asthma patients (n=74). The top 15 genera (C) and species (D) in the sputum microbiomes were shown. (E) Gut microbiome alpha-diversity in the HC (n=14) and asthma patients (n=28) was measured by counting the OTUs or using the Shannon and Simpson indices. (F) Gut microbiome beta-diversity in the HC (n=14) and asthma patients (n=28) was measured by using the weighted normalized UniFrac distance matrix. (G, H) Phi-charts showing the relative microbial abundances in the gut of HC (n=14) and asthma patients (n=28). The top 15 genera (G) and species (H) in the gut microbiomes were shown. The healthy control and asthma groups were compared in terms of alpha-diversity data (the OTU, Shannan index, and Simpson index data) by Mann-Whitney U test or unpaired t-tests. Each dot represents one human sample.
We also conducted similar analyses with the fecal microbiome. Again, the HC and asthma patients did not differ significantly in terms of alpha-diversity (Fig. 4E), composition (Fig. 4F), or relative abundance of genera or species (Fig. 4G and H).
Diversity of airway microbiome correlated with NCR+ILC3s and normal lung function
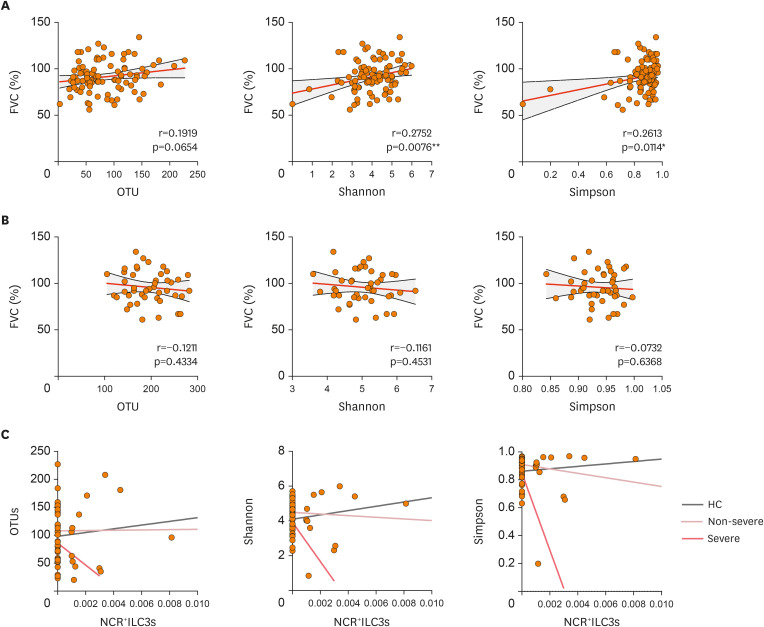
Since grouping subjects according to strict disease thresholds can obscure relationships between study variables, we then used correlation analyses to determine whether airway or gut microbiome diversity (as measured by using the OTUs, the Shannon and Simpson indices) can affect lung functions. Importantly, we observed that FVC correlated positively with both the Shannon and Simpson indices in the sputum (Fig. 5A). Such correlations were not observed with fecal microbiome diversity (Fig. 5B). Thus, high lung microbiome diversity may be critical for maintaining lung function. By contrast, gut microbiome diversity does not seem to participate in lung function.
Figure 5. Correlations between microbiome diversity, sputum NCR+ILC3 frequencies, and asthma severity. (A, B) Correlations between FVC and microbial alpha-diversity in the sputum (A) or gut (B). (C) Correlations between sputum NCR+ILC3 frequencies and sputum microbial alpha-diversity in HC (gray linear regression line), non-severe asthma patients (pink), and severe asthma patients (red). The correlations between FVC and microbial alpha-diversity were determined with Pearson or Spearman r correlation tests depending on data normality. The correlations between sputum NCR+ILC3 frequencies and sputum microbiome alpha-diversity were determined with Pearson's r correlation test. Each dot represents one human sample.
