Abstract
Although most studies describing coronavirus disease 2019 vaccine responses have focused on antibodies, there is increasing evidence that T cells play a critical role. Here the authors evaluated T-cell responses in seronegative donors before and after vaccination to define responses to the severe acute respiratory syndrome coronavirus 2 reference strain as well as to mutations in the variant strains Alpha/B.1.1.7 and Beta/B.1.351. The authors observed enhanced T-cell responses to reference and variant spike strains post-vaccination.
Key Words: COVID-19 variants, SARS-CoV-2, T-cell responses to coronavirus, vaccination
Introduction
The Alpha/B.1.1.7 (UK) [1] and Beta/B.1.351 (South African) [2] strains, with multiple mutations in the spike protein (the structural protein serving as the antigen for almost all coronavirus disease 2019 [COVID-19] vaccines), have been spreading globally. These two mutant strains are among the variants of concern spreading across the United States, with (as of April 2021) 5567 reported cases of Alpha/B.1.1.7 across 50 jurisdictions and 180 cases of Beta/B.1.351 across 26 jurisdictions [3]. The mutations within the spike protein are within the receptor binding domain, which influences virus attachment and entry into cells, with some studies suggesting increased binding affinity and, consequently, increased infectivity and transmission [4]. Recently, different studies have suggested that there is diminished neutralization of the two strains even after vaccination [5]. Sera from vaccinated individuals have neutralization titers several logs lower than those seen with the reference strain [6,7]. This raises appreciable concerns regarding the efficacy of current vaccine strategies to control the pandemic.
T-cell responses are a critical complementary immune reaction to antibody responses. Indeed, in individuals with combined variable immune deficiency, rates of COVID-19 hospitalization and mortality parallel those of the general population, suggesting that in these B-cell-deficient individuals, T-cell response plays a key immunologic role in viral control [8]. Previous studies have already determined the ability of T cells derived from convalescent individuals to recognize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [9], and the presence of virus-specific T cells correlates with protection against severe COVID-19 and with disease severity and recovery [10].
However, although antibody responses have been well characterized in individuals who have received most of the approved vaccines [6,7,[11], [12], [13]], there is currently a paucity of data available regarding the breadth and cross-reactivity of T-cell responses to mutant viral strains post-vaccination. A study by Tarke et al. [14] suggests that variants have a negligible impact on response post-vaccination; however, the antibody and T-cell status of these donors prior to vaccination is unknown, and it is unclear whether prior asymptomatic infections contributed to the post-vaccine response.
Hence, the authors sought to define vaccine-specific T-cell responses in documented seronegative donors who had no prior history of SARS-CoV-2 and evaluated their response to SARS-CoV-2 structural proteins pre- versus post-vaccination. This allowed interpretation of responses to be assigned solely to the vaccine and not to pre-existing immunity following COVID-19 infection.
Methods
Donors
Blood was obtained from seronegative donors with no history of SARS-CoV2 infection after obtaining informed consent approved by the institutional review board (Pro00004033) of the Children's National Hospital (Washington, DC, USA). The age, sex and ethnicity of the donors are shown in supplementary Table 1. All donors had a documented absence of COVID-19 infection by antibody testing (National Institutes of Health) immediately prior to the blood draw (Figure 1 A) and in the months before vaccination became available (see supplementary Figure 1). Peripheral blood mononuclear cells (PBMCs) obtained prior to and after vaccination with each dose of the Pfizer BNT162b2 messenger RNA (mRNA) vaccine (New York, NY, USA) were isolated by density gradient centrifugation using Lymphoprep (STEMCELL Technologies, Cambridge, MA, USA). HLA typing of the healthy donor PBMCs was performed by Kashi Clinical Laboratories (Portland, OR, USA) or The Sequencing Center (Fort Collins, CO, USA).
