Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces coronavirus disease 2019 (COVID-19) and may increase the risk of adverse outcomes in lung cancer patients. In this study, we investigated the expression and function of mucin 1 (MUC1) after SARS-CoV-2 infection in the lung epithelial cancer cell line Calu-3. MUC1 is a major constituent of the mucus layer in the respiratory tract and contributes to pathogen defense. SARS-CoV-2 infection induced MUC1 C-terminal subunit (MUC1-C) expression in a STAT3 activation-dependent manner. Inhibition of MUC1-C signaling increased apoptosis-related protein levels and reduced proliferation-related protein levels; however, SARS-CoV-2 replication was not affected. Together, these results suggest that increased MUC1-C expression in response to SARS-CoV-2 infection may trigger the growth of lung cancer cells, and COVID-19 may be a risk factor for lung cancer patients.
Keywords: Calu-3, Lung epithelial cancer, Mucin 1, SARS-CoV-2, STAT3
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a lineage B betacoronavirus that is responsible for the current coronavirus disease 2019 (COVID-19) pandemic. COVID-19 first appeared in Wuhan City, China in December 2019 (1, 2). By March 17, 2021, the World Health Organization reported that SARS-CoV-2 had infected 120,383,919 people and resulted in the death of 2,664,386 individuals since January 2020 (https://covid19.who.int).
Clinical reports suggest that age, gender, and comorbidities are risk factors for morbidity, complications, and mortality of COVID-19 patients (3). Adults ≥ 65 years old show more severe illness and higher mortality rates (3). Comorbidities that increase the risk of COVID-19 include diabetes, obesity, chronic kidney disease, solid organ transplant, and cancer. Indeed, a recent report from Wuhan University demonstrated that cancer patients were hospitalized at a two-fold higher rate than the general population (4, 5). Given the clinical complexity of cancer, it is critical to develop management plans for cancer patients that contract COVID-19 (6). As lung cancer is the leading cause of cancer death worldwide (7) and SARS-CoV-2 infects the respiratory track, COVID-19 is a key concern for lung cancer patients (8, 9).
Mucins are high molecular weight glycoproteins that are prominently expressed in respiratory, gastrointestinal, and reproductive tracts. The human mucin (MUC) family primarily consists of secreted and transmembrane mucins, which function as physical barriers to pathogens (10). MUC1, a type I transmembrane mucin, is expressed as a single protein that is auto-cleaved at the SEA (sea-urchin sperm protein, enterokinase and agrin) domain to form two heterodimerizing subunits: MUC1-N and MUC1-C (11, 12). Whereas MUC1 expression is localized to the apical surface of normal epithelia, MUC1 is overexpressed in the entire membrane of malignant tumors, such as breast, pancreatic, colon, and lung. Furthermore, MUC1-C is a well-known oncoprotein that is involved in various signaling pathways, including signal transducer and activator of transcription 3 (STAT3), glycogen synthase kinase 3 beta (GSK-3β), and β-catenin, which are implicated in the maintenance of cancer cells (13-16). MUC1, along with the secretory mucin, MUC5AC, is overexpressed in non-small cell lung cancer and seems to be closely associated with cancer progression (17, 18). Based on these observations, MUC1 has been studied as a potential therapeutic target in many cancers, including lung cancer (17-19).
Here, we investigated MUC1-C expression and function after SARS-CoV-2 infection in the human airway epithelial cell line Calu-3 as a model for lung cancer cells. We confirmed involvement of STAT3 in SARS-CoV-2-induced MUC1-C expression and investigated the effect of MUC1-C inhibition on viral replication and cell viability.
