Abstract
ErbB3-binding protein 1 (EBP1) is a multifunctional protein associated with neural development. Loss of Ebp1 leads to upregulation of the gene silencing unit suppressor of variegation 3-9 homolog 1 (Suv39H1)/DNA (cytosine 5)-methyltransferase (DNMT1). EBP1 directly binds to the promoter region of DNMT1, repressing DNA methylation, and hence, promoting neural development. In the current study, we showed that EBP1 suppresses histone methyltransferase activity of Suv39H1 by promoting ubiquitin-proteasome system (UPS)-dependent degradation of Suv39H1. In addition, we showed that EBP1 directly interacts with Suv39H1, and this interaction is required for recruiting the E3 ligase MDM2 for Suv39H1 degradation. Thus, our findings suggest that EBP1 regulates UPS-dependent degradation of Suv39H1 to govern proper heterochromatin assembly during neural development.
Keywords: ErbB3-binding protein 1, MDM2, Suppressor of variegation 3-9 homolog 1, Ubiquitin-proteasome system
INTRODUCTION
ErbB3-binding protein 1 encodes two alternatively spliced EBP1 isoforms, p48 EBP1 and p42 EBP1. The p48 EBP1 isoform is 54 amino acids longer than p42 EBP1 at its N-terminus. Both EBP1 isoforms are constitutively expressed in all tissues and cells, including cells that do not express the ERBB3 receptor, and only p42 EBP1, and not p48 EBP1, binds to ERBB3 (1, 2). Embryonic development inherently involves many distinct cellular activities, including cell proliferation, survival, and differentiation (2-6), and unlike p42 EBP1, p48 EBP1 is expressed throughout embryonic tissues including brain, contributing to epigenetic control by suppressing the gene silencing unit suppressor of variegation 3-9 homolog 1 (Suv39H1)/DNA (cytosine 5)-methyltransferase (DNMT1) (7, 8).
Despite lacking E3 ligase activity, EBP1 has been implicated in the ubiquitin proteasome system (UPS) for degradation of its binding partner by linking an E3 ligase to its target protein. For instance, p48 EBP1 physically associates with MDM2 (also known as HDM2) and enhances the interaction between MDM2 and p53, promoting p53 degradation in glioblastoma cells of patients with poor clinical outcome (4). Moreover, p48 EBP1 sustains Akt-dependent MDM2 phosphorylation, confining MDM2 to the nucleus, and thereby preventing self-ubiquitination of MDM2 via upregulation of Akt activity (9). Similarly, p42 EBP1 interacts with regulatory subunit, p85 subunit of phosphatidyl inositol 3-kinase (PI3K) that results in the UPS-dependent degradation of the p85 subunit by recruiting the E3 ligase carboxy terminus of heat shock protein 70 (Hsp70) interacting protein (CHIP) (5), which accounts for the tumor suppressor activity of p42 EBP1. Despite growing evidence of the roles of the EBP1 isoforms in UPS-dependent protein degradation for the regulation of diverse cellular activities over the past decade, it is not known whether EBP1 contributes to transcriptional regulation exclusively by epigenetic control or also via UPS-dependent protein suppression during embryonic development.
Methylation of lysine 9 on histone H3 (H3K9) generates silent domains, a process which is critical for heterochromatin assembly, and is sufficient for the initiation of gene repression (10, 11). Spurred by our finding of dramatic Suv39H1-mediated histone methylation changes in EBP1-deficient cells during embryonic development, we then questioned whether Suv39H1 protein level is also affected by p48 EBP1. We previously reported that gene expression of Suv39H1 and H3K9 trimethylation was upregulated in the absence of EBP1; similarly, in the current study, we not only found transcriptional repression of Suv39H1 by p48 EBP1, but also found that the protein level of Suv39H1 was markedly increased in EBP1-deficient mouse brain and Ebp1(−/−) mouse embryonic fibroblasts (MEFs). Furthermore, the observed change in Suv39H1 protein level was due to avoidance from ubiquitination and subsequent degradation as a result of the loss of EBP1. We also found that an interaction between EBP1 and Suv39H1 is required for Suv39H1 ubiquitination, and EBP1 enhances MDM2-mediated Suv39H1 ubiquitination. Thus, during development and apart from repression of Suv39H1, p48 EBP1 regulates UPS-dependent degradation of Suv39H1 to govern proper heterochromatin assembly during embryonic development.
