Abstract
Background
Brain metastasis was one of the factors leading to the poor long-term prognosis of patients with lung adenocarcinoma (LUAD).
Methods
The expression levels of immune genes in LUAD and LUAD brain metastases tissues were analyzed in GSE161116 dataset using the GEO2R, and the levels of differential immune genes in normal lung and LUAD tissues were verified. The biological functions and signaling mechanisms of the differential immune genes were explored via Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis. Cox regression analysis was used to screen the prognostic factors of LUAD patients, and a risk model was constructed. The role of the model was checked in the development of LUAD via receiver operating characteristic analysis, gene set enrichment analysis, and Cox regression analysis.
Results
Differentially expressed genes (DEGs) in brain metastasis were involved in the adaptive immune response, B cell differentiation, leukocyte migration, NF-kB signaling pathway, among others. The expression levels of TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 were independent factors affecting the poor prognosis of LUAD patients via Cox regression analysis and Akaike information criterion. In the constructed risk model, the overall survival of LUAD patients in the high-risk group was poor. The risk model was significantly related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients. In addition, the risk model score was an independent risk factor that affected the poor prognosis of LUAD patients. TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 of the risk factors had diagnostic significance in LUAD brain metastasis and LUAD. The risk model participated in cytokinetic process, cell cycle, citrate cycle TCA cycle, etc. The risk model score was correlated with the levels of B cells memory, mast cells resting, macrophages M0, mast cells activated, neutrophils, eosinophils, T cells gamma delta, and immune cell markers.
Conclusions
The risk model based on the LUAD brain metastasis immune factors TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 was related to the diagnosis, poor prognosis, and immune infiltrating cells of LUAD patients, and is expected to provide a reference for the development of treatment strategies for LUAD patients.
Keywords: Risk model, lung adenocarcinoma (LUAD), poor prognosis, overall survival (OS), immune
Introduction
In recent years, cancer immunotherapy has been applied in the treatment of a variety of cancer patients, which has improved the prognosis of patients and has had a profound impact on the current treatment patterns (1-6). For example, Liu et al. reported that the overall response rate of patients was 77.8% (N=7/9), and the complete remission rate was 55.6% (N=5/9) with CD19-CAR T cell treatment of B-cell lymphoma (BCL). The CD19-CAR T cells were safe and effective in the treatment of patients with refractory and relapsed BCL (1). Buderath et al. reported that PD-1-positive ovarian cancer (OC) infiltrating immune cells were associated with long overall survival (OS). PD-1- and PD-L1-positive OC infiltrating immune cells were related to lymph node metastasis and tumor grade (2). These findings indicate that immunotherapy for cancer patients has shown great therapeutic value, and is also a hotspot in current research. However, immunotherapy is effective for few cancer patients. Therefore, the development of new immunotherapy targets would improve the prognosis of cancer patients, highlighting the urgent need for further research.
In addition, a variety of immune cells and abnormal expression of immune genes are involved in the process of disease progression (7-10). For instance, fingolimod therapy could reduce CXCR4 expression and B cell migration. B cells of patients treated with fingolimod could induce the decrease of T cell pro-inflammatory cytokines, and increase the expression of transforming growth factor beta (TGFβ) B and T cells (7). TGF-β1 stimulation led to the up-regulation of L1CAM expression in a colonic intestinal epithelial cell line, NCM460, which enhanced cell motility and resistance to apoptosis. The expression of L1CAM in inflammatory bowel disease (IBD) patients was elevated. The expression level of L1CAM increased with the duration of the disease and the presence of CD33+ macrophages. In co-cultured NCM460 cells, inhibiting the TGF-β1 signaling pathway could eliminate the expression of Slug and L1CAM, which resulted in decreased cell migration and apoptosis resistance (8). However, the characteristics of the tumor immune microenvironment and immune factors in lung adenocarcinoma (LUAD) progression have not been fully revealed. Song et al. reported on the expression profile of immune factors in the primary tumors and brain metastases of non-small cell lung cancer (11), then the role and regulation mechanism of immune microenvironment molecules were identified in the progression of LUAD via the data of Song et al. and LUAD gene expression profiles from the Cancer Genome Atlas (TCGA) database. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-935).
Methods
Differentially expressed immune genes
There were 9 LUAD and LUAD brain metastases tissues in the Gene Expression Omnibus (GEO) database GSE161116 dataset (Figure S1A). Among them, there were 10 EGFR mutant and 8 wild-type patients (Figure S1B,C). There were 11 female patients and 7 male patients (Figure S1D). The expression levels of immune genes involved in LUAD brain metastasis were analyzed in the GSE161116 dataset using the GEO2R and screening criteria: adjusted P < 0.05, and log (fold change) >1 or <−1. In addition, we downloaded the transcriptome data of the type HTSeq-FPKM on the TCGA website. Among them, there were 59 normal lung tissues and 535 LUAD tissues. The expression data of brain metastasis immune genes of the GSE161116 dataset were extracted in the TCGA database. The expression levels of these immune genes were verified via the R limma package in normal lung and LUAD tissues. Screening criteria: adjusted P<0.05, and log (fold change) >1 or <−1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Biological functions and signaling mechanisms
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis have often been used to explore the biological functions and signaling mechanisms of multiple genes. The Metascape website contains an online website with GO annotation and KEGG analysis. Metascape was used to analyze the biological functions and signaling mechanisms involved in the differentially expressed immune genes of brain metastasis in LUAD tissues. Screening criteria: race type was human, and P<0.05.
The protein and protein interaction (PPI) network
The PPI network showed the possible interaction relationship between the 2 genes. The String database was used to construct the relationships between the differentially expressed immune proteins of brain metastasis in LUAD tissue, and the screening criterion was a combined score >0.4. Cytoscape software was used to display the interaction network between the differentially expressed immune proteins of brain metastasis, and the CytoHubba plug-in maximal clique centrality (MCC) score was used to screen the hub genes in the PPI network.
Risk model
The clinical data of 522 LUAD patients were obtained from the TCGA database (available online: https://cdn.amegroups.cn/static/public/jtd-21-935-01.pdf), and the differentially expressed immune gene expression levels of brain metastasis were sorted and merged with the clinical prognostic data of LUAD patients. Univariate Cox regression analysis was used to evaluate the prognostic genes of LUAD patients, with a screening criterion of P<0.05. On this basis, multivariate Cox regression analysis and Akaike information criterion (AIC) screened the independent genes affecting the prognosis of LUAD, and a risk model was constructed.
Identification of independent prognostic genes
The GEO GSE161116 dataset of 9 LUAD and 9 LUAD brain metastasis samples and GPL19965 annotation information were downloaded, and the expression levels of the prognostic genes were analyzed in LUAD and LUAD brain metastasis tissues by gender and EGFR mutation groups. The expression levels of the prognostic genes were explored in normal lung and LUAD tissues from the TCGA database. The diagnostic value of prognostic genes was identified in LUAD and LUAD brain metastasis via receiver operating characteristic (ROC) analysis. The value of prognostic genes in the prognosis of LUAD patients was verified in the PrognoScan database.
The clinical value of the risk model in LUAD
Kaplan-Meier (K-M) survival analysis and heat map visualization showed the relationship between high- and low-risk groups and risk genes, as well as the poor prognosis and clinicopathological characteristics of LUAD patients. In addition, univariate and multivariate Cox regression analyses were performed to evaluate the impact of high- and low-risk scores on the prognosis of LUAD patients.
Gene set enrichment analysis (GSEA)
GSEA was a method of analyzing gene expression information. In our study, we divided the gene expression data of 535 LUAD patients from the TCGA database into high- and low-risk groups based on the median expression values of risk scores, and the biological processes (BP) and signaling mechanisms involved in the high-risk groups were explored using GSEA. The enrichment was performed with 1,000 cycles, and nominal (NOM) P<0.05 was the screening criterion.
The value of the risk model in the LUAD microenvironment
The scores of immune cells and the microenvironment were calculated in the tissues of 535 LUAD patients with the TCGA database via CIBERSORT. The scores of the risk model were sorted and merged with the immune cells and immune cell marker data of LUAD patients. Pearson correlation analysis was used to analyze the relationship between the risk model scores and the immune cells and immune cell markers of LUAD patients. The screening criterion was P<0.05.