RESULTS
SARS-CoV-2 infection induces MUC1-C expression in Calu-3 cells
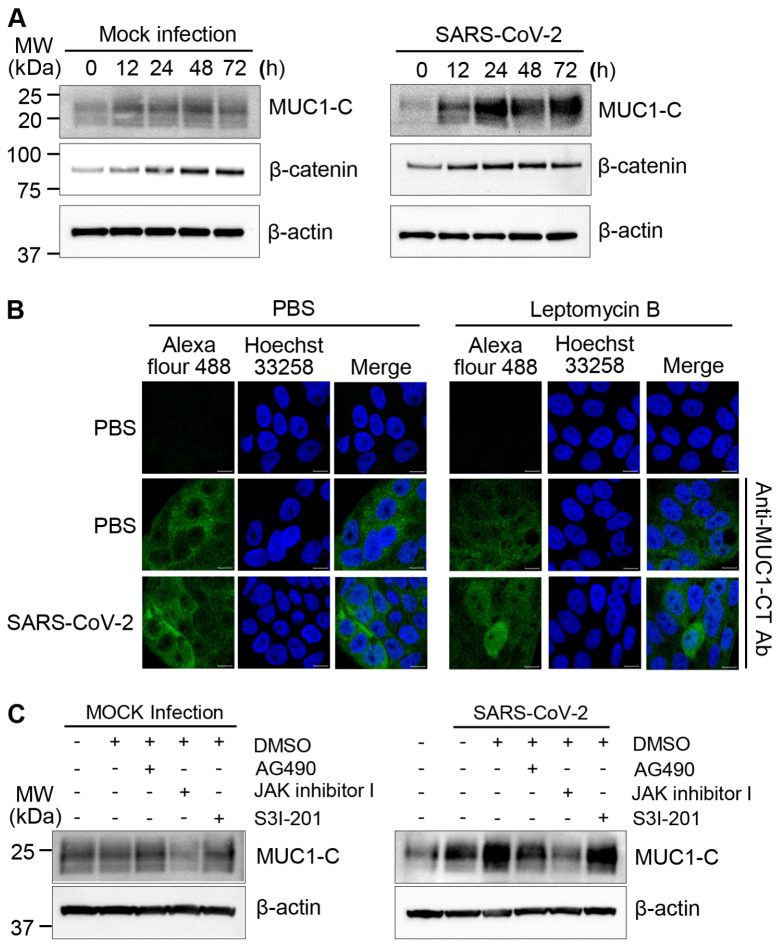
Many COVID-19 patients have difficulty breathing partially due to excessive mucus formation (20). As MUC1 is a major constituent of the mucus layer in the respiratory system, we investigated the impact of SARS-CoV-2 infection on MUC1 expression in Calu-3 cells using an antibody that recognizes the intracellular C-terminal region of MUC1. As controls, Calu-3 cells were cultured without infection (mock infection). We found that MUC1-C expression was slightly elevated during culture in mock-infected cells and was markedly induced by SARS-CoV-2 (Fig. 1A). Specifically, we observed that SARS-CoV-2 infection promptly induced MUC1-C expression at 12 h. Moreover, SARS-CoV-2-induced MUC1-C expression continued to rise up to 24 h and was maintained at peak levels until 72 h. As β-catenin is one of the known downstream targets activated by MUC1, we investigated expression levels of β-catenin. However, we did not observe any differences between SARS-CoV-2 and mock infections. In addition to its role at the cell membrane, MUC1-C can function as an oncogene by translocating to the nucleus to activate transcription (13, 14). To determine if MUC1-C was imported to the nucleus after SARS-CoV-2 infection, we treated Calu-3 cells with Leptomycin B to inhibit nuclear export. We found that a small proportion of MUC1-C was present in nuclei after SARS-CoV-2 infection (Fig. 1B). In contrast, no MUC1-C signal was observed in the nuclei of uninfected control cells. These results suggest that MUC1-C signaling activity is substantially altered in Calu-3 cells after SARS-CoV-2 infection.
Fig. 1.
MUC1-C expression in SARS-CoV-2-infected Calu-3 cells. (A) Calu-3 cells were mock-infected as a control or infected with SARS-CoV-2 at a MOI of 0.5 for the indicated time periods. Cell lysates were prepared, and western blotting was performed with the indicated antibodies. β-actin was analyzed as a loading control. (B) Calu-3 cells were mock-infected or infected with SARS-CoV-2 at a MOI of 0.5. After 1 h of incubation, the viral medium was replaced with DMEM containing 2% FBS. After 45 h of incubation, the cells were treated with PBS or 20 nM Leptomycin B for 3 h. To evaluate localization of MUC1-C, cells were stained with anti-MUC1-CT (green) and Hoechst 33258 to visualize nuclei (blue). (C) Calu-3 cells were pretreated with 0.1% DMSO, 25 μM AG490, 1 μM JAK inhibitor I, or 20 μM S3I-201 for 30 min. The cells were washed with PBS, and then mock-infected or infected with SARS-CoV-2 in PBS at a MOI of 0.5. Cell lysates were prepared at 48 h and analyzed by western blotting.