RESULTS
EBP1 regulates Suv39H1
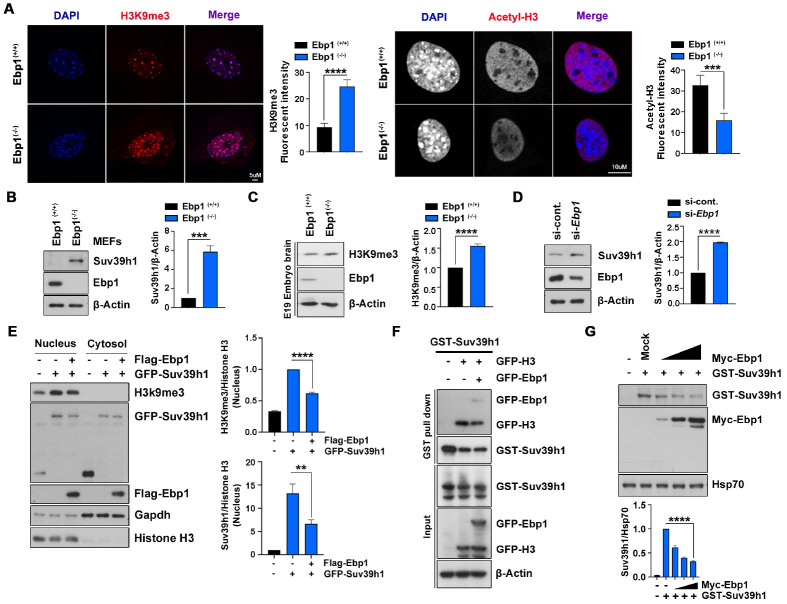
Our recent study of genetic ablation of Ebp1 in mice demonstrated that the loss of Ebp1 elicits upregulation of Suv39H1, which encodes a histone-lysine N-methyltransferase, that not only affects mammalian gene expression, but also its methyltransferase activity during embryonic development. In accordance with enriched H3K9 trimethylation visualized at 4’,6-diamidino-2-phenylindole (DAPI)-dense heterochromatin, we found that the protein level of Suv39H1 was notably increased in Ebp1(−/−) MEFs compared with Ebp1(+/+) MEFs, and that protein aggregation was enriched at regions of euchromatin as identified by high levels of H3 acetylation (Fig. 1A, B). In addition, we clarified that H3K9 trimethylation was increased in embryo brain lysate of Ebp1(−/−) mice (Fig. 1C). Depletion of Ebp1 also enhanced protein levels of Suv39H1 (Fig. 1D). These findings indicate that, in addition to transcriptional regulation, EBP1 may regulate Suv39H1 at the protein level.
Fig. 1.
EBP1 regulates Suv39H1. (A) Ebp1(−/−) and Ebp1(+/+) MEFs were immunostained with H3K9 me3 (left) and acetyl-H3 (right) antibodies for heterochromatin and euchromatin respectively, and counterstained with DAPI. Each bar graph represents the level of H3K9 me3 and acetyl-H3. Scale bars = 5 μm (left) and 10 μm (right). (B) Ebp1(−/−) MEFs have increased levels of Suv39H1 protein. Endogenous protein levels were detected by immunoblotting as indicated. The bar graph shows Suv39H1 protein normalized to β-actin protein abundance. (C) Mouse embryos isolated at E19 from an Ebp1 knockout mouse. Ebp1(+/+) and Ebp1(−/−) embryo brains were used to compare levels of H3K9 me3. Endogenous levels of H3K9 me3 and EBP1 in brain lysates were detected with specific antibodies as indicated. (D) HEK293T cells were transfected using si-control and si-Ebp1 and immunoblotting was performed as indicated. The bar graph shows the quantification of Suv39H1 protein level. (E) Subcellular fractionation was performed using HEK293T cells transfected with GST-Suv39H1 and Flag-EBP1. Histone H3 and GAPDH antibodies for the nucleus and cytosol, respectively. The bar graphs indicate the levels of H3K9 me3 (upper) and GFP-Suv39H1 (lower) normalized to nuclear histone H3. (F) GST-Suv39H1 and GFP-tagged H3 and EBP1 plasmids were transfected to HEK293T cells and GST pull- down assay was performed. (G) HEK293T cells were transfected to GST-Suv39H1 and gradient Myc-EBP1. Immunoblots (upper) and bar graph (lower) show decreasing Suv39H1 protein level. **P < 0.01, ***P < 0.001, ****P < 0.0001. Images shown here are representative at least 3 independent experiments and values in these figures represent means ± SEM from more than 3 independent experiments.