Since we noted that low sputum NCR+ILC3 frequencies are associated with severe asthma (Fig. 2B-E), we next assessed whether airway microbiome diversity correlates with NCR+ILC3 frequencies in the lung. Importantly, the NRC+ILC3 frequencies in the sputum correlated positively with airway microbiome diversity in the HC (Fig. 5C and Table 2). This supports the notion that high NCR+ILC3 cell numbers maintain high airway microbiome diversity. However, this correlation turned slightly negative in the non-severe asthma patients and became very strongly negative in the severe asthma patients (Fig. 5C and Table 2). Thus, as asthma worsened, the relationship between NCR+ILC3 frequencies and microbial diversity in the airways changed: microbial diversity dropped as NCR+ILC3 frequencies rose, and this change became most pronounced in severe asthma. Such changes were not observed for total ILCs, M1, M2, or AMs in the sputum (Table 2). Thus, the microbial diversity: NCR+ILC3 axis could serve as an essential indicator of asthma severity.
Table 2. Correlation coefficients and the corresponding P-values showing the relationships between immune cell frequencies in the sputum and the alpha-diversity of the microbial communities in the airway in HC and asthma patients.
| Alpha diversity in sputum | Immune cells | Correlation Coefficient | p-value | p-value (adjusted) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC | Non-severe | Severe | All | HC | Non-severe | Severe | All | HC | Non-severe | Severe | All | ||
| Simpson Index | Total ILCs | −0.0254 | −0.3046 | −0.2118 | −0.2153 | 0.9377 | 0.2345 | 0.3204 | 0.1216 | 0.9377 | 0.7035 | 0.4806 | 0.4809 |
| Shannon Index | Total ILCs | 0.0972 | −0.3322 | −0.4063 | −0.2734 | 0.7637 | 0.1927 | 0.0488 | 0.0476 | 0.9377 | 0.7035 | 0.3134 | 0.4809 |
| Simpson Index | NCR−ILC3s | −0.2054 | −0.2686 | −0.1700 | −0.1842 | 0.5218 | 0.2972 | 0.4272 | 0.1868 | 0.9377 | 0.7877 | 0.5243 | 0.4809 |
| Shannon Index | NCR−ILC3s | −0.0851 | −0.3454 | −0.2621 | −0.1898 | 0.7926 | 0.1746 | 0.2161 | 0.1735 | 0.9377 | 0.7035 | 0.4405 | 0.4809 |
| Simpson Index | NCR+ILC3s | 0.0723 | −0.2562 | −0.4402 | −0.1254 | 0.8233 | 0.3209 | 0.0313 | 0.3711 | 0.9377 | 0.7877 | 0.3134 | 0.5893 |
| Shannon Index | NCR+ILC3s | 0.2640 | −0.1991 | −0.3515 | 0.0037 | 0.4070 | 0.4436 | 0.0921 | 0.9792 | 0.9377 | 0.7899 | 0.3134 | 0.9792 |
| Simpson Index | M1 | −0.0957 | −0.3135 | −0.1920 | −0.2059 | 0.7674 | 0.2204 | 0.3688 | 0.1390 | 0.9377 | 0.7035 | 0.5241 | 0.4809 |
| Shannon Index | M1 | −0.0470 | −0.1888 | −0.3508 | −0.2210 | 0.8846 | 0.4681 | 0.0929 | 0.1117 | 0.9377 | 0.7899 | 0.3134 | 0.4809 |
| Simpson Index | M2 | −0.0265 | −0.1002 | −0.1141 | −0.0842 | 0.9349 | 0.7019 | 0.5954 | 0.5487 | 0.9377 | 0.8390 | 0.6698 | 0.7408 |
| Shannon Index | M2 | 0.0940 | −0.0212 | −0.1210 | −0.0365 | 0.7714 | 0.9356 | 0.5734 | 0.7952 | 0.9377 | 0.9356 | 0.6698 | 0.8912 |
| Simpson Index | AM | 0.0293 | −0.2318 | −0.0953 | −0.0889 | 0.9280 | 0.3708 | 0.6577 | 0.5265 | 0.9377 | 0.7899 | 0.7046 | 0.7408 |
| Shannon Index | AM | 0.1114 | −0.1506 | −0.2228 | −0.0941 | 0.7304 | 0.5639 | 0.2954 | 0.5028 | 0.9377 | 0.8013 | 0.4692 | 0.7408 |
The coefficients were calculated both separately for each patient/control group and for all subjects.