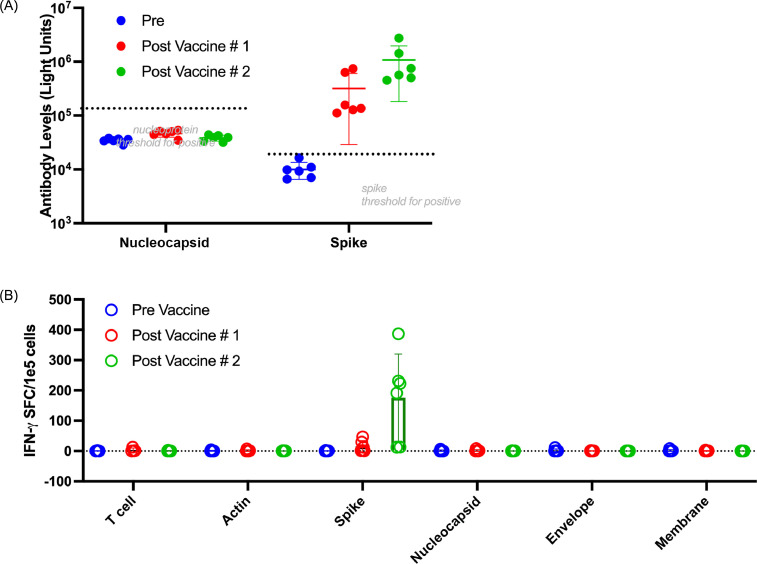
Fig. 1.
Antibodies and T cells from seronegative donors demonstrate enhanced reactivity to spike post-vaccination. (A) Antibody to nucleocapsid and spike measured in six donors immediately before vaccination (blue circles), 3 weeks after the first vaccine and before administration of the second vaccine (red circles) and 3–4 weeks after administration of the second vaccine (green circles). (B) T-cell responses as measured by IFN-γ ELISpot after 10- to 11-day expansion of PBMCs with COVID-19 non-structural antigens spike, membrane, nucleocapsid and envelope to zoom in on specific T cells measured immediately before vaccination (blue circles), 3 weeks after the first vaccine and before administration of the second vaccine (red circles) and 3–4 weeks after administration of the second vaccine (green circles). (Color version of figure is available online).
SARS-CoV-2 peptides
SARS-CoV-2 structural peptide libraries consisting of 15-mer peptides overlapping by 11 amino acids and spanning the entire sequence of viral structural proteins (nucleocapsid, spike, envelope and membrane) were designed from the SARS-CoV-2 reference sequence (NC_045512.2) (A&A Peptide, San Diego, CA, USA) (see supplementary Table 6) and Alpha/B.1.1.7 variant (see supplementary Table 7) and Beta/B.1.1351 variant (TC Peptide Lab, San Diego, CA, USA) to expand T cells and to subsequently identify reactivity on enzyme-linked immunospot (ELISpot) and intracllular cytokine stain. All peptides were reconstituted at a working concentration of 1 µg/µL in dimethyl sulfoxide and stored at –80°C until further use.
Antibody testing
Antibodies were determined as previously described [15,17]. Plasma was obtained from samples up to 24 h following blood draw by centrifugation at 1000 g for 15 min. Samples were incubated with spike and nucleocapsid proteins fused to Gaussia and Renilla luciferase enzymes. Luciferase activity was measured in light units with a 165 Centro LB 960 microplate luminometer (Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany). Seropositive thresholds of 125 000 and 45 000 light units based on known positive samples for spike and nucleocapsid, respectively, were used.
Expansion of SARS-CoV-2 T cells
As shown in Figure 1, PBMCs were pulsed with an overlapping peptide mix (as described by Keller et al. [15]) of viral structural proteins: nucleocapsid, spike, envelope and membrane. As shown in Figure 2 , PBMCs were separately pulsed with a mix of (i) overlapping peptides spanning the reference spike (see supplementary Table 6), (ii) peptides spanning mutated sequences in spike for Alpha variant B.1.1.7 (see supplementary Table 7) and (iii) peptides spanning mutated sequences in spike for Beta variant B.1.351. Cells were then resuspended with IL-15 at 5 ng/mL and IL-7 at 10 ng/mL (R&D Systems, Minneapolis, MN, USA) in CTL medium consisting of Click's medium (Irvine Scientific, Santa Ana, CA, USA), RPMI (GE Healthcare, Logan, UT, USA) and human AB serum (Gemini BioProducts, West Sacramento, CA, USA) and supplemented with GlutaMAX (Gibco, Grand Island, NY, USA) as previously described [15]. Cells were fed regularly and split when confluent. Cells were harvested on day 10 or day 11 and evaluated for antigen specificity by ELISpot.