To further assess the role of EBP1 in Suv39H1 regulation, we transfected GFP-tagged Suv39H1 with or without EBP1 and performed cellular fractionation analysis. As anticipated, in the nuclear fraction, we found that the protein level of Suv39H1 was decreased, and taken in accordance with reduced H3K9 methylation in the presence of Flag-EBP1 expression, reflects decreased enzymatic activity of Suv39H1 (Fig. 1E). Moreover, we found decreased binding of Suv39H1 to histone H3 under the condition of Ebp1 overexpression (Fig. 1F). However, there was neither detectable H3K9 methylation nor alteration of Suv39H1 levels in the cytosolic fraction, indicating this event occurs in the nucleus. Commensurate with this finding, we also found that increased expression of Ebp1 reduces protein levels of Suv39H1 in a dose-dependent manner, further indicating that EBP1 influences methyltransferase activity of Suv39H1 by reducing its protein level (Fig. 1G).
EBP1 physically interacts with Suv39H1
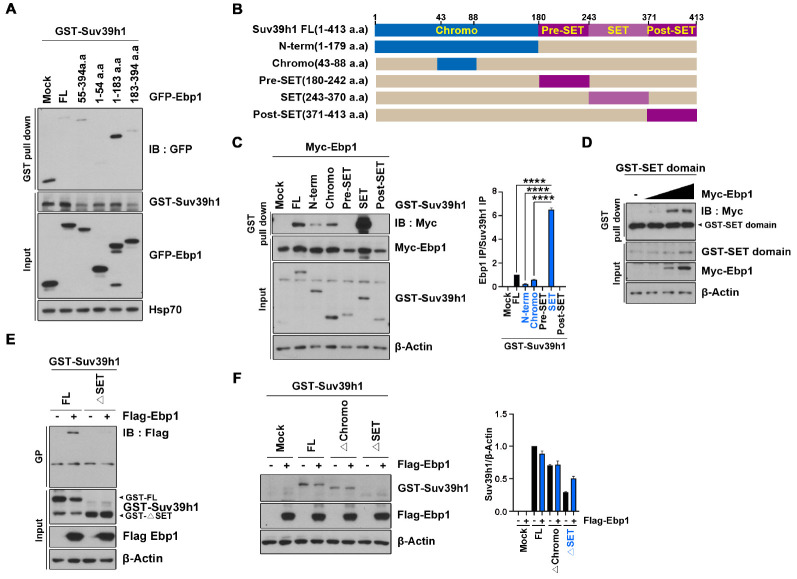
To determine whether EBP1 physically interacts with Suv39H1, we cotransfected GST-Suv39H1 with various fragments of GFP-EBP1 in HEK293T cells. GST pull-down experiments revealed that EBP1 directly interacts with Suv39H1, and its N-terminus 54 amino acid residues are indispensable for the interaction between EBP1 and Suv39H1 (Fig. 2A). To map the specific region of Suv39H1 that binds to EBP1, we generated a series of Suv39H1 fragments (Fig. 2B) and performed mapping analysis. We found that EBP1 weakly binds to full-length Suv39H1 and to the N-terminal domain (amino acids 1-179) that includes an essential chromatin organization modifier domain (chromodomain), which is a major reader of histone methylation tags in proteins, at amino acids 44-88. However, EBP1 robustly bound to the SET domain of Suv39H1, which is a conserved site for lysine methylation (Fig. 2C). To identify the residues involved in this interaction, we first verified the association between EBP1 and the SET domain of Suv39H1 by demonstrating increased binding in response to increased Ebp1 expression (Fig. 2D), and found that a deletion mutant of Suv39H1 lacking the SET domain (ΔSET) completely lost its binding ability to EBP1 (Fig. 2E). These findings show that EBP1 predominantly binds to the SET domain of Suv39H1, thereby accounting for the effect of EBP1 in the regulation of methyltransferase activity.