Non-severe, patients with non-severe asthma; Severe, patients with severe asthma.
DISCUSSION
The current study suggests that there is a relationship between the NCR+ILC3s in the healthy lung and the airway microbiome and that perturbation of this NCR+ILC3: microbiome axis may contribute to the severity of asthma. Up to now, most asthma therapeutics are based on a previously well-known type 2, allergic asthma. Therefore, there is a clinically unmet need to clarify non-allergic asthma, which is resistant to classical treatments such as corticosteroids (41). However, clustering asthmatics based on clinical factors, including smoking, disease onset, or atopy, is inaccurate in adult asthmatics. Therefore, predicting and managing asthma exacerbations is clinically essential for successful asthma treatment (42). In this regard, identifying immune cell profiles and microbiome can be intrinsic predictive markers of asthma.
Many studies have shown that the gut microbiome shapes the development of asthma. For example, Herbst et al. (43) showed that when germ-free (GF) mice, which have never been exposed to pathogenic or nonpathogenic microorganisms, were sensitized intraperitoneally with ovalbumin and then challenged with the same Ag intranasally, they display exaggerated TH2 inflammation in the airways compared to specific pathogen-free (SPF) mice. Similarly, several studies show that when the gut microbiota of mice are disrupted by antibiotics, especially in the perinatal period, the mice exhibit enhanced susceptibility to allergic asthma (44,45). Besides, antibiotic use in pregnancy and early life in humans associates with an increased risk of recurrent wheeze and asthma in childhood (46,47). However, our current results suggest that the lung microbiome is a better indicator than the gut microbiome, which reflects asthma status. The gut microbiome does not participate in the relationship between NCR+ILC3 frequencies in the lung and asthma severity, rather, the lung microbiome plays this role. This is consistent with a growing number of studies that show that the lung microbiome participates in the clinical outcomes of asthma (27,48,49). The study by Huang et al. (49) on asthma patients who were treated with a standard dose of ICS but continued to have symptoms showed that the degree of bronchial hyperresponsiveness associated with the bacterial composition in the airways. Similarly, Green et al. (50) reported that the relative abundance of Moraxella catarrhalis, Haemophilus, or Streptococcus species in the sputum of ICS-resistant asthma patients correlated with worse lung function. Interestingly, they also showed that high numbers of these microorganisms in the lung are associated with higher sputum neutrophil counts. This suggests that the lung microbiome may participate in neutrophilic airway inflammation, which is a severe asthma phenotype that is characterized by frequent exacerbations and resistance to current asthma treatments. Although further studies using standardized sample collection and analysis protocols are needed to confirm these observations, these studies suggest that changes in the structure and function of the human airway microbiome may contribute to asthma heterogeneity. All of these gut/lung microbiome-related observations together suggest that interventions in the microbiome could help manage susceptibility to asthma in early life, TH2-low asthma, particularly neutrophilic asthma, and steroid-resistant asthma (25). However, it should be noted that there are still little data regarding the precise impact of such interventions on the composition and function of the human microbiome, and even less is understood about their putative ability to effect clinically meaningful outcomes in asthma patients. Further studies on the relationship between the human microbiota and lung-specific immunity are warranted.