Statistical analysis
The differentially expressed genes of brain metastases were analyzed in the GSE161116 dataset via the GEO2R analysis, and the gene expression level was identified in the LUAD tissue of the TCGA database via the use the R limma package. Univariate and multivariate Cox regression analysis screened the factors affecting the prognosis of LUAD, and constructed the risk model. ROC analysis was used to determine the diagnostic value of prognostic genes in LUAD and LUAD brain metastases. The relationship between risk model scores and LUAD immune cells and immune cell markers were explored via the Pearson correlation analysis. P<0.05 was regarded as a sign of statistical significance.
Results
Differentially expressed immune genes in LUAD brain metastasis
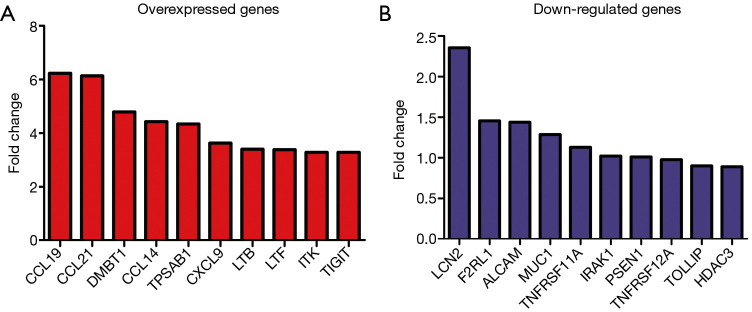
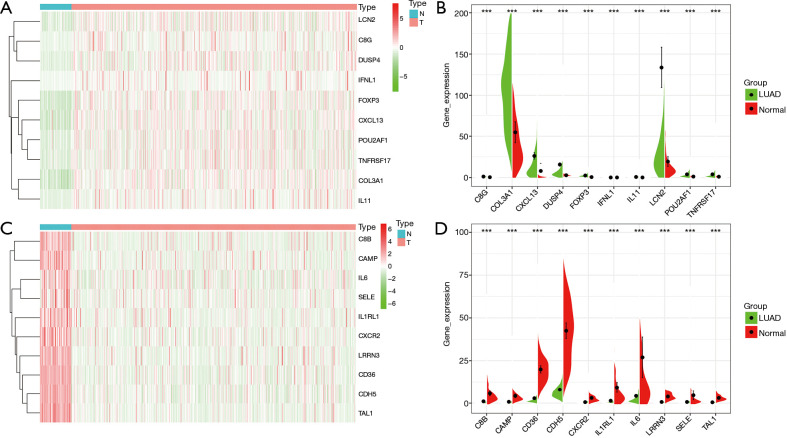
Compared with LUAD tissues, there were 298 differentially expressed genes (DEGs) in LUAD brain metastasis tissues (Figure 1 and available online: https://cdn.amegroups.cn/static/public/jtd-21-935-02.pdf). Among them, there were 270 highly expressed genes and 7 lowly expressed genes in the screening criteria with log (fold change) >1 or <−1 (available online: https://cdn.amegroups.cn/static/public/jtd-21-935-02.pdf). Compared with normal lung tissues, 97 immune genes of LUAD brain metastasis were abnormally expressed in LUAD tissues (Figure 2 and Table 1). Among them, there were 35 highly-expressed genes and 62 lowly-expressed genes.
Figure 1.
Top 10 DEGs in LUAD brain metastasis tissues. (A) Overexpressed immune genes; (B) down-regulated immune genes. DEGs, differentially expressed genes; LUAD, lung adenocarcinoma.
Figure 2.
Top 10 DEGs of brain metastasis in LUAD tissues. (A,B) Overexpressed immune genes; (C,D) lowly expressed immune genes. DEGs, differentially expressed genes; LUAD, lung adenocarcinoma; ***, P<0.001.
Table 1. The 97 immune genes of brain metastasis were abnormally expressed in LUAD tissues.
| Gene | logFC | Gene | logFC |
|---|---|---|---|
| IL5RA | −1.935544008 | LTA | 1.029192307 |
| CXCL12 | −1.11625871 | C8B | −2.503444527 |
| SERPING1 | −1.254772313 | THY1 | 1.469872699 |
| ENG | −1.481935175 | PDCD1 | 1.124336529 |
| CXCR2 | −2.430598894 | TNFSF12 | −1.069045544 |
| S100A12 | −1.859356435 | CTLA4 | 1.18824909 |
| SLAMF7 | 1.287415758 | IL12RB2 | 1.632032293 |
| POU2AF1 | 1.720086457 | CD36 | −2.768036549 |
| HCK | −1.165212969 | IRAK1 | 1.35421843 |
| TIGIT | 1.339436197 | SPN | −2.114601644 |
| CD244 | −1.190002432 | CCL23 | −2.375043085 |
| SH2D1B | −1.673271291 | TNFSF4 | 1.389908131 |
| EGR1 | −1.730851751 | SELE | −2.76186385 |
| CCL19 | 1.249183307 | CXCL9 | 1.4624964 |
| DUSP4 | 2.514985106 | MME | −2.346562181 |
| CLEC6A | −1.501689577 | IL18R1 | −1.310835154 |
| TNFRSF13C | 1.106785317 | ETS1 | −1.009611373 |
| TNFRSF11A | 1.340590987 | IRF4 | 1.110619628 |
| PRF1 | −1.138331152 | LILRA5 | −1.884862824 |
| TPSAB1 | −1.589114687 | TNFRSF9 | 1.54614469 |
| MEFV | −1.50059253 | IL1RL1 | −2.700417127 |
| FCER1G | −1.085643516 | EGR2 | −1.708277642 |
| A2M | −1.703151458 | PLA2G1B | −2.048853019 |
| IFNL1 | 1.901621178 | C8G | 1.813201339 |
| LILRB3 | −1.209333681 | ||
| FCGR3A | −1.116555199 | ITGAL | −1.045741499 |
| CXCL13 | 1.707906442 | CTSG | −1.704944547 |
| KLRF1 | −1.416602094 | IL6 | −2.664089273 |
| LCN2 | 2.78939134 | IFNG | 1.399852741 |
| TNFRSF17 | 1.99794901 | LAG3 | 1.413191105 |
| JAM3 | −1.047962968 | BST1 | −1.011723917 |
| TLR4 | −1.37511463 | CAMP | −2.468673216 |
| CD79A | 1.622713923 | THBD | −2.304370096 |
| PECAM1 | −2.164796954 | CD19 | 1.413990294 |
| CCL18 | −1.039124738 | TLR10 | 1.369103797 |
| CDH5 | −2.406255545 | CYBB | −1.155101404 |
| JAK3 | 1.263929458 | MS4A1 | 1.112359924 |
| CD27 | 1.441085273 | IL7R | −1.85923146 |
| LTF | 1.438494755 | BTK | −1.21746455 |
| CD34 | −1.528396601 | CXCL2 | −2.240927179 |
| MS4A2 | −1.69248968 | CFD | −2.212692861 |
| CCL14 | −2.268425282 | F2RL1 | 1.447418839 |
| RRAD | −1.04531552 | LRRN3 | −2.488365743 |
| CEACAM8 | −1.952591862 | C7 | −1.60908323 |
| TAL1 | −2.625803635 | CFP | −1.910704296 |
| CMA1 | −1.586930888 | FOS | −1.547672977 |
| FOXP3 | 1.982929634 | C6 | −1.906589961 |
| IL11 | 2.914495535 | COL3A1 | 2.685880542 |
| CD83 | −1.518456051 | IL34 | −1.129101453 |
LUAD, lung adenocarcinoma.