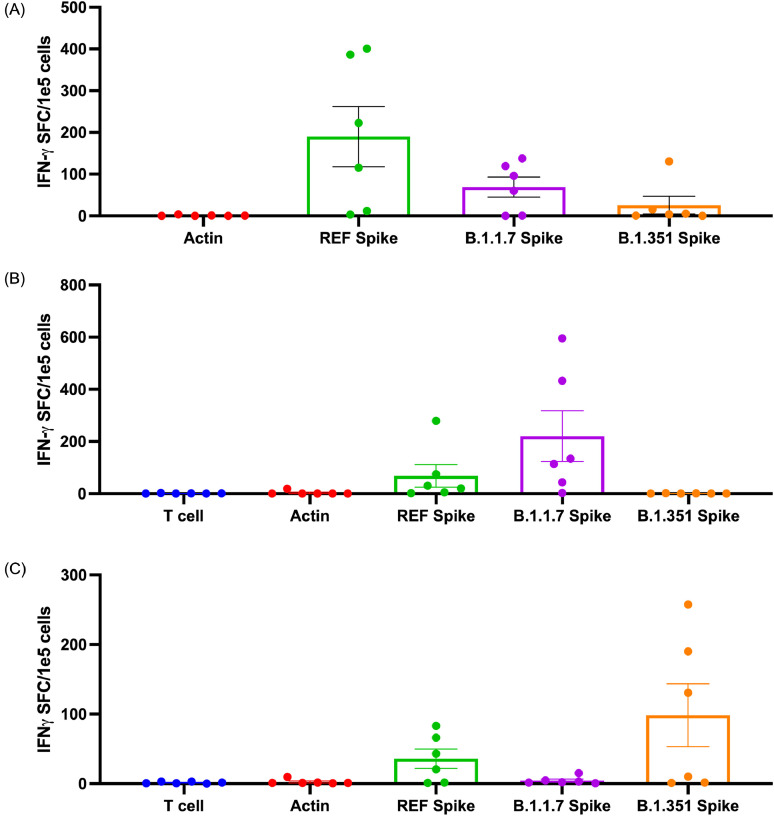
Fig. 2.
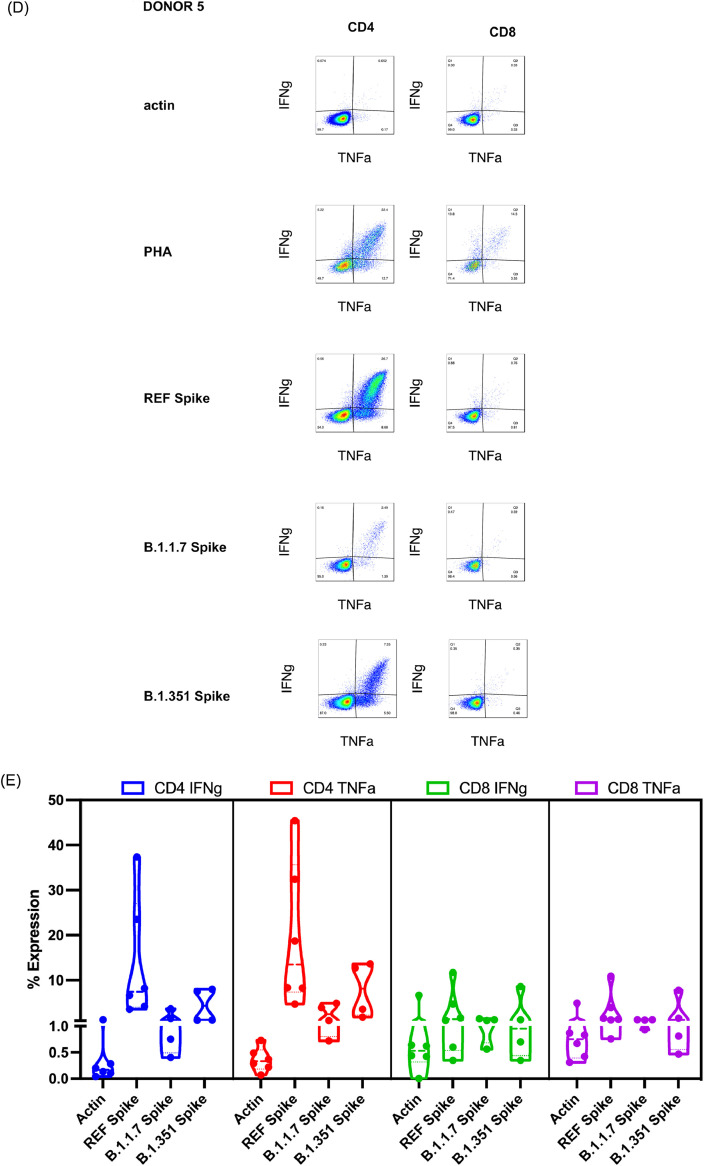
T cells exhibit cross-reactivity against B.1.1.7 and B.1.351 variants. (A) T-cell responses as measured by IFN-γ ELISpot after 10- to 11-day expansion of PBMCs with COVID-19 reference spike. Bars depict mean and error bars show standard deviation. Reference spike responses were significantly different from actin responses (P = 0.0016 following Friedman's test and Dunn's multiple comparisons test). Alpha/B.1.1.7 responses and Beta/B.1.351 responses were not significantly different from actin responses. (B) T-cell responses as measured by IFN-γ ELISpot after 10- to 11-day expansion of PBMCs with COVID-19 Alpha/B.1.1.7 variant spike peptides. Bars depict mean and error bars show standard deviation. Alpha/B.1.1.7 responses were significantly different from actin responses (P = 0.0024 following Friedman's test and Dunn's multiple comparisons test). Reference responses and Beta/B.1.351 responses were not significantly different from actin responses. (C) T-cell responses as measured by IFN-γ ELISpot after 10- to 11-day expansion of PBMCs with COVID-19 Beta/B.1.351 variant spike peptides. Bars depict mean and error bars show standard deviation. Beta/B.1351 responses were significantly different from actin responses (P = 0.0219 following Friedman's test and Dunn's multiple comparisons test). Reference responses and Alpha/B.1.1.7 responses were not significantly different from actin responses. (D) Representative intracellular stain of a donor showing responses to negative control (actin), positive control (PHA) and three different spike sequences. Reference spike containing the complete sequence of reference peptides for the spike protein, Alpha/B.1.1.7 variant spike peptides containing the mutated sequences present in the Alpha/B.1.1.7 variant that differ from the reference strain (see supplementary Table 7) and Beta/B.1.351 variant spike peptides containing the mutated sequences present in the Beta/B.1.351 variant that differ from the reference strain. (E) Summary of responses measured as percent of CD4+ or CD8+ T cells showing IFN-γ or TNF-α secretion against actin (irrelevant self-antigen), reference spike containing the complete sequence of reference peptides for the spike protein, Alpha/B.1.1.7 spike containing the mutated sequences present in the Alpha/B.1.1.7 variant that differ from the reference and Beta/B.1.351 spike containing the mutated sequences present in the Beta/B.1.351 variant that differ from the reference. All T cells were expanded for 10–11 days from PBMCs against reference spike peptides (see supplementary Table 6). REF, reference. (Color version of figure is available online).
Anti-IFN-γ ELISpot assay