Along with the airway microbiome, the recently discovered ILCs are predominantly present at mucosal sites and provides novel insights into the regulation and function of the host−microbial mutualism. Among ILCs, the association between ILC3s and the microbiome is of interest. Studies comparing SPF and GF mice reported a decrease in the number of intestinal lamina propria NCR+ILC3s in GF mice (51,52,53). The relationship between NCR+ILC3 cell numbers and the microbiome is also displayed by the study of Guo et al. (54) using RORcCre×Id2fl/fl mice: these mice lack ILC3s and demonstrate greatly impaired resistance to colonization with the intestinal mouse pathogen Citrobacter rodentium. Similarly, mice lacking the AhR in ILC3s, which have reduced ILC3s, also carried more segmented filamentous bacteria (SFB) (55). Therefore, the ILC3s may be involved in the SFB colonization that can induce TH17-inflammatory responses (56).
In the current study, we did not observe any differences between HC and asthma patients in terms of lung or gut microbial diversity or phylum composition. However, we did find that decreased lung function (FVC) is associated with reductions in both the frequency of NCR+ILC3s in the sputum and the microbial diversity in the lung. These data suggest that low NCR+ILC3 numbers in the lung may lead to low microbial diversity and this increases asthma severity. This notion is supported by several studies on the role of NCR+ILC3s and their hallmark cytokine, IL-22, in protecting the host from intestinal infections. Thus, Satoh-Takayama et al. (53) showed that by secreting IL-22, NCR+ILC3s promote intestinal barrier integrity and the proliferation of intestinal epithelium. Zenewicz et al. (57) then showed that IL-22-deficient mice exhibit dysbiosis of the colonic microbiota that enhances their susceptibility to experimentally induced colitis. Since studies on the relationship between asthma, commensal microbiota, and immune cells (especially ILCs) are very scarce, greater research effort on this issue is warranted. In particular, it will be important to examine this relationship in specific asthma phenotypes/endotypes since several studies show that asthma patients have different lung and gut microbial profiles than HC (58,59,60,61).
In summary, we investigated the airway microbiome of HC and asthma patients with differing disease severity. While the HC and asthma patients did not differ in terms of microbial diversity at the genus and species level, airway microbial diversity correlated positively with lung function. Lung function also correlated positively with NCR+ILC3 frequencies in the sputum. Moreover, NCR+ILC3s were significantly decreased with the severity of asthma and specifically interacted with microbiome diversity in the sputum. Thus, these results suggest that NCR+ILC3 may play a critical role in maintaining a healthy microbiota in the airways. Notably, the decreased NCR+ILC3 frequencies and microbial diversity in the airway could serve as candidate markers of severe asthma. Further studies validating the causal relationships in the lung microbiome-immune cell axis may provide critical clues regarding microbiome-based therapeutic approaches for refractory asthma.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (2019R1A2C2087574 and SRC 2017R1A5A1014560).
Abbreviations
- ACT
asthma control test
- AD
atopic dermatitis
- ACQ
asthma control questionnaire
- AR
atopic rhinitis
- AM
alveolar macrophage
- BMI
body mass index
- FEV1
forced expiratory volume in 1 second
- FVC
forced vital capacity
- GF
germ-free
- HC
healthy controls
- ICS
inhaled corticosteroid
- ILC
innate lymphoid cell
- NCR
natural cytotoxicity receptor
- OTU
operational taxonomic unit
- PCoA
principal component analysis
- SFB
segmented filamentous bacteria
- SPF
specific pathogen-free
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Kang HR, Kim HY.
- Data curation: Ham J, Choi S, Park J, Kim Y, Baek MG.
- Investigation: Ham J, Kim J, Choi S, Park J.
- Project administration: Kang HR, Kim HY, Yi H, Won S, Yang S, Bae YS, Chung DH.
- Resources: Kang HR, Sohn KH, Won S, Yi H.
- Supervision: Kim HY.
- Writing - original draft: Ham J, Kim HY.
- Writing - review & editing: Ham J, Choi S, Park J, Kim HY, Kim J, Sohn K, Kang HR, Yi H, Won S, Yang S, Bae YS, Chung DH.