The biological functions, signaling mechanisms, and the PPI network involved in the differentially expressed immune genes of LUAD brain metastasis
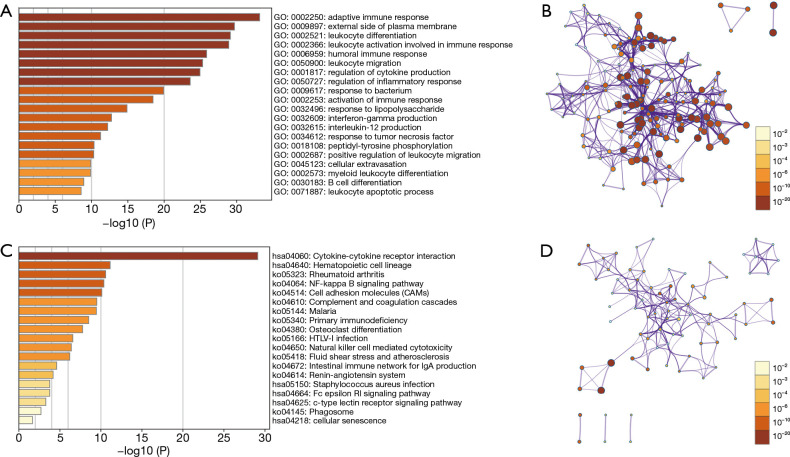
GO annotation showed that 97 differentially expressed immune genes of brain metastasis were involved in the adaptive immune response, B cell differentiation, myeloid leukocyte differentiation, cellular extravasation, positive regulation of leukocyte migration, interleukin-12 production, activation of immune response, regulation of inflammatory response, regulation of cytokine production, leukocyte migration, leukocyte activation involved in immune response, and leukocyte differentiation, among others (Figure 3A,B). KEGG pathway analysis found that these genes might be involved in the regulation of cellular senescence, c-type lectin receptor signaling pathway, renin-angiotensin system, natural killer cell mediated cytotoxicity, primary immunodeficiency, complement and coagulation cascades, cell adhesion molecules (CAMs), NF-kappa B (NF-kB) signaling pathway, and other mechanisms (Figure 3C,D). Figure S2 shows the relationships between the 97 differentially expressed immune proteins of brain metastasis. The hub genes in the PPI network were FOXP3, CTLA4, IL6, CD19, IFNG, CD27, PRF1, IL7R, TNFRSF9, and TNFSF4 (Figure S2B and Table 2).
Figure 3.
The biological functions and signaling mechanisms of 97 DEGs of brain metastasis via GO and KEGG analysis. (A,B) GO; (C,D) KEGG. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 2. Hub genes of the PPI network.
| Name | Description | Score |
|---|---|---|
| FOXP3 | Forkhead box P3 | 6.73E+07 |
| CTLA4 | Cytotoxic T-lymphocyte associated protein 4 | 6.72E+07 |
| IL6 | Interleukin 6 | 6.68E+07 |
| CD19 | CD19 molecule | 6.64E+07 |
| IFNG | Interferon gamma | 6.56E+07 |
| CD27 | CD27 molecule | 6.53E+07 |
| PRF1 | Perforin 1 | 5.97E+07 |
| IL7R | Interleukin 7 receptor | 5.34E+07 |
| TNFRSF9 | TNF receptor superfamily member 9 | 5.15E+07 |
| TNFSF4 | TNF superfamily member 4 | 5.12E+07 |
PPI, protein and protein interaction.
Construction of the risk model
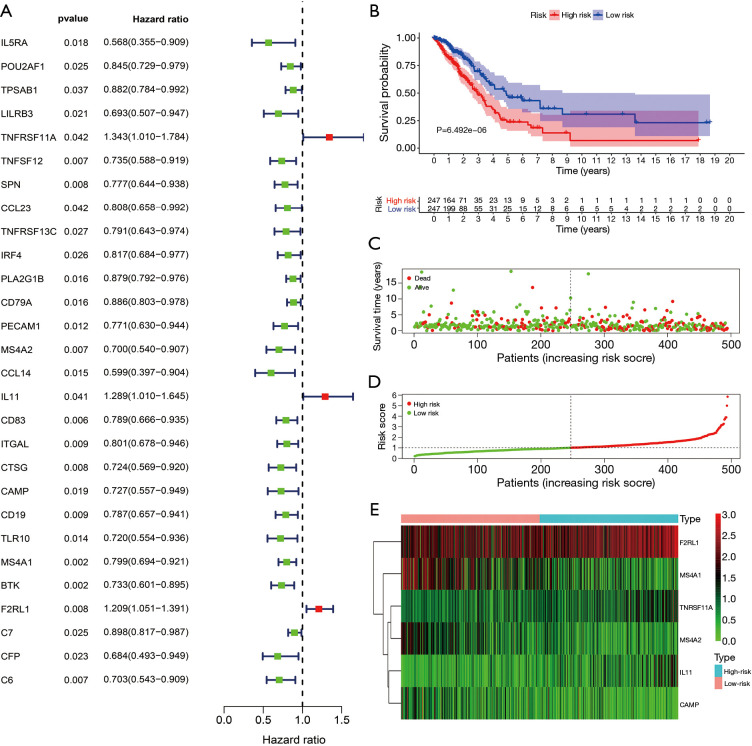
The univariate Cox regression analysis revealed that MS4A1, BTK, CD83, TNFSF12, MS4A2, C6, F2RL1, CTSG, SPN, CD19, ITGAL, PECAM1, TLR10, CCL14, CD79A, C7, PLA2G1B, IL5RA, CAMP, LILRB3, CFP, POU2AF1, IRF4, TNFRSF13C, TPSAB1, IL11, CCL23, and TNFRSF11A expression levels were the influencing factors for the poor prognosis of LUAD patients (Figure 4A and Table 3). The multivariate Cox regression analysis and AIC screening showed that the expression levels of TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 were independent factors affecting the prognosis of LUAD patients (Table 4), and the brain metastasis immune gene risk model was constructed (available online: https://cdn.amegroups.cn/static/public/jtd-21-935-03.pdf).
Figure 4.
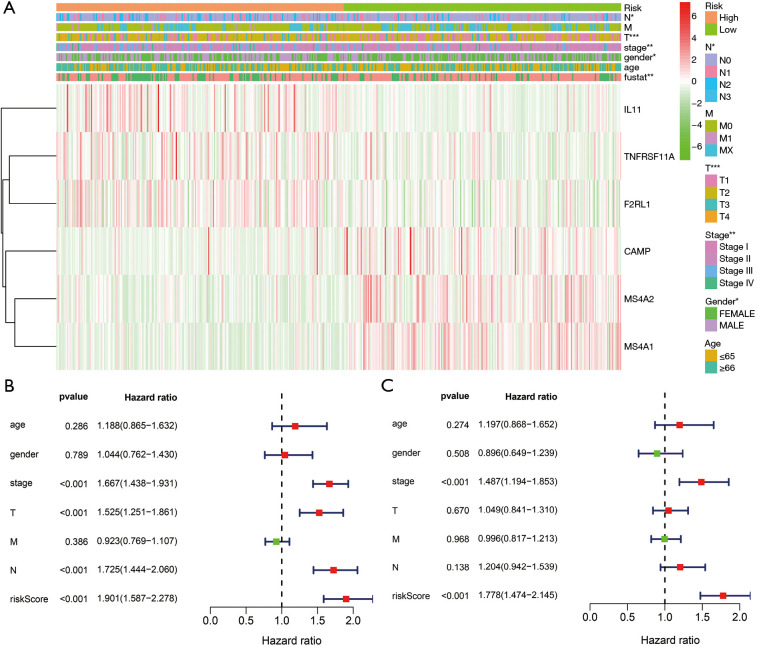
Construction and application of the risk model. (A) Univariate Cox regression analysis showed the influencing factors of the poor prognosis of LUAD patients; (B) K-M survival analysis showed that LUAD patients in the high-risk group had shorter OS; (C,D) survival times and risk scores of LUAD patients; (E) immune genes were related to the high- and low-risk groups. LUAD, lung adenocarcinoma; OS, overall survival; K-M, Kaplan-Meier.