IFN-γ ELISpot assay was performed as previously described, with some modifications [15]. Antigen specificity of expanded T cells from PBMCs grown in the presence of SARS-CoV-2 peptide mixes (A&A Peptide, San Diego, CA, USA) was evaluated by stimulating expanded cells with SARS-CoV-2 peptide mixes and measuring IFN-γ production by ELISpot assay. T cells were plated at 1 × 105 cells/well with no peptide mixes, actin PepMix (negative control), phytohemagglutinin (PHA) (positive control) and each of the individual SARS-CoV-2 peptide mixes (1 μg/peptide/well). Antigen specificity of expanded T cells from PBMCs grown in the presence of peptides spanning spike (see supplementary Table 6), peptides spanning B.1.1.7 (Alpha variant)-mutated proteins (see supplementary Table 7) or peptides spanning B.1.351 (Beta variant) was evaluated by stimulating expanded cells with spike, B.1.1.7 or B.1.351 peptide mixes [15] and measuring IFN-γ production by ELISpot assay. T cells were plated at 1 × 105 cells/well with no peptide mixes, actin PepMix (negative control), PHA (positive control) and each of the individual spike/variant peptide mixes (1 μg/peptide/well). Spot-forming cells were independently counted by ZellNet Consulting (Fort Lee, NJ, USA). Responses that were at least 10 spot-forming cells/1 × 105 T cells and greater than 2-fold the background level of non-stimulated T cells or T cells stimulated with actin were considered positive responses (in keeping with definitions used in the authors’ previous work).
Intracellular cytokine stain
Ex vivo-expanded T cells were frozen after 10 days of culture and thawed the day before intracellular cytokine staining in the presence of 40 U/mL IL-2. Intracellular cytokine stain was performed as previously described, with some modifications [15]. Briefly, T cells were stimulated with actin (negative control), spike peptides (peptides spanning spike) (see supplementary Table 6), peptides spanning B.1.1.7 (Alpha variant)-mutated proteins (see supplementary Table 7) or peptides spanning B.1.351 (Beta variant) or PHA (positive control) at a concentration of 1 μg/mL, in the presence of CD28/49d antibody (BD FastImmune; BD Biosciences, Franklin Lakes, NJ, USA) and brefeldin A (BD FastImmune), and incubated at 37°C in 5% carbon dioxide. After 6 h, T cells were then washed in 2% fetal bovine serum with phosphate-buffered saline and surface stained with fluorochrome-conjugated antibodies to CD3, CD4 and CD8. Cells were then fixed, permeabilized using BD Cytofix/Cytoperm solution (BD Biosciences) and stained with anti-IFN-γ and anti-TNF-α. T cells were fixed until acquired the following day and were then analyzed on a BD Cytoflex, with data analysis performed using FlowJo software (FlowJo, LLC, Ashland, OR, USA).