References
- 1.Busse WW, Lemanske RF., Jr Asthma. N Engl J Med. 2001;344:350–362. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 2.Croisant S. Epidemiology of asthma: prevalence and burden of disease. Adv Exp Med Biol. 2014;795:17–29. doi: 10.1007/978-1-4614-8603-9_2. [DOI] [PubMed] [Google Scholar]
- 3.Desai M, Oppenheimer J. Elucidating asthma phenotypes and endotypes: progress towards personalized medicine. Ann Allergy Asthma Immunol. 2016;116:394–401. doi: 10.1016/j.anai.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 5.Peters SP. Asthma phenotypes: nonallergic (intrinsic) asthma. J Allergy Clin Immunol Pract. 2014;2:650–652. doi: 10.1016/j.jaip.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–584. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HY, Umetsu DT, Dekruyff RH. Innate lymphoid cells in asthma: Will they take your breath away? Eur J Immunol. 2016;46:795–806. doi: 10.1002/eji.201444557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Ryu S, Kim HY. Innate lymphoid cells in tissue homeostasis and disease pathogenesis. Mol Cells. 2021;44:301–309. doi: 10.14348/molcells.2021.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo Y, Jeong D, Chung DH, Kim HY. The roles of innate lymphoid cells in the development of asthma. Immune Netw. 2014;14:171–181. doi: 10.4110/in.2014.14.4.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 11.Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. 2015;45:2171–2182. doi: 10.1002/eji.201545598. [DOI] [PubMed] [Google Scholar]
- 12.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Sugimoto C, Arainga M, Alvarez X, Didier ES, Kuroda MJ. In vivo characterization of alveolar and interstitial lung macrophages in rhesus macaques: implications for understanding lung disease in humans. J Immunol. 2014;192:2821–2829. doi: 10.4049/jimmunol.1302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 15.van der Veen TA, de Groot LE, Melgert BN. The different faces of the macrophage in asthma. Curr Opin Pulm Med. 2020;26:62–68. doi: 10.1097/MCP.0000000000000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal NR, King LS, D’Alessio FR. Diverse macrophage populations mediate acute lung inflammation and resolution. Am J Physiol Lung Cell Mol Physiol. 2014;306:L709–L725. doi: 10.1152/ajplung.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z, Zhu L. Update on the role of alternatively activated macrophages in asthma. J Asthma Allergy. 2016;9:101–107. doi: 10.2147/JAA.S104508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Chang Y, Bae B, Sohn KH, Cho SH, Chung DH, Kang HR, Kim HY. Innate immune crosstalk in asthmatic airways: innate lymphoid cells coordinate polarization of lung macrophages. J Allergy Clin Immunol. 2019;143:1769–1782.e11. doi: 10.1016/j.jaci.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 19.Ver Heul A, Planer J, Kau AL. The human microbiota and asthma. Clin Rev Allergy Immunol. 2019;57:350–363. doi: 10.1007/s12016-018-8719-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budden KF, Shukla SD, Rehman SF, Bowerman KL, Keely S, Hugenholtz P, Armstrong-James DP, Adcock IM, Chotirmall SH, Chung KF, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019;7:907–920. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- 21.Losol P, Choi JP, Kim SH, Chang YS. The role of upper airway microbiome in the development of adult asthma. Immune Netw. 2021;21:e19. doi: 10.4110/in.2021.21.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faner R, Sibila O, Agustí A, Bernasconi E, Chalmers JD, Huffnagle GB, Manichanh C, Molyneaux PL, Paredes R, Pérez Brocal V, et al. The microbiome in respiratory medicine: current challenges and future perspectives. Eur Respir J. 2017;49:1602086. doi: 10.1183/13993003.02086-2016. [DOI] [PubMed] [Google Scholar]
- 23.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang YJ. Asthma microbiome studies and the potential for new therapeutic strategies. Curr Allergy Asthma Rep. 2013;13:453–461. doi: 10.1007/s11882-013-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hufnagl K, Pali-Schöll I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42:75–93. doi: 10.1007/s00281-019-00775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 30.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, editors. Vegan: Community Ecology Package. R Package Version 2.5-7. Vienna: R Foundation; 2020. [Google Scholar]