MUC1-C expression is dependent on STAT3 activation in Calu-3 cells after SARS-CoV-2 infection
We have previously reported that SARS-CoV-2 infection induces STAT3 phosphorylation in Calu-3 cells (21), and others have demonstrated that STAT3 influences MUC1 expression in lung cancer cells (22). Therefore, we investigated the effect of STAT3 inhibitors, including AG490, JAK inhibitor I, and S3I-201, on MUC1-C expression. In mock-infected Calu-3 cells, basal expression of MUC1-C was reduced by treatment with JAK inhibitor I but not by treatment with AG490 or S3I-201. Importantly, JAK inhibitor I also markedly reduced MUC1-C expression in SARS-CoV-2-infected Calu-3 cells (Fig. 1C). These result support that increased MUC1 expression after SARS-CoV-2 infection is dependent on JAK1/3 activation and subsequent STAT3 activation.
MUC1-C contributes to cell survival in Calu-3 cells after SARS-CoV-2 infection
To investigate the contribution of MUC1-C to cell survival and proliferation in SARS-CoV-2-infected Calu-3 cells, we inhibited the dimerization and functional activity of MUC1-C with the cell-penetrating peptide GO-201. GO-201 treatment increased the levels of cleaved PARP and cleaved caspase-3, suggesting that GO-201 induces apoptosis after SARS-CoV-2-infection (Fig. 2A). Furthermore, the expression of c-Myc and cyclin D1 were reduced, implying decreased proliferation after GO-201 treatment in SARS-CoV-2-infected Calu-3 cells (Fig. 2A). These findings appear to be specific to inhibition of MUC1-C because we did not observe any evident effect of the control peptide CP-1 (2 μM). However, we observed that higher concentration of control peptide CP-1 (5 μM) has some GO-201-like effects suggesting non-specific inhibition of cell-permeable peptides at high concentration. Inhibition of MUC1 signaling did not prominently affect the expression of β-catenin in SARS-CoV-2-infected Calu-3 cells. Moreover, neither GO-201 nor CP-1 treatment had any effect in mock-infected Calu-3 cells (Fig. 2B). These results suggest that MUC1-C signaling contributes to cell survival and the maintenance of Calu-3 cells after infection with SARS-CoV-2.
Fig. 2.
Inhibition of MUC1-C signaling enhances apoptotic protein expression in SARS-CoV-2-infected Calu-3 cells. Calu-3 cells were mock-infected (A) or infected with SARS-CoV-2 at a MOI of 0.5 (B). After infection, the cells were treated with 2 μM of cell-penetrating peptides GO-201 or CP-1. Cell lysates were prepared 48 h after infection, and western blotting was performed with the indicated antibodies. β-actin was analyzed as a loading control.
Effect of MUC1-C signaling on SARS-CoV-2 replication in Calu-3 cells
To investigate the possible functions of MUC1-C in SARS-CoV-2 replication, we treated Calu-3 cells with GO-201 or CP-1 and measured virus replication by real-time PCR and plaque formation assays. We amplified RdRP, which is synthesized by SARS-CoV-2, to estimate the viral copy number. We found that treatment with GO-201 induced 10-fold higher copy numbers of viral RNAs compared to CP-1 treatment (Fig. 3A). Additionally, we assessed the viral titer by standard plaque formation assay; however, we did not find any significant differences in the number of plaques between treatment groups (Fig. 3B). Considering that GO-201 induces apoptosis of Calu-3 cells, it is likely that the higher viral copy numbers of the GO-201-treated samples resulted from the infection-incompetent virus RNAs released by the dead cells. Taken together, the effect of MUC1-C signaling on SARS-CoV-2 replication may be limited in Calu-3 cells.