Table 3. Univariate Cox regression analysis showed the influencing genes of the poor prognosis of LUAD patients.
| Gene | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|
| IL5RA | 0.567969541 | 0.354788527 | 0.909244169 | 0.018460133 |
| POU2AF1 | 0.845127424 | 0.729284998 | 0.979370705 | 0.02528008 |
| TPSAB1 | 0.882145437 | 0.784237159 | 0.992277097 | 0.036696354 |
| LILRB3 | 0.693103481 | 0.507451451 | 0.94667664 | 0.021197708 |
| TNFRSF11A | 1.342513034 | 1.010304964 | 1.783957628 | 0.042291011 |
| TNFSF12 | 0.734870574 | 0.587851237 | 0.918658882 | 0.006832572 |
| SPN | 0.776883091 | 0.643736666 | 0.937568681 | 0.008487492 |
| CCL23 | 0.808140618 | 0.658461621 | 0.991844076 | 0.04151735 |
| TNFRSF13C | 0.79122038 | 0.642776791 | 0.973945697 | 0.02717529 |
| IRF4 | 0.817449694 | 0.684234106 | 0.976601424 | 0.026362416 |
| PLA2G1B | 0.879355908 | 0.791992086 | 0.976356743 | 0.016033967 |
| CD79A | 0.886163139 | 0.803165593 | 0.977737487 | 0.016010351 |
| PECAM1 | 0.771046003 | 0.630105179 | 0.943512222 | 0.011585317 |
| MS4A2 | 0.700052865 | 0.540280972 | 0.907072503 | 0.006978937 |
| CCL14 | 0.598738403 | 0.396642108 | 0.903806399 | 0.014632385 |
| IL11 | 1.289066215 | 1.010263193 | 1.644810697 | 0.041143312 |
| CD83 | 0.789017057 | 0.66590724 | 0.93488684 | 0.006183644 |
| ITGAL | 0.800765117 | 0.677978583 | 0.945789128 | 0.008890182 |
| CTSG | 0.72383541 | 0.569448846 | 0.920078607 | 0.008278353 |
| CAMP | 0.72714556 | 0.55717819 | 0.948961526 | 0.018995652 |
| CD19 | 0.786520514 | 0.657125374 | 0.941394964 | 0.008831838 |
| TLR10 | 0.719753444 | 0.553719116 | 0.935573661 | 0.013984072 |
| MS4A1 | 0.799304568 | 0.693873229 | 0.920755788 | 0.001909762 |
| BTK | 0.733470106 | 0.600763588 | 0.895491015 | 0.002334972 |
| F2RL1 | 1.209148957 | 1.051086802 | 1.390980459 | 0.007883053 |
| C7 | 0.89802593 | 0.817405862 | 0.98659749 | 0.025018563 |
| CFP | 0.683871416 | 0.492614918 | 0.949382765 | 0.023188729 |
| C6 | 0.702597335 | 0.543092969 | 0.908947535 | 0.007218133 |
LUAD, lung adenocarcinoma.
Table 4. The multivariate Cox regression analysis and AIC screening showed that the immune genes were independent factors affecting the prognosis of LUAD patients.
| Gene | coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| TNFRSF11A | 0.35878476 | 1.431588634 | 1.081923122 | 1.894262147 | 0.012037371 |
| MS4A2 | −0.333382786 | 0.716495877 | 0.542458352 | 0.946370058 | 0.018863415 |
| IL11 | 0.284973997 | 1.329727451 | 1.038356185 | 1.702859886 | 0.023931533 |
| CAMP | −0.242800806 | 0.784427751 | 0.613385392 | 1.003165229 | 0.053017128 |
| MS4A1 | −0.163327726 | 0.849312801 | 0.730494719 | 0.98745715 | 0.033660229 |
| F2RL1 | 0.177638067 | 1.194392953 | 1.044013084 | 1.366433572 | 0.009672824 |
AIC, Akaike information criterion; LUAD, lung adenocarcinoma.
The clinical role of the risk model
K-M survival analysis showed that LUAD patients in the high-risk group had shorter OS (Figure 4B). Figure 4C,D,E shows the relationships between high- and low-risk groups and risk genes and the prognosis of LUAD patients. In addition, the risk score was significantly related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients (Figure 5A). Univariate and multivariate Cox regression analyses showed that the risk model score was an independent risk factor that affected the poor prognosis of LUAD patients (Figure 5B,C).
Figure 5.
The clinical value of the risk model in LUAD. (A) Clinicopathological characteristics; (B,C) prognostic factors. *, P<0.05; **, P<0.01; ***, P<0.001. LUAD, lung adenocarcinoma.
The role of independent prognostic gene expression levels in the progression of LUAD
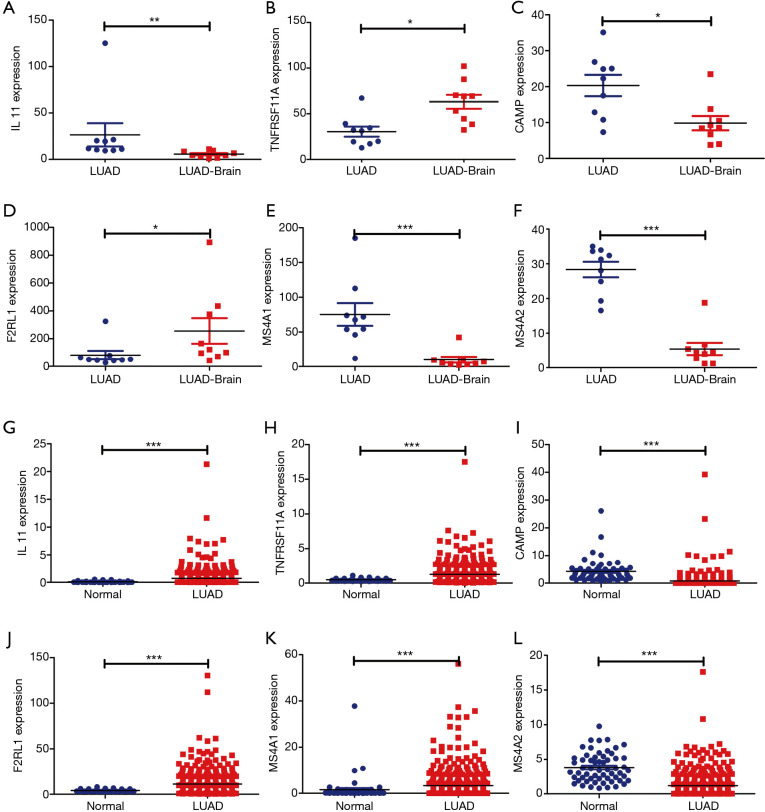
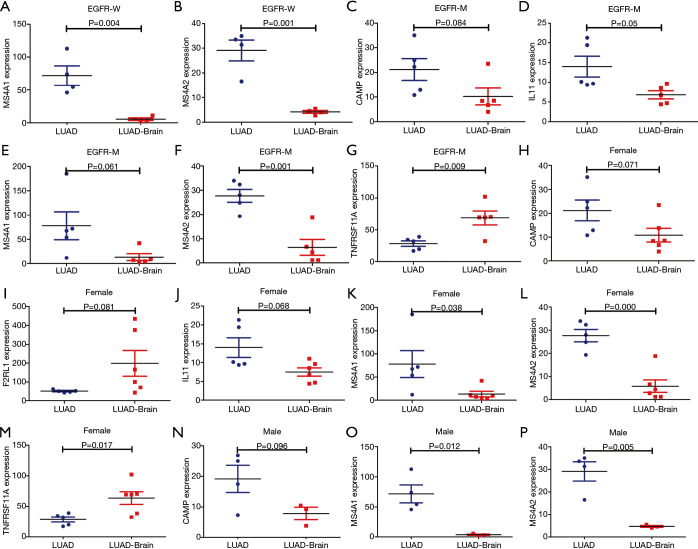
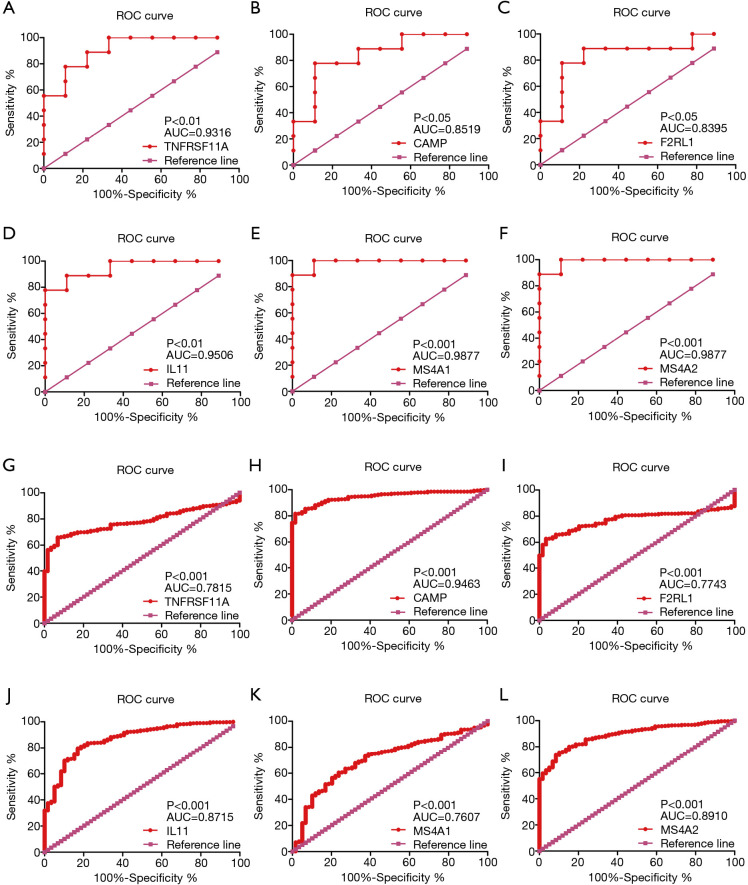
In the GSE161116 dataset, the expression levels of TNFRSF11A and F2RL1 in LUAD brain metastasis tissues were significantly higher than those of LUAD tissues, while the expression levels of IL11, CAMP, MS4A2, and MS4A1 in LUAD brain metastasis tissues were significantly lower than those of LUAD tissues (Figure 6A,B,C,D,E,F). In the TCGA database, compared with normal lung tissues, the expression levels of IL11, TNFRSF11A, F2RL1, and MS4A1 in LUAD tissues were significantly increased, and the expression levels of CAMP and MS4A2 in LUAD tissues were significantly decreased (Figure 6G,H,I,J,K,L). Subtype analysis showed that the expression levels of MS4A1 were significantly decreased, and the expression level of MS4A2 was significantly increased in EGFR wild-type LUAD brain metastasis tissues. The expression levels of IL11 and MS4A2 were significantly decreased, and the expression level of TNFRSF11A was significantly increased in EGFR-mutant LUAD brain metastasis tissues. The expression levels of MS4A1 