- 35.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. A lego system for conditional inference. Am Stat. 2006;60:257–263. [Google Scholar]
- 36.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a class of permutation tests: the coin package. J Stat Softw. 2008;28:1–23. [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 38.Jia Y, Fang X, Zhu X, Bai C, Zhu L, Jin M, Wang X, Hu M, Tang R, Chen Z. IL-13+ type 2 innate lymphoid cells correlate with asthma control status and treatment response. Am J Respir Cell Mol Biol. 2016;55:675–683. doi: 10.1165/rcmb.2016-0099OC. [DOI] [PubMed] [Google Scholar]
- 39.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134:671–678.e4. doi: 10.1016/j.jaci.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantazi E, Powell N. Group 3 ILCs: Peacekeepers or troublemakers? What's your gut telling you?! Front Immunol. 2019;10:676. doi: 10.3389/fimmu.2019.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDowell PJ, Heaney LG. Different endotypes and phenotypes drive the heterogeneity in severe asthma. Allergy. 2020;75:302–310. doi: 10.1111/all.13966. [DOI] [PubMed] [Google Scholar]
- 42.Ivanova JI, Bergman R, Birnbaum HG, Colice GL, Silverman RA, McLaurin K. Effect of asthma exacerbations on health care costs among asthmatic patients with moderate and severe persistent asthma. J Allergy Clin Immunol. 2012;129:1229–1235. doi: 10.1016/j.jaci.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 43.Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J, McCoy K, Marsland BJ, Harris NL. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 44.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dom S, Droste JH, Sariachvili MA, Hagendorens MM, Oostveen E, Bridts CH, Stevens WJ, Wieringa MH, Weyler JJ. Pre- and post-natal exposure to antibiotics and the development of eczema, recurrent wheezing and atopic sensitization in children up to the age of 4 years. Clin Exp Allergy. 2010;40:1378–1387. doi: 10.1111/j.1365-2222.2010.03538.x. [DOI] [PubMed] [Google Scholar]
- 47.Feldman MF, Bird JA. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. Pediatrics. 2013;132:S5–S6. doi: 10.1016/j.jpeds.2012.09.049. [DOI] [PubMed] [Google Scholar]
- 48.Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, Good JT, Jr, Gelfand EW, Martin RJ, Leung DY. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381.e1-3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, Carroll MP, Bruce KD, Howarth PH. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Hölscher C, et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Guo X, Liang Y, Zhang Y, Lasorella A, Kee BL, Fu YX. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. 2015;42:731–743. doi: 10.1016/j.immuni.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fazlollahi M, Lee TD, Andrade J, Oguntuyo K, Chun Y, Grishina G, Grishin A, Bunyavanich S. The nasal microbiome in asthma. J Allergy Clin Immunol. 2018;142:834–843.e2. doi: 10.1016/j.jaci.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Li F, Liang B, Liang Y, Chen S, Mo X, Ju Y, Zhao H, Jia H, Spector TD, et al. A metagenome-wide association study of gut microbiota in asthma in UK adults. BMC Microbiol. 2018;18:114. doi: 10.1186/s12866-018-1257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldman DL, Chen Z, Shankar V, Tyberg M, Vicencio A, Burk R. Lower airway microbiota and mycobiota in children with severe asthma. J Allergy Clin Immunol. 2018;141:808–811.e7. doi: 10.1016/j.jaci.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 61.Begley L, Madapoosi S, Opron K, Ndum O, Baptist A, Rysso K, Erb-Downward JR, Huang YJ. Gut microbiota relationships to lung function and adult asthma phenotype: a pilot study. BMJ Open Respir Res. 2018;5:e000324. doi: 10.1136/bmjresp-2018-000324. [DOI] [PMC free article] [PubMed] [Google Scholar]