Fig. 2.
EBP1 physically interacts with Suv39H1. (A) GST-Suv39H1 and GFP-EBP1 fragments were transfected to HEK293T cells, and GST pull-down assay was performed after 24 h transfection. (B) A schematic image of Suv39H1 domains. (C) Cells were transfected with a series of GST-Suv39H1 fragments and Myc-EBP1. Cell lysates were used for GST pull-down assay. An anti-Myc antibody were used to indicate the binding affinity. (D) A gradient increase of Myc-EBP1 plasmid was transfected with GST-SET domain into HEK293T cells. GST pull-down assay was performed. (E) HEK293T cells were transfected with GST-Suv39H1 WT and ΔSET constructs with Flag-EBP1. GST pull-down assay was conducted to the cell lysates and the interaction was confirmed using an anti-Flag antibody. (F) GST-Suv39H1 constructs and Flag-EBP1 were transfected to HEK293T cells. Cell lysates were immunoblotted after 24 h transfection. **P < 0.01, ***P < 0.001, ****P < 0.0001. Images shown here are represen-tative at least 3 independent experiments and values in these figures represent means ± SEM from more than 3 independent experiments.
As Ebp1 expression alters Suv39H1 protein levels and EBP1 strongly binds to the SET domain of Suv39H1, we next determined whether the binding of EBP1 to Suv39H1 is involved in the regulation of Suv39H1 protein level. Cotransfection of Flag-EBP1 with GST-tagged wildtype (WT)-Suv39H1 or a deletion mutant lacking either the catalytic SET domain (ΔSET) or the chromodomain (Δchromo) was performed, and we found that Ebp1 expression decreased Suv39H1 protein level in both the WT and Δchromo mutant. In contrast, we found that the ΔSET Suv39H1 mutant lost the ability to associate with EBP1, and in fact resulted in increased levels of Suv39H1 protein (Fig. 2F), indicating that EBP1 stably regulates Suv39H1 protein levels by direct interaction with the SET domain.
EBP1 controls Suv39H1 stability via UPS
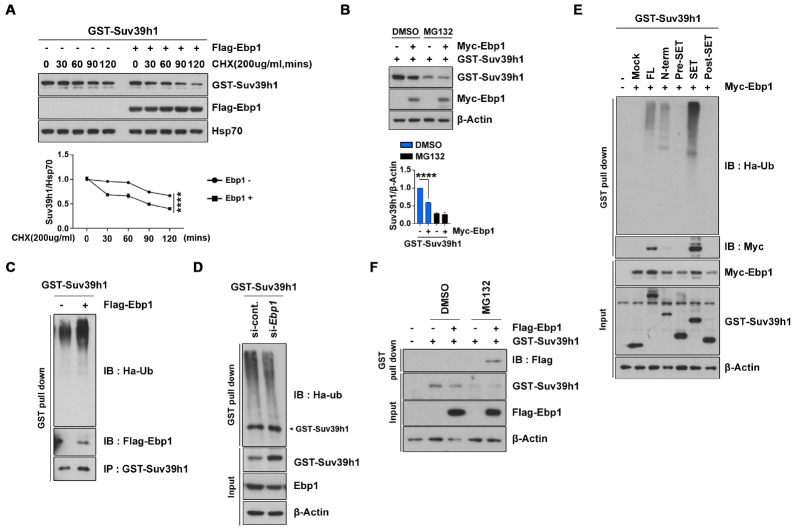
To explore the molecular mechanisms underlying EBP1-induced reduction of Suv39H1, we first examined the half-life of Suv39H1, and found that it was markedly decreased in Ebp1-expressing cells compared to control cells. Further, we found that Suv39H1 protein levels were unaltered for up to an hour after treatment of cycloheximide (CHX), which is an inhibitor of eukaryotic translation (Fig. 3A). As we previously reported that EBP1 links to a binding partner in UPS (4, 5), we next examined whether proteasomal degradation is involved in the observed reduction of Suv39H1. Pretreatment of the proteasome inhibitor MG132 efficiently protects Suv39H1 from EBP1-mediated degradation, indicating that EBP1 decreases Suv39H1 protein abundance by promoting ubiquitination-dependent proteasomal degradation (Fig. 3B). Moreover, GST pull-down experiments using cells cotransfected with GST-Suv39H1 and HA-tagged ubiquitin showed that Suv39H1 was remarkably ubiquitinated in the presence of Ebp1 expression compared with control vector- expressing cells (Fig. 3C). In contrast, silencing of Ebp1 by siRNA inhibited Suv39H1 ubiquitination (Fig. 3D). Thus, EBP1 facilitates degradation of Suv39H1 by the ubiquitin-proteasome pathway.