Fig. 3.
MUC1-C signaling modestly influences the replication of SARS-CoV-2. Calu-3 cells (n = 3, 6-well plate) were infected with SARS-CoV-2 at a MOI of 0.1 prior to treatment with PBS, 2 μM GO-201, or 2 μM CP-1. Viral particles were collected from cell culture supernatants 48 h after infection, and virus quantification was performed by qRT-PCR analysis of the SARS-CoV-2 RdRP gene (A) and plaque formation assay (B). Significance was determined by comparison to the untreated control or CP-1 control. **P < 0.01; ns, not significant.
DISCUSSION
MUC1 is upregulated in lung cancer cells and is hyper-secreted in the airway mucus of COVID-19 patients, suggesting that it may play a role in both conditions (22). To understand the risk of lung cancer patients infected with SARS-CoV-2, we investigated the expression and function of MUC1-C in the lung cancer cell line Calu-3. Our results demonstrate that induction of MUC1-C in response to SARS-CoV-2 infection may contribute to the sustained growth of lung cancer cells instead of offering protection from SARS-CoV-2.
In normal epithelial cells, highly glycosylated MUC1-N interacts and forms heterodimers with the membrane subunit MUC1-C and functions as a barrier to pathogens (11, 23). However, in lung cancer cells, MUC1-N is released from the membrane by proteases, such as TNF-α converting enzyme and Matrix metalloproteinase-14 (24, 25). Furthermore, MUC1 expression is polarized to the luminal membrane in normal epithelial cells but is indiscriminately expressed throughout the membrane in lung cancer cells (26, 27). These observations suggest that the protective function of MUC1 is markedly reduced in lung cancer cells compared to normal epithelial cells. Therefore, we expect that malignant cells of lung cancer patients are more susceptible to SARS-CoV-2 infection (Fig. 4).
Fig. 4.
Modeling the influence of MUC1-C on cell survival in SARS-CoV-2-infected Calu-3 cells. In lung cancer cells, MUC1-N domains are frequently released, which may weaken the protective function of MUC1 and sensitize cells to SARS-CoV-2 infection. Due to lack of interaction with MUC1-N, MUC1-C is internalized and interacts with transcription factors, such as β-catenin and phospho-STAT3. The MUC1 complex translocates to the nucleus and activates the transcription of several genes, such as MUC1, c-Myc, and cyclin D1, which contribute to the survival and proliferation of the infected cells. Several questions remain, including the mechanism of JAK1/3 activation after SARS-CoV-2 infection, and the putative function of viral proteins. Altogether, we suggest that SARS-CoV-2 infection may aggravate lung cancer via induction of MUC1 in a JAK/STAT3 activation-dependent manner.
MUC1 is upregulated upon infection with respiratory viruses, such as respiratory syncytial virus and human metapneumovirus, in A549 epithelial cells (28, 29). Consistent with these reports, we found STAT3-dependent induction of MUC1 in Calu-3 cells after SARS-CoV-2 infection. Induction of MUC1 during SARS-CoV-2 infection may contribute to pathogen defense by reducing efficacious infection or as an immunomodulator in normal and cancerous epithelial cells. In contrast, some reports have suggested that high MUC1 expression may be problematic. For example, MUC1 and MUC5AC proteins are hyper-secreted in the airway mucus of COVID-19 patients and may contribute to dyspnea (22). Of note, these observations show induction of MUC1 in normal epithelial cells during SARS-CoV-2 infection. We expect more complicated outcomes in lung cancer patients due to the diverse roles of MUC1-C as a pathogen barrier and an oncogene in cancer cells. MUC1-C interacts with other transcription factors, such as β-catenin, STAT3, and TP53 (13-15), and induces the expression of downstream genes, including MUC1, c-Myc, and cyclin D1 (Fig. 4). As we have previously demonstrated that SARS-CoV-2 infection induces STAT3 activation in Calu-3 cells (22), we hypothesize that MUC1-C induction and STAT3 activation create a positive feedback loop that further induces the expression of MUC1 in Calu-3 cells after SARS-CoV-2 infection. Moreover, we found that inhibition of MUC1-C signaling reduces the expression of proliferation-related proteins (c-Myc and cyclin D1) and enhances the expression of apoptosis-related proteins after SARS-CoV-2 infection in Calu-3 cells. However, the effect of MUC1 signaling on virus replication was not clear. These results suggest that induction of MUC1 expression during SARS-CoV-2 infection may trigger sustained growth of lung cancer cells instead of suppression of viral infection and replication.