and MS4A2 were decreased significantly, and the expression level of TNFRSF11A increased significantly in female LUAD brain metastasis tissues. The expression levels of MS4A1 and MS4A2 in male LUAD brain metastasis tissues decreased significantly (Figure 7). ROC analysis of the GSE161116 dataset data showed that the areas under the curves (AUCs) of TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 for the diagnosis of LUAD brain metastasis were 0.9316, 0.8519, 0.8395, 0.9506, 0.9877, and 0.9877, respectively (Figure 8A,B,C,D,E,F). The AUCs of TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 for LUAD diagnosis were 0.7815, 0.9463, 0.7743, 0.8715, 0.7607, and 0.8910, respectively (Figure 8G,H,I,J,K,L). This indicated that TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 had diagnostic significance in the diagnosis of LUAD brain metastasis and LUAD. In addition, K-M survival analysis of the GSE31210 and Jacob-00182-UM datasets showed that the expression levels of TNFRSF11A and MS4A2 were significantly correlated with the OS and relapse free survival (RFS) in LUAD patients. K-M survival analysis of the Jacob-00182-UM dataset showed that the MS4A1 expression level was significantly correlated with the OS in LUAD patients. K-M survival analysis of the Jacob-00182-CANDF and GSE31210 datasets showed that the IL11 expression level was significantly correlated with the OS in LUAD patients. K-M survival analysis of the GSE13213 and MICHIGAN-LC datasets showed that the expression levels of F2RL1 and CAMP were significantly correlated with the OS in LUAD patients (Table 5).
Figure 6.
Independent prognostic immune genes were abnormally expressed in LUAD brain metastases and LUAD tissues. (A) IL11; (B) TNFRSF11A; (C) CAMP; (D) F2RL1; (E) MS4A1; (F) MS4A2; (G) IL11; (H) TNFRSF11A; (I) CAMP; (J) F2RL1; (K) MS4A1; (L) MS4A2. *, P<0.05; **, P<0.01; ***, P<0.001. LUAD, lung adenocarcinoma.
Figure 7.
Subtype analysis showed that the independent prognostic immune genes were abnormally expressed in LUAD and LUAD brain metastases. LUAD, lung adenocarcinoma.
Figure 8.
The independent prognostic immune genes had diagnostic value for LUAD and LUAD brain metastasis via ROC analysis. (A) TNFRSF11A; (B) CAMP; (C) F2RL1; (D) IL11; (E) MS4A1; (F) MS4A2; (G) TNFRSF11A; (H) CAMP; (I) F2RL1; (J) IL11; (K) MS4A1; (L) MS4A2. LUAD, lung adenocarcinoma; ROC, receiver operating characteristic.
Table 5. K-M survival analysis showed that the expression levels of the immune genes were significantly correlated with the OS and PFS in LUAD patients in the PrognoScan database.
| Gene and dataset | Prognosis | Probe ID | N | P | COX P | HR (95% CI) |
|---|---|---|---|---|---|---|
| TNFRSF11A-GSE31210 | OS | 207037_at | 204 | 0.00634647 | 0.00898712 | 2.38 (1.24–4.55) |
| TNFRSF11A-GSE31210 | OS | 238846_at | 204 | 0.168781 | 0.0472737 | 1.65 (1.01–2.72) |
| TNFRSF11A-GSE31210 | RFS | 238846_at | 204 | 0.0270956 | 0.0154956 | 1.60 (1.09–2.34) |
| TNFRSF11A-GSE31210 | RFS | 207037_at | 204 | 0.0180054 | 0.0221418 | 1.84 (1.09–3.10) |
| MS4A2-GSE31210 | OS | 207496_at | 204 | 0.00987325 | 0.00296026 | 0.54 (0.36–0.81) |
| MS4A2-GSE31210 | OS | 207497_s_at | 204 | 0.0173826 | 0.0307801 | 0.58 (0.35–0.95) |
| MS4A2-Jacob-00182-UM | OS | 207496_at | 178 | 0.086174 | 0.0336075 | 0.79 (0.63–0.98) |
| MS4A2-GSE31210 | RFS | 207496_at | 204 | 0.00155053 | 0.0032846 | 0.64 (0.48–0.86) |
| MS4A1-Jacob-00182-UM | OS | 210356_x_at | 178 | 0.259787 | 0.0342218 | 0.69 (0.49–0.97) |
| MS4A1-Jacob-00182-UM | OS | 217418_x_at | 178 | 0.168491 | 0.0421624 | 0.76 (0.59–0.99) |
| IL11-Jacob-00182-CANDF | OS | 206924_at | 82 | – | 0.0146906 | 1.96 (1.14–3.36) |
| IL11-GSE31210 | OS | 206924_at | 204 | 0.046182 | 0.248705 | 1.28 (0.84–1.95) |
| F2RL1-GSE13213 | OS | A_23_P58835 | 117 | 0.0159142 | 0.0256215 | 1.29 (1.03–1.61) |
| CAMP-MICHIGAN-LC | OS | Z38026_at | 86 | 0.00882622 | 0.207221 | 0.61 (0.28–1.32) |
K-M, Kaplan-Meier; LUAD, lung adenocarcinoma; OS, overall survival; PFS, progression-free survival.
The biological functions and signaling mechanisms involved in the risk model
In the risk model, GSEA analysis found that the risk model was involved in the biological functions of establishment of mitotic spindle localization, establishment of mitotic spindle orientation, regulation of translational initiation in response to stress, cytokinetic process, and transcription by RNA polymerase I, among others (Figure S3 and available online: https://cdn.amegroups.cn/static/public/jtd-21-935-04.pdf). The risk model was involved in the pentose phosphate pathway, glycolysis gluconeogenesis, Pathogenic Escherichia coli infection, pyrimidine metabolism, cell cycle, citrate cycle TCA cycle, bladder cancer, and fructose and mannose metabolism (Figure S4).