Fig. 3.
EBP1 controls Suv39H1 stability via UPS. (A) HEK293T cells were transfected with GST-Suv39H1 and Flag-EBP1. Cells were treated with 200 μg/mL cycloheximide (CHX) at indicated time points. Suv39H1 protein level at each point was normalized to Hsp70 antibody abundance. (B) GFP-Suv39H1 and Flag-EBP1 were transfected into HEK293T cells, and cells were then treated with 10 μM DMSO and MG132. After 8 h treatment, cells were lysed and immunoblotted. (C) PC12 cells were transfected with GST-Suv39H1, Flag-EBP1, and HA-Ubiquitin (HA-Ub), and then treated with 10 μM MG132 for 8 h followed by GST pull-down assay. Ubiquitinated Suv39H1 was detected by an anti-HA antibody. (D) PC12 cells were transfected with GST-Suv39H1 and si-Ebp1. After 48 h transfection, cell lysates were subjected to GST pull-down assay. (E) HEK293T cells were transfected by the indicated plasmids. GST pull-down assay was performed. (F) GST-Suv39H1 and Flag-EBP1 were transfected to HEK293T cells, followed by treatment of 10 μM DMSO and MG132 for 8 h. Cell lysates were conducted to GST pull-down assay. **P < 0.01, ***P < 0.001, ****P < 0.0001. Images shown here are representative at least 3 independent experiments and values in these figures represent means ± SEM from more than 3 indepen-dent experiments.
To further evaluate the specificity of Suv39H1 degradation by EBP1, we examined ubiquitination of various truncated forms of Suv39H1. Similar to that found in our protein binding analysis, Suv39H1 ubiquitination was robust in the SET domain, exhibiting a strong binding ability with EBP1, and the full WT protein and chromodomain of Suv39H1 were also ubiquitinated albeit at lower levels. In contrast, areas flanking the SET domain did not interact with EBP1 and were not ubiquitinated (Fig. 3E). Further, the interaction between EBP1 and Suv39H1 was enhanced by MG132 exposure (Fig. 3F). These findings indicate that EBP1 binding is required for Suv39H1 ubiquitination.
EBP1 enhances MDM2 activity on Suv39H1
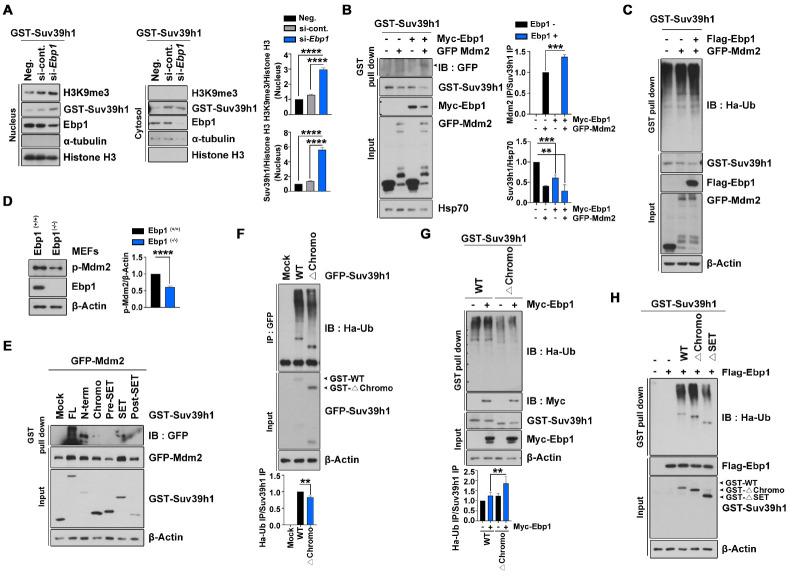
Next, we hypothesized that EBP1 recruits an E3 ligase for Suv39H1 in the nucleus based on the following: 1) our cellular fractionation analysis after EBP1 depletion revealed that stabilization of Suv39H1 was prominently in the nucleus, 2) we also found enhanced methyltransferase activity as shown by H3K9 trimethylation (Fig. 4A), and 3) EBP1 does not possess E3 ligase activity. Considering our previous findings that EBP1 physically associates with MDM2 and enhances the interaction between MDM2 and p53 (4), and that EBP1 confines MDM2 in the nucleus by sustaining MDM2 phosphorylation and inhibiting self-ubiquitination (9), in addition with the finding that Suv39H1 is a substrate of MDM2 (12), we investigated whether EBP1 contributes to MDM2-dependent Suv39H1 degradation. Overexpression of MDM2 elicits a >50% reduction of Suv39H1, and that this effect was facilitated by co-expression with EBP1 up to a 70% reduction, although Ebp1 expression alone also exhibits marked reduction of Suv39H1 (Fig. 4B). Moreover, Suv39H1 ubiquitination was highly promoted by co-expression of MDM2 with EBP1 (Fig. 4C). Consistent with our previous finding that EBP1 maintains MDM2 phosphorylation, the level of phospho-MDM2 was drastically decreased in Ebp1(−/−) MEFs compared with Ebp1(+/+) MEFs (Fig. 4D), and thus, E3 ligase activity of MDM2 may also be decreased.
Fig. 4.
EBP1 enhances MDM2 activity on Suv39H1. (A) HEK293T cells were transfected with GST-Suv39H1, si-Ebp1, and control. After 48 h transfection, cells were lysed and subjected to subcellular fractionation. The nuclear fraction was normalized to histone H3 and the cytosolic fraction to α-tubulin abundance. The bar graphs show the level of nuclear H3K9 me3 (upper) and Suv39H1 (lower) protein. (B) HEK293T cells were transfected with the indicated plasmids. GST pull-down assay and immunobloting was performed as indicated. (C) GST-Suv39H1, Flag-EBP1, GFP-MDM2, and HA-Ub were used to transfect to HEK293T cells for 24 h, after which transfected cells were treated with 10 μM MG132 for 8 h. Cell lysates were used for GST pull-down assay. The level of Suv39H1 ubiquitination was confirmed with an anti-HA antibody. (D) Ebp1-expressing whole-body mouse E13.5 MEF cells were used to determine phospho-MDM2 (p-MDM2) protein level. The bar graph shows p-MDM2 abundance. (E) Indicated plasmids transfected to HEK293T cells and GST pull-down assay was performed. (F) GFP-Suv39H1 WT and Δchromo constructs were transfected to PC12 cells. Cells were then treated with 10 μM MG132 for 8 h and lysed for immunoprecipitation with an anti-GFP antibody. (G) PC12 cells were transfected by GST-Suv39H1 WT and Δchromo, Myc-EBP1, and HA-Ub plasmids. After treatment of 10 μM MG132 for 8 h, cells were lysed and used for GST pull-down assay. The level of ubiquitination was detected using an anti-HA antibody. The bar graph indicates the ubiquitination level of the Suv39H1 constructs normalized to that of the Suv39H1 pull-down control. (H) GST-Suv39H1 WT, Δchromo, and ΔSET constructs, as well as Flag-EBP1 and HA-Ub were transfected to PC12 cells. GST pull-down assay was performed and the HA-Ub blot shows the ubiquitination level of the Suv39H1 constructs in the presence of EBP1. **P < 0.01, ***P < 0.001, ****P < 0.0001. Images shown here are representative at least 3 independent experiments and values in these figures represent means ± SEM from more than 3 independent experiments.