There is evidence that COVID-19 poses a risk for lung cancer patients. A recent investigation of 102 COVID-19 patients with lung cancer demonstrated that COVID-19 was severe in lung cancer patients (62% hospitalized, 25% died), but the severity of COVID-19 was related more closely to patient-specific features, such as age, genetic variation, and smoking status, than cancer-specific characteristics (30). The conclusions that can be drawn from this study are limited by its relatively small scale and short time frame. Together with our present study, these findings indicate that extensive and long-term investigation is required to determine whether COVID-19 is an important risk factor for lung cancer patients. Nevertheless, our findings suggest that MUC1 expression is induced by SARS-CoV-2 and may contribute to the increased risk of lung cancer cells upon SARS-CoV-2. Importantly, we show that these effects can be targeted by inhibition of JAK-dependent STAT3 activation which may have important clinical implications for lung cancer patients who contract COVID-19.
MATERIALS AND METHODS
Cell culture and virus
The human airway epithelial cell line, Calu-3, and African green monkey kidney cell line, Vero E6 cell, were obtained from the Korean Cell Line Bank. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific), 25 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were incubated in atmospheric conditions of 95% air and 5% CO2 at 37°C. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, NCCP No. 43326) was provided by the National Culture Collection for Pathogens (Osong, Korea).
Virus amplification and quantification
SARS-CoV-2 amplification and quantification was performed as described previously with minor modifications (21, 31, 32), and detailed information is provided in the Supplementary Material. SARS-CoV-2 amplification and cell culture procedures were performed in biosafety level 3 (BSL-3) conditions.
Antibodies and inhibitors
To detect the cytoplasmic tail of MUC1-C in cells by western blotting, we purchased a commercially available rabbit anti-MUC1-CT antibody from Abcam (Catalog No. ab109185). Anti-bodies to poly-ADP ribose polymerase (PARP, Catalog No. 9542S), cleaved caspase-3 (Catalog No. 9661S), c-Myc (Catalog No. 5605S), cyclin D1 (Catalog No. 2978S), and β-catenin (Catalog No. 8480S) were purchased from Cell Signaling Technology. Anti-β-actin (Catalog No. A5316) was obtained from Sigma-Aldrich. AG490, a JAK2 kinase inhibitor (Catalog No. T3434), and S3I-201, a STAT3 dimerization inhibitor (Catalog No. SML0330), were purchased from Sigma-Aldrich. JAK inhibitor I (Catalog No. 420099), a potent ATP-competitive inhibitor of JAK1, JAK2, and JAK3, was purchased from Calbiochem. All inhibitors were dissolved in dimethyl sulfoxide (DMSO, Catalog No. 10378-73, Kanto Chemical). For inhibitor studies, cells were pretreated with 0.1% DMSO, 25 μM AG490, 20 μM S3I-201, or 1 μM JAK inhibitor I for 30 min. After pretreatment, cells were infected with SARS-CoV-2 at a MOI of 0.5 for 1 h and then cultured in DMEM containing 2% FBS for 48 h. GO-201, a MUC1-C inhibitor (d-RRRRRRRRR-CQCRRK NYGQLDIFP), and a control peptide (CP-1, d-RRRRRRRRR-AQARRKNYGQLDIFP) were synthesized by Anygen as described previously (33). Calu-3 cells were infected with SARS-CoV-2 at a MOI of 0.1. After viral infection, cells were treated with PBS or 2 μM of GO-201 or CP-1 and incubated for another 48 h.