The risk model was related to the LUAD immune microenvironment
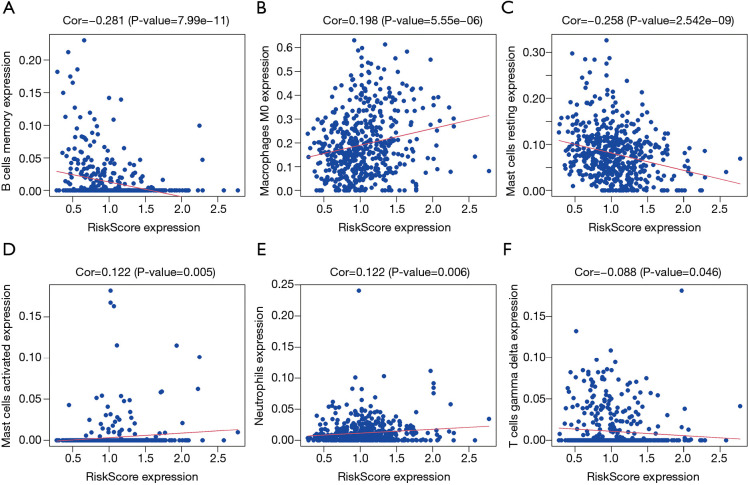
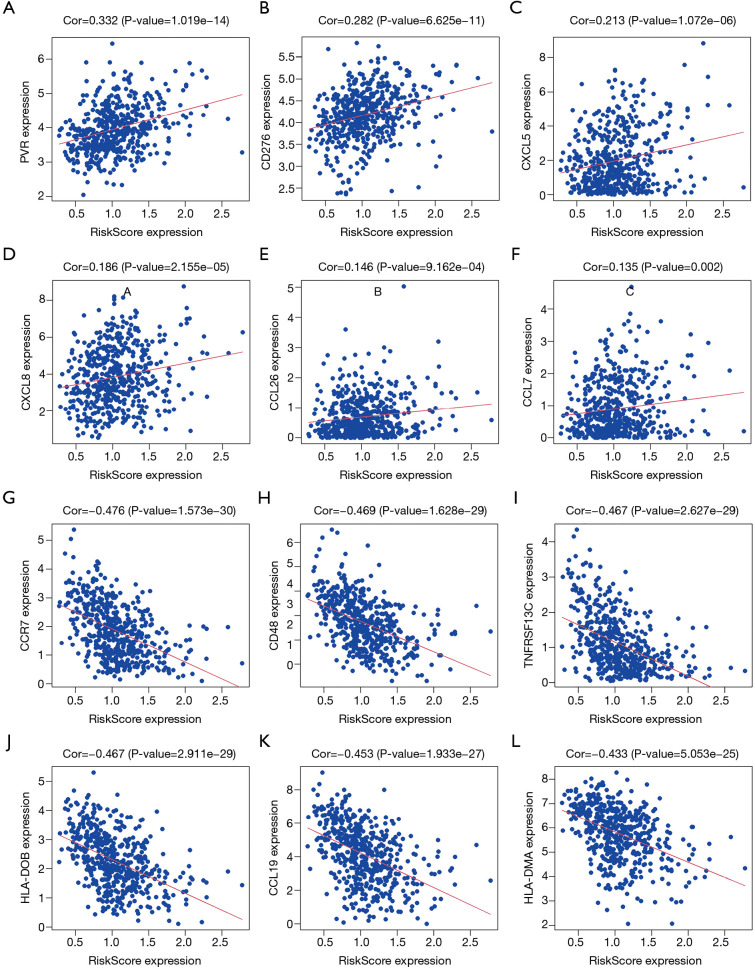
Correlation analysis revealed that the risk score was significantly correlated with LUAD B cells memory, mast cells resting, macrophages M0, mast cells activated, neutrophils, eosinophils, and T cells gamma delta levels (Figure 9). Further analysis found that the risk score was significantly correlated with the levels of LUAD immune cell markers CCR7, CD48, TNFRSF13C, HLA-DOB, CCL19, HLA-DMA, HLA-DPB1, CCR4, CD27, HLA-DOA, BTLA, LTA, HLA-DMB, HLA-DRA, HLA-DQA1, HLA-DPA1, and HLA-DRB1, among others (Figure 10 and Table 6).
Figure 9.
The risk score was significantly correlated with the LUAD immune microenvironment. (A) B cells memory; (B) macrophages M0; (C) mast cells resting; (D) mast cells activated; (E) neutrophils; (F) T cells gamma delta. LUAD, lung adenocarcinoma.
Figure 10.
The risk score was significantly correlated with LUAD immune cell markers. (A) PVR; (B) CD276; (C) CXCL5; (D) CXCL8; (E) CCL26; (F) CCL7; (G) CCR7; (H) CD48; (I) TNFRSF13C; (J) HLA-DOB; (K) CCL19; (L) HLA-DMA. LUAD, lung adenocarcinoma.
Table 6. The risk score was significantly correlated with the levels of LUAD immune cell markers.
| Gene | Cor |
|---|---|
| XCL1 | −0.162 |
| CSF1R | −0.254 |
| CCR5 | −0.317 |
| CXCL12 | −0.284 |
| CCL3 | −0.13 |
| PVR | 0.332 |
| CCL7 | 0.135 |
| CXCR2 | −0.157 |
| IL6R | −0.231 |
| CCL17 | −0.257 |
| CXCR4 | −0.377 |
| TIGIT | −0.343 |
| CD244 | −0.309 |
| NT5E | 0.121 |
| CD48 | −0.469 |
| TNFSF14 | −0.226 |
| HAVCR2 | −0.248 |
| CCL2 | −0.114 |
| CCL19 | −0.453 |
| CD80 | −0.22 |
| CD276 | 0.282 |
| CXCR3 | −0.312 |
| CD86 | −0.254 |
| TNFRSF25 | −0.26 |
| HLA-DRB1 | −0.383 |
| HLA-DQA1 | −0.393 |
| HLA-DQB1 | −0.323 |
| HLA-DMB | −0.414 |
| HLA-DPB1 | −0.431 |
| CCL22 | −0.206 |
| PDCD1LG2 | −0.174 |
| TNFRSF18 | −0.133 |
| TNFRSF4 | −0.225 |
| TNFRSF14 | −0.29 |
| TNFSF13B | −0.309 |
| HLA-DPA1 | −0.391 |
| TNFRSF13C | −0.467 |
| TNFRSF9 | −0.126 |
| CTLA4 | −0.337 |
| PDCD1 | −0.28 |
| CXCR6 | −0.329 |
| CCL21 | −0.134 |
| CCL23 | −0.301 |
| CXCL9 | −0.205 |
| CCR2 | −0.372 |
| HLA-B | −0.225 |
| CCR1 | −0.22 |
| CCL4 | −0.199 |
| CXCL10 | −0.111 |
| CCR4 | −0.422 |
| LTA | −0.417 |
| CCR7 | −0.476 |
| ENTPD1 | −0.249 |
| BTLA | −0.418 |
| CXCL13 | −0.368 |
| XCL2 | −0.266 |
| HLA-E | −0.307 |
| TNFSF15 | −0.146 |
| TNFRSF17 | −0.368 |
| TNFSF13 | −0.171 |
| CX3CR1 | −0.253 |
| TMEM173 | −0.257 |
| IL10 | −0.295 |
| HLA-C | −0.183 |
| CXCL8 | 0.186 |
| HLA-DMA | −0.433 |
| CCL18 | −0.263 |
| TGFBR1 | 0.111 |
| CD27 | −0.421 |
| CCL26 | 0.146 |
| TNFSF9 | 0.101 |
| HLA-DQA2 | −0.222 |
| CD40 | −0.221 |
| HLA-DOB | −0.467 |
| CCR8 | −0.222 |
| HLA-DRA | −0.398 |
| CCL13 | −0.133 |
| IDO1 | −0.157 |
| ICOS | −0.341 |
| HLA-F | −0.235 |
| CD96 | −0.371 |
| LAG3 | −0.235 |
| TNFRSF8 | −0.107 |
| CXCL14 | −0.18 |
| B2M | −0.182 |
| LGALS9 | −0.182 |
| IL2RA | −0.133 |
| CXCL2 | −0.144 |
| CXCL17 | −0.32 |
| CD28 | −0.357 |
| HLA-DOA | −0.42 |
| TAPBP | −0.11 |
| CXCL5 | 0.213 |
| CX3CL1 | −0.163 |
| HLA-A | −0.159 |
LUAD, lung adenocarcinoma.