In agreement with a previous finding that MDM2 binds to the chromodomain of Suv39H1 and induces subsequent ubiquitination of Suv39H1 (12), our protein binding analysis showed that MDM2 binds to the chromodomain of Suv39H1, and in fact, exhibits a strong interaction with the chromodomain containing the N-terminal domain (amino acids 1-179) of Suv39H1, but not the SET domain or any other domain of the protein (Fig. 4E). In addition, our ubiquitination experiments showed that the Δchromo mutant exhibited approximately 60% of the ubiquitination found for WT-Suv39H1 (Fig. 4F). However, this decrease in ubiquitination was reversed by overexpression of Ebp1 (Fig. 4G, H). Intriguingly, overexpression of Ebp1 not only proceeded ubiquitination of Δchromo protein, but also decreased ubiquitination of ΔSET domain protein (Fig. 4H), reflecting enhanced protein stability of the ΔSET domain Suv39H1 mutant (Fig. 2F). These findings indicate that an interaction between EBP1 and Suv39H1 is required for MDM2-mediated Suv39H1 ubiquitination.
DISCUSSION
In the current study, we uncovered an additional role of EBP1 for the regulation of SUV39H1-mediated H3K9 methylation in a post-translational modification process. In Ebp1(−/−) mouse brain and Ebp1(−/−) MEFs, we found a significant increase in histone H3K9 trimethylation. Further, in the absence of EBP1, histone methyltransferase Suv39H1 activity was stable, there was less interaction with the E3 ligase MDM2, and there was decreased ubiquitination. The observed increase of Suv39H1 and subsequent H3K9 trimethylation at chromatin sites in Ebp1(−/−) MEFs correlate with a shift in the expression of a variety of cell cycle and neurodevelopmental genes, resulting in failures during cell cycle progression and deficits leading to neural death. Therefore, the physiological relevance of these changes in Suv39H1 protein stability by EBP1 could contribute to a better understanding of the epigenome during development.
We have shown that EBP1 controls Suv39H1 protein levels by enhancing polyubiquitination with the E3 ligase MDM2, which is an interesting finding considering the role of MDM2 as the main regulator of p53 and its involvement in p53 pathway modulation in various cellular contexts (13). In fact, Suv39H1 forms a complex with MDM2 and p53 that negatively regulates p53 function (14). In p53-activating conditions, binding of Suv39H1 to MDM2 is inhibited. Previously, we have shown that EBP1 forms a complex with MDM2 and p53, facilitating p53 degradation as well as inhibition of p53 function in certain types of cancer cells (4). In the absence of EBP1, the level of phospho-MDM2 is decreased (Fig. 4D), a finding which is consistent with reduced ubiquitination of Suv39H1 (Fig. 3D), and reflects the increased binding affinity of Suv39H1 and MDM2 under conditions of p48 EBP1 overexpression (Fig. 4B). Although EBP1 promotes access of MDM2 to target proteins, such as p53 and Suv39H1, and enhances the ubiquitination and degradation of target proteins, it has not yet been determined whether EBP1-mediated degradation of Suv39H1 is a spatio-temporal event during development as H3K9 modification appears to have a critical role in embryogenesis. Furthermore, this importance is highlighted by the finding that mutations in genes of H3K9-modifying enzymes cause severe embryonic growth defects (15) as well as our finding that aberrant upregulation of Suv39H1 contributes to embryonic lethality (8). In addition, whether EBP1 links MDM2 to p53 or Suv39H1 with any preference to the presence of p53 expression, and whether regulation of Suv39H1 levels by EBP1 ultimately modulates the p53 pathway, is unknown and should be addressed in the future.
Chromosome instability is involved in tumor initiation and progression. Suv39H1 deficiency impairs H3K9 methylation at pericentromeric heterochromatin and leads to chromosome instability. For instance, Suv39H1-deficient mice exhibit spontaneous B cell lymphoma and meiosis defects (16). We found that overexpression of Ebp1 reduces Suv39H1-mediated H3K9 trimethylation, whereas depletion of EBP1 reverses it (Fig. 1D and Fig. 4A). Therefore, EBP1 appears to act as a negative regulator of chromosome stability not only as a transcriptional repressor, but also by negatively regulating translational modification of Suv39H1, which may explain the tumorigenic potential of EBP1 in acute leukemia and many types of cancer cells (4, 17, 18). Ebp1 is normally expressed in most brain regions at embryonic day 10.5 (E10.5), with expression encompassing entire organs and tissues at E11.5. Expression of p48 EBP1 is also high during embryogenesis and gradually decreases after the postnatal period. However, in cancer cells including glioblastoma, lung cancer, and acute leukemia, p48 EBP1 protein is highly expressed, whereas the shorter isoform p42 EBP1 is rarely detectable (18). These observations suggest that an increased level of EBP1 in cancer could suppress either transcription of Suv39H1 or protein stability, thereby promoting aberrant gene expression that contributes to cancerous cell growth. Further, a study has shown that H3K9 methylation is associated with aberrant gene silencing in cancer cells (19). Thus, mechanisms of epigenetic regulation such as histone modification involved in gene regulation may not only simply occur in a single way, but may also be strictly controlled under different circumstances or conditions.