Western blotting
SARS-CoV-2-infected Calu-3 cells were lysed with cell lysis buffer (20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 100 mM NaF, 2 mM Na3VO4, and 1% NP-40) and the cell lysates were run on SDS-PAGE and subsequently transferred onto a nitrocellulose membrane as described previously (22, 34), and detailed information is provided in the Supplementary Material.
Confocal images
MUC1-C expression and localization was confirmed by confocal imaging as described previously (35, 36). Briefly, Calu-3 cells were cultured for 24 h on poly-L-lysine-coated glass cover slips in 12-well culture plates. The cells were washed with PBS and infected with SARS-CoV-2 at an MOI of 0.5. After 1 h, viral supernatants were removed and replaced with 2 ml of DMEM containing 2% FBS. The cells were incubated for 45 h at 37°C in a CO2 incubator prior to treatment with PBS or Leptomycin B (LMB, Cell Signaling Technology, 20 nM), an inhibitor of chromosomal region maintenance 1 (CRM1) that is a nuclear export protein. After a 3 h treatment, the cells were prepared for immunofluorescent imaging as follows. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 3% BSA, and incubated with rabbit anti-MUC1-CT antibody for 2 h at room temperature. After washing the cells with PBST (0.1% Triton X-100 in PBS) containing 1% BSA, the cells were incubated with Alexa Fluor 488-conjugated secondary antibody (Catalog No. A32790, Thermo Fisher Scientific) for 1 h at room temperature. Nuclei were stained with Hoechst 33258 (Thermo Fisher Scientific). Images were collected using a confocal laser scanning microscope system (CLSM, LSM 710, Carl Zeiss).
Quantitative real-time RT-PCR
Viral particles were obtained from virus-infected cell culture supernatants, and quantification of the RNA-dependent RNA polymerase (RdRP) gene of SARS-CoV-2 was performed as described previously (21, 37), and detailed information is provided in the Supplementary Material.
Statistical analysis
Results are shown as the mean ± standard deviation. The statistical significance of differences between two samples was evaluated using Student’s t-test, and P < 0.05 was considered the threshold for statistical significance.
Supplementary Materials
ACKNOWLEDGEMENTS
This research was supported by grants from the National Research Foundation (NRF-2016M3A9B6916708, NRF-2020M 3A9I2107294) funded by the Ministry of Science and ICT in the Republic of Korea.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X, Wang X, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J, Ouyang W, Chua MLK. SARS-CoV-2 transmission in cancer patients of a tertiary hospital in Wuhan. JAMA Oncol. 2020;6:1108–1110. doi: 10.1001/jamaoncol.2020.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidaway P. COVID-19 and cancer: What we know so far. Nat Rev Clin Oncol. 2020;17:336. doi: 10.1038/s41571-020-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bersanelli M. Controversies about COVID-19 and anticancer treatment with immune checkpoint inhibitors. Immunotherapy. 2020;12:269–273. doi: 10.2217/imt-2020-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 8.Russano M, Vincenzi B, Tonini G, Tonini G, Santini D. Coronavirus disease 2019 or lung cancer: What should we treat? J Thorac Oncol. 2020;15:e105–106. doi: 10.1016/j.jtho.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Huang Y, Xie C. The treatment and outcome of a lung cancer patient infected with severe acute respiratory syndrome coronavirus-2. J Thorac Oncol. 2020;15:e63–64. doi: 10.1016/j.jtho.2020.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 11.Levitin F, Stern O, Weiss M, et al. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 12.Macao B, Johansson DG, Hansson GC, Härd T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 13.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad R, Rajabi H, Kosugi M, et al. MUC1-C oncoprotein promotes STAT3 activation in an autoinductive regulatory loop. Sci Signal. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 17.Awaya H, Takeshima Y, Yamasaki M, Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am J Clin Pathol. 2004;121:644–653. doi: 10.1309/U4WGE9EBFJN6CM8R. [DOI] [PubMed] [Google Scholar]