Discussion
Lung cancer has a high recurrence rate and distant metastasis rate, threatening human health. LUAD is one of the common subtypes of lung cancer. In the process of LUAD metastasis, genes, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs), among others, play roles as carcinogenic or tumor suppressor factors (12-16). For example, the up-regulation of polypeptide N-acetylgalactosaminyltransferase 6 (GALNT6) expression was associated with LUAD lymph node metastasis and poor prognosis. In LUAD cells, overexpression of GALNT6 could promote epithelial-mesenchymal transition (EMT), migration, and invasion, while the results of GALNT6 silencing were significantly the opposite. Silencing GALNT6 could prolong the survival time of xenograft mice (12). Circ-CAMK2A was overexpressed in LUAD, and the high expression level of CAMK2A was closely related to the lymph node metastasis, distant metastasis, advanced clinical stage, and poor prognosis of LUAD patients. Silencing of CAMK2A inhibited the migration and invasion of LUAD cells, while overexpression of CAMK2A had the opposite effect. Furthermore, overexpression of CAMK2A enhanced LUAD metastasis in vivo. CAMK2A could up-regulate the expression of fibronectin 1 by sponging miR-615-5p, thereby increasing the expression of MMP2 and MMP9 to promote LUAD metastasis (13). Mex3a was significantly up-regulated in LUAD tissues, and increased expression of Mex3a was related to the poor prognosis and metastasis of LUAD. Interference with the expression of Mex3a significantly inhibited the migration, invasion, and metastasis of LUAD cells. Inhibition of LAMA2 expression could partly reverse the decrease in Mex3a expression and cause cell migration and invasion. Mex3a and LAMA2 could affect the PI3K-AKT signaling pathway to participate in LUAD metastasis (14). In this study, the GSE161116 dataset was used to analyze the differentially expressed immune genes involved in the process of LUAD brain metastasis, and the TCGA database verified that 97 immune genes were differentially expressed in both LUAD tissue and LUAD brain metastasis tissue.
Immune genes of LUAD brain metastasis were involved in the adaptive immune response, B cell differentiation, myeloid leukocyte differentiation, cellular extravasation, positive regulation of leukocyte migration, interleukin-12 production, activation of immune response, regulation of inflammatory response, regulation of cytokine production, leukocyte migration, leukocyte activation involved in immune response, leukocyte differentiation, and other biological functions, and might be involved in the regulation of cellular senescence, c-type lectin receptor signaling pathway, natural killer cell mediated cytotoxicity, CAMs, primary immunodeficiency, and NF-κB signaling pathway, among others. Studies have reported that the adaptive immune response, the NF-κB signaling pathway, and the IL12 signaling mechanism, among others, play important roles in the progression of LUAD (17-20). For example, IL33 inhibited the growth of LUAD by regulating dendritic cells (DCs). Knockdown of CYLD expression in DC cells could inhibit NF-κB signaling transduction and inhibit the proliferation and function of CD4 T cells induced by DCs (17). There was also a negative correlation between WNT1 and T cell abundance. Abnormal expression of WNT1 could affect the growth of LUAD, which was related to the level of DCs and T cells. WNT1 could promote CC/CXC chemokine transcription silencing, T cell rejection, and cross tolerance in DCs. The WNT target gene was up-regulated in DCs, and the level of WNT1 target gene decreased when WNT1 was silenced, accompanied by enhanced T cell toxicity (20). These findings indicate that these genes play important roles in the occurrence and metastasis of LUAD.
The risk score model was a common method to predict the poor prognosis of cancer patients (21,22). A risk model was constructed through Cox regression analysis and AIC screening. K-M survival analysis showed that LUAD patients in the high-risk group had poorer OS. The risk score was significantly related to the gender, clinical stage, T stage, lymph node metastasis, and survival status of LUAD patients, and was an independent risk factor affecting the poor prognosis of LUAD patients. Among them, our risk model genes were TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1. ROC analysis showed that the AUCs of TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 in the diagnosis of LUAD brain metastasis and LUAD were all greater than 0.5, and were statistically significant. K-M survival analysis showed that TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 were related to the prognosis of LUAD patients. Currently, studies have found that miR-3150b-3p was down-regulated in cervical cancer cells. MiR-3150b-3p attenuated the proliferation, migration, and invasion of HeLa and SiHa cells. Overexpression of TNFRSF11A could eliminate the anti-tumor effect of miR-3150b-3p (23). Furthermore, IL11 was upregulated in endometrial cancer, and in AN3CA cells, IL11 could cause cell adhesion and migration through STAT3 (24). Protease-activated receptor 2 (PAR) is also known as F2RL1. F2RL1 activation could promote cancer cell migration. F2RL1 activation could also reduce the expression level of miR-205. MiR-205 inhibition promoted cancer cell migration, while overexpression of miR-205 prevented PAR-mediated cell migration (25). Recently, IL11 was shown to promote A549 cell growth and EMT, which might be related to the activation of the STAT3/HIF-1α/EMT signaling pathway (26). The same genes could play different roles in different cancers. For example, TNFRSF11A was overexpressed in colorectal cancer (CRC) tissues. TNFRSF11A overexpression was positively correlated with metastasis, high TNM stage, and poor prognosis of CRC patients. Overexpression of TNFRSF11A could promote CRC cell migration and invasion in vitro and in vivo (27). In glioma cells, overexpression of TNFRSF11A could lead to reduced lesion formation and increased apoptotic activity (28). Therefore, the roles of TNFRSF11A, MS4A2, CAMP, MS4A1, and F2RL1 in LUAD progression need further verification.
Research by Song et al. showed that the immune microenvironment and its molecular abnormalities were involved in NSCLC brain metastasis (11,29). A total of 535 LUAD patients in the TCGA were scored on immune cells and the microenvironment, and it was found that the risk model score was significantly correlated with LUAD B cells memory, mast cells resting, macrophages M0, mast cells activated, neutrophils, eosinophils, and T cells gamma delta levels. In addition, the risk model score was correlated with the levels of immune cell markers CCR7, CD48, TNFRSF13C, HLA-DOB, CCL19, HLA-DMA, HLA-DPB1, CCR4, CD27, HLA-DOA, BTLA, LTA, HLA-DMB, HLA-DRA, HLA-DQA1, HLA-DPA1, and HLA-DRB1. This further showed that our risk model based on the immune genes TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 was related to LUAD immune infiltrating cells, and played an important role in the development and immune interaction of LUAD.
In this study, the commonly databases of GEO and TCGA were used to screen and evaluate the roles of LUAD brain metastasis-related immune genes in the cancer progression, and the risk model was constructed to evaluate the prognosis of patients. Our research had the advantages of a wide variety of methods and a large sample size. However, our research had the certain limitations. The GEO dataset lacked prognostic data, and could not show the value of brain metastasis-related immune target molecules in the prognosis of patients with EGFR mutation. In addition, our research results needed to be confirmed by collecting LUAD tissue specimens. In general, the expression levels of TNFRSF11A, MS4A2, IL11, CAMP, MS4A1, and F2RL1 were independent factors affecting the poor prognosis of LUAD patients. The risk model was significantly related to the gender, clinical stage, T stage, lymph node metastasis, and survival time of LUAD patients. The risk genes TNFRSF11A, CAMP, F2RL1, IL11, MS4A1, and MS4A2 had diagnostic value in the diagnosis of LUAD brain metastasis and LUAD. The risk model participated in the regulation of translational initiation in response to stress, cytokinetic process, and cell cycle, among others. The risk model score was significantly correlated with the levels of LUAD immune cells. However, these research results need to be further confirmed by basic experiments. In general, our risk model is expected to provide a reference for the development of treatment strategies for LUAD patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The National Natural Science Foundation of China (No. 81960532), and The Science and Technology Project of Guizhou Province (Supported by Guizhou Science and Technology Combined [2021] General 073).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-935
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-935). The authors have no conflicts of interest to declare.