Overall, our results demonstrate that p48 EBP1 is a key regulator of Suv239H1 through UPS-dependent degradation by modulating accessibility of the E3 ligase MDM2 during embryonic development. These findings provide a molecular explanation for the role of EBP1 in the regulation of epigenetic control and furthers a possible role of EBP1 in the regulation of chromosome instability.
MATERIALS AND METHODS
Materials and methods are available in the supplemental materials.
Supplementary Materials
ACKNOWLEDGEMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government [Ministry of Science, Information and Communication Technology and Future Planning (MSIP)] (2016R1A5A2945889 and 2020R 1A2C2003268).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Yoo JY, Wang XW, Rishi AK, et al. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Brit J Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc Natl Acad Sci U S A. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn JY, Liu X, Liu Z, et al. Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 2006;25:2083–2095. doi: 10.1038/sj.emboj.7601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim CK, Nguyen TLX, Joo KM, et al. Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer Res. 2010;70:9730–9741. doi: 10.1158/0008-5472.CAN-10-1882. [DOI] [PubMed] [Google Scholar]
- 5.Ko HR, Kim CK, Lee SB, et al. P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP. Cell Death Dis. 2014;5:e1131–e1131. doi: 10.1038/cddis.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang I, Ahn JY. Dysregulation of epigenetic control contributes to Schizophrenia-like behavior in Ebp1+/− mice. Int J Mol Sci. 2020;21:2609. doi: 10.3390/ijms21072609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko HR, Hwang I, Ahn SY, Chang YS, Park WS, Ahn JY. Neuron-specific expression of p48 Ebp1 during murine brain development and its contribution to CNS axon regeneration. BMB Rep. 2017;50:126–131. doi: 10.5483/BMBRep.2017.50.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko HR, Hwang I, Jin EJ, et al. Roles of ErbB3-binding protein 1 (EBP1) in embryonic development and gene-silencing control. Proc Natl Acad Sci U S A. 2019;116:24852–24860. doi: 10.1073/pnas.1916306116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CK, Lee SB, Nguyen TLX, et al. Long isoform of ErbB3 binding protein, p48, mediates protein kinase B/Akt-dependent HDM2 stabilization and nuclear localization. Exp Cell Res. 2012;318:136–143. doi: 10.1016/j.yexcr.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Boros J, Arnoult N, Stroobant V, Collet JF, Decottignies A. Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1α at chromatin. Mol Cell Biol. 2014;34:3662–3674. doi: 10.1128/MCB.00205-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Towbin BD, González-Aguilera C, Sack R, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 12.Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 13.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Li Z, Zwolinska AK, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J. 2010;29:2538–2552. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dambacher S, Hahn M, Schotta G. Epigenetic regulation of development by histone lysine methylation. Heredity. 2010;105:24–37. doi: 10.1038/hdy.2010.49. [DOI] [PubMed] [Google Scholar]
- 16.Peters AHFM, O'Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen LXT, Zhu L, Lee Y, Ta L, Mitchell BS. Expression and role of the ErbB3-binding protein 1 in acute myelogenous leukemic cells. Clin Cancer Res. 2016;22:3320–3327. doi: 10.1158/1078-0432.CCR-15-2282. [DOI] [PubMed] [Google Scholar]
- 18.Ko HR, Chang YS, Park WS, Ahn JY. Opposing roles of the two isoforms of ErbB3 binding protein 1 in human cancer cells. Int J Cancer. 2016;139:1202–1208. doi: 10.1002/ijc.30165. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.