- 18.Kaira K, Okumura T, Nakagawa K, et al. MUC1 expression in pulmonary metastatic tumors: a comparison of primary lung cancer. Pathol Oncol Res. 2012;18:439–447. doi: 10.1007/s12253-011-9465-9. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, Wang X. Critical roles of mucin-1 in sensitivity of lung cancer cells to tumor necrosis factor-alpha and dexamethasone. Cell Biol Toxicol. 2017;33:361–371. doi: 10.1007/s10565-017-9393-x. [DOI] [PubMed] [Google Scholar]
- 20.Lu W, Liu X, Wang T, et al. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical Ill COVID‐19 patients. J Med Virol. 2021;93:582–584. doi: 10.1002/jmv.26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park BK, Kim D, Park S, et al. Differential signaling and virus production in Calu-3 cells and Vero cells upon SARS-CoV-infection. Biomol Ther (Seoul) 2021;29:273–281. doi: 10.4062/biomolther.2020.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, McConnell MJ, Yu B, et al. MUC1 is a downstream target of STAT3 and regulates lung cancer cell survival and invasion. Int J Oncol. 2009;35:337–345. [PMC free article] [PubMed] [Google Scholar]
- 23.Lindén SK, Sheng YH, Every AL, et al. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5:e1000617. doi: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386–3394. doi: 10.1074/jbc.M208326200. [DOI] [PubMed] [Google Scholar]
- 25.Thathiah A, Carson DD. Mt1-Mmp mediates muc1 shedding independent of tace/adam17. Biochem J. 2004;382:363–373. doi: 10.1042/BJ20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaira K, Nakagawa K, Ohde Y, et al. Depolarized MUC1 expression is closely associated with hypoxic markers and poor outcome in resected non-small cell lung cancer. Int J Surg Pathol. 2012;20:223–232. doi: 10.1177/1066896911429296. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Dinwiddie DL, Harrod KS, Jiang Y, Kim KC. Anti-inflammatory effect of MUC1 during respiratory syncytial virus infection of lung epithelial cells in vitro. Am J Physiol Lung Cell Mol Physiol. 2010;298:L558–563. doi: 10.1152/ajplung.00225.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baños-Lara Mdel R, Piao B, Guerrero-Plata A. Differential mucin expression by respiratory syncytial virus and human metapneumovirus infection in human epithelial cells. Mediators Inflamm. 2015;2015:347292. doi: 10.1155/2015/347292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Rizvi H, Preeshagul IR, et al. COVID-19 in patients with lung cancer. Ann Oncol. 2020;31:1386–1396. doi: 10.1016/j.annonc.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandeel M, Yamamoto M, Al-Taher A, et al. Small molecule inhibitors of Middle East respiratory syndrome coronavirus fusion by targeting cavities on heptad repeat trimers. Biomol Ther (Seoul) 2020;28:311–319. doi: 10.4062/biomolther.2019.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park BK, Maharjan S, Lee SI, et al. Generation and characterization of a monoclonal antibody against MERS-CoV targeting the spike protein using a synthetic peptide epitope-CpG-DNA-liposome complex. BMB Rep. 2019;52:397–402. doi: 10.5483/BMBRep.2019.52.6.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raina D, Ahmad R, Joshi MD, et al. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin MJ, Kim DW, Choi YJ, et al. PEP-1-GLRX1 protein exhibits anti-inflammatory effects by inhibiting the activation of MAPK and NF-κB pathways in Raw 264.7 cells. BMB Rep. 2020;53:106–111. doi: 10.5483/BMBRep.2020.53.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Kim D, Kim JN, et al. A mucin1 C-terminal subunit-directed monoclonal antibody targets overexpressed mucin1 in breast cancer. Theranostics. 2018;8:78–91. doi: 10.7150/thno.21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu G, Maharjan S, Kim D, et al. A novel monoclonal antibody targets mucin1 and attenuates growth in pancreatic cancer model. Int J Mol Sci. 2018;19:2004. doi: 10.3390/ijms19072004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park BK, Kim J, Park S, et al. MERS-CoV and SARS-CoV-2 replication can be inhibited by targeting the interaction between the viral spike protein and the nucleocapsid protein. Theranostics. 2021;11:3853–3867. doi: 10.7150/thno.55647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.