(English Language Editor: C. Betlazar-Maseh)
References
- 1.Liu X, Zhang Y, Li K, et al. A novel dominant-negative PD-1 armored anti-CD19 CAR T cell is safe and effective against refractory/relapsed B cell lymphoma. Transl Oncol 2021;14:101085. 10.1016/j.tranon.2021.101085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buderath P, Mairinger F, Mairinger E, et al. Prognostic significance of PD-1 and PD-L1 positive tumor-infiltrating immune cells in ovarian carcinoma. Int J Gynecol Cancer 2019;29:1389-95. 10.1136/ijgc-2019-000609 [DOI] [PubMed] [Google Scholar]
- 3.Xiao L, Li L, Chen G, et al. The lymphocyte-to-monocyte ratio could predict the efficacy of PD-1 inhibitors in patients with advanced cancer. Transl Cancer Res 2020;9:4111-20. 10.21037/tcr-20-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kono Y, Saito H, Miyauchi W, et al. Increased PD-1-positive macrophages in the tissue of gastric cancer are closely associated with poor prognosis in gastric cancer patients. BMC Cancer 2020;20:175. 10.1186/s12885-020-6629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander PG, McMillan DC, Park JH. A meta-analysis of CD274 (PD-L1) assessment and prognosis in colorectal cancer and its role in predicting response to anti-PD-1 therapy. Crit Rev Oncol Hematol 2021;157:103147. 10.1016/j.critrevonc.2020.103147 [DOI] [PubMed] [Google Scholar]
- 6.Amanam I, Mambetsariev I, Gupta R, et al. Role of immunotherapy and co-mutations on KRAS-mutant non-small cell lung cancer survival. J Thorac Dis 2020;12:5086-95. 10.21037/jtd.2020.04.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenfeld-Kan S, Staun-Ram E, Miller A. Fingolimod reduces CXCR4-mediated B cell migration and induces regulatory B cells-mediated anti-inflammatory immune repertoire. Mult Scler Relat Disord 2019;34:29-37. 10.1016/j.msard.2019.06.016 [DOI] [PubMed] [Google Scholar]
- 8.Schäfer H, Struck B, Feldmann EM, et al. TGF-β1-dependent L1CAM expression has an essential role in macrophage-induced apoptosis resistance and cell migration of human intestinal epithelial cells. Oncogene 2013;32:180-9. 10.1038/onc.2012.44 [DOI] [PubMed] [Google Scholar]
- 9.Shi L, Xi J, Xu X, et al. MiR-148a suppressed cell invasion and migration via targeting WNT10b and modulating β-catenin signaling in cisplatin-resistant colorectal cancer cells. Biomed Pharmacother 2019;109:902-9. 10.1016/j.biopha.2018.10.080 [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Ding H, He Y, et al. NLRC5 mediates cell proliferation, migration, and invasion by regulating the Wnt/β-catenin signalling pathway in clear cell renal cell carcinoma. Cancer Lett 2019;444:9-19. 10.1016/j.canlet.2018.11.024 [DOI] [PubMed] [Google Scholar]
- 11.Song SG, Kim S, Koh J, et al. Comparative analysis of the tumor immune-microenvironment of primary and brain metastases of non-small-cell lung cancer reveals organ-specific and EGFR mutation-dependent unique immune landscape. Cancer Immunol Immunother 2021;70:2035-48. 10.1007/s00262-020-02840-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song J, Liu W, Wang J, et al. GALNT6 promotes invasion and metastasis of human lung adenocarcinoma cells through O-glycosylating chaperone protein GRP78. Cell Death Dis 2020;11:352. 10.1038/s41419-020-2537-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Zhang G, Qiu H, et al. The novel circular RNA circ-CAMK2A enhances lung adenocarcinoma metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell Mol Biol Lett 2019;24:72. 10.1186/s11658-019-0198-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Liang J, Li H, Han J, et al. Mex3a interacts with LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT pathway. Cell Death Dis 2020;11:614. 10.1038/s41419-020-02858-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Yu R, Ling X, et al. COPB2 promotes metastasis and inhibits apoptosis of lung adenocarcinoma cells through functioning as a target of miR-216a-3p. Transl Cancer Res 2020;9:2648-59. 10.21037/tcr.2020.02.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Hu A, Liang Y, et al. Genome-wide analysis of long non-coding RNA expression profile in lung adenocarcinoma compared to spinal metastasis. Ann Transl Med 2020;8:1516. 10.21037/atm-20-7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Q, Lei Y, Huang Y, et al. CYLD expression in dendritic cells involved in the immunoregulation of pulmonary adenocarcinoma via NF-κB pathway. Artif Cells Nanomed Biotechnol 2020;48:137-42. 10.1080/21691401.2019.1699820 [DOI] [PubMed] [Google Scholar]
- 18.Airoldi I, Di Carlo E, Cocco C, et al. IL-12 can target human lung adenocarcinoma cells and normal bronchial epithelial cells surrounding tumor lesions. PLoS One 2009;4:e6119. 10.1371/journal.pone.0006119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt L, Eskiocak B, Kohn R, et al. Enhanced adaptive immune responses in lung adenocarcinoma through natural killer cell stimulation. Proc Natl Acad Sci U S A 2019;116:17460-9. 10.1073/pnas.1904253116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerdidani D, Chouvardas P, Arjo AR, et al. Wnt1 silences chemokine genes in dendritic cells and induces adaptive immune resistance in lung adenocarcinoma. Nat Commun 2019;10:1405. 10.1038/s41467-019-09370-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Deng J, Lai S, et al. A risk score model with five long non-coding RNAs for predicting prognosis in gastric cancer: an integrated analysis combining TCGA and GEO datasets. PeerJ 2021;9:e10556. 10.7717/peerj.10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, Zhong F, Yi X, et al. Application of an Autophagy-Related Gene Prognostic Risk Model Based on TCGA Database in Cervical Cancer. Front Genet 2021;11:616998. 10.3389/fgene.2020.616998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Wang L, Li X. MiR-3150b-3p inhibits the proliferation and invasion of cervical cancer cells by targeting TNFRSF11a. J Investig Med 2020;68:1166-70. 10.1136/jim-2020-001284 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Lay V, Yap J, Sonderegger S, et al. Interleukin 11 regulates endometrial cancer cell adhesion and migration via STAT3. Int J Oncol 2012;41:759-64. 10.3892/ijo.2012.1486 [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Yang L, Ma Y, et al. MicroRNA-205 Mediates Proteinase-Activated Receptor 2 (PAR2) -Promoted Cancer Cell Migration. Cancer Invest 2017;35:601-9. 10.1080/07357907.2017.1378671 [DOI] [PubMed] [Google Scholar]
- 26.Peng N, Lu M, Kang M, et al. Recombinant Human IL-11 Promotes Lung Adenocarcinoma A549 Cell Growth and EMT through Activating STAT3/HIF-1α/EMT Signaling Pathway. Anticancer Agents Med Chem 2020. [Epub ahead of print]. doi: . 10.2174/1871520621666201207091248 [DOI] [PubMed] [Google Scholar]
- 27.Liang Q, Wang Y, Lu Y, et al. RANK promotes colorectal cancer migration and invasion by activating the Ca2+-calcineurin/NFATC1-ACP5 axis. Cell Death Dis 2021;12:336. 10.1038/s41419-021-03642-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von dem Knesebeck A, Felsberg J, Waha A, et al. RANK (TNFRSF11A) is epigenetically inactivated and induces apoptosis in gliomas. Neoplasia 2012;14:526-34. 10.1596/neo.12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilariño N, Bruna J, Bosch-Barrera J, et al. Immunotherapy in NSCLC patients with brain metastases. Understanding brain tumor microenvironment and dissecting outcomes from immune checkpoint blockade in the clinic. Cancer Treat Rev 2020;89:102067. 10.1016/j.ctrv.2020.102067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as