Epitope analysis
Predicted HLA class I epitopes were derived from artificial neural network prediction by NetMHCpan-4.1 [18]. Spike sequences—those for the reference strain were derived from the reference sequence (NC_045512) at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/nuccore/1798174254), whereas those for the mutant strains were derived from published sequences in Pango (https://cov-lineages.org/)—were inputted with no pre-determined peptide length selection, alleles of the recruited donors (where no specific type was available, the HLA supertype representative was used) and a threshold of 0.5% rank for strong binders. Predicted HLA class II epitopes were derived from the Immune Epitope Database and Analysis Resource [19]. Sequences were inputted as peptides with the 2.22 prediction method recommended by the Immune Epitope Database, selecting for the full HLA reference set, with select alpha and beta chains analyzed separately where applicable. Percentile ranks <2 were considered strong binders, and percentile ranks between 2 and 10 were considered weak binders.
Discussion
The authors evaluated T cells derived from six donors who had no detectable antibody response (National Institutes of Health) to spike and nucleocapsid (median IgG antibody levels of 35 094 light units for nucleocapsid and 9569 light units for spike [n = 6], all below seropositive thresholds of 125 000 and 45 000 light units, respectively) (Figure 1A) immediately prior to vaccination. All donors also had documented absence of COVID-19 infection by antibody testing in the months before vaccination became available (see supplementary Figure 1). For five of the six donors, there was also documented negative COVID-19 polymerase chain reaction/antigen testing, with three of the six donors showing negative results within the week preceding the blood draw obtained immediately before vaccination #1.
The authors have previously shown that amplification of specific responses can be done by culturing PBMCs in cytokines and antigen for 10 days [15]. Hence, the authors evaluated SARS-CoV-2-specific T-cell responses in the peripheral blood immediately after blood collection (day 0) and after a 10-day expansion step to amplify the response (see supplementary Figure 1). With this approach, the authors demonstrated reliable amplification of the response post-vaccination, similar to what we have observed in antigen-exposed, convalescent donors. However, even after this expansion step, no detectable responses were observed in the authors’ pre-vaccine donor cohort (Figure 1B; also see supplementary Figure 2). By contrast, after one dose of vaccine, increased T-cell responses to spike were observed, and this was further enhanced after two doses of vaccine in all donors.
Because current circulating SARS-CoV-2 variants threaten to prolong the pandemic and observations of decreased neutralization activity from antibodies have been made [6,12], the authors next evaluated potential spike T-cell responses to the currently circulating major variants in the United States (Alpha/B.1.1.7 and Beta/B.1.351) in the post-vaccine donor cohort. Epitope predictions of both class I and class II binding suggest the potential for cross-reactivity since epitopes from the variant are predicted to bind to the same HLA alleles as their respective reference peptides (see supplementary Tables 2, 3) in the authors’ donor pool. As shown in supplementary Tables 2 and 3, some variant epitopes are predicted to be even stronger binders than the reference homologous epitopes. Regions of the mutations in variants are also a rich source of other HLA-binding epitopes not present in the reference strain (see supplementary Tables 4, 5).
The authors confirmed the potential cross-reactivity of post-vaccine T-cell responses following observations that T-cell products stimulated with the reference spike (see supplementary Table 6) were also able to recognize the Alpha/B.1.1.7 variant (see supplementary Table 7) or Beta/B.1.351 variant in all but one donor (donor three) (Figure 2A; also see supplementary Figure 3), which is in contrast to the variable antibody responses reported in the literature [[5], [6], [7],12,16]. To verify cross-reactivity, the authors next sought to amplify the response by specifically expanding T cells using the mutant sequences within the Alpha/B.1.1.7 and Beta/B.1.351 variant spike peptide libraries (see supplementary Table 7) as stimulating antigens and determined responses against reference spike peptides (see supplementary Table 6). In Alpha/B.1.1.7 variant spike-stimulated T cells, the authors observed reactivity to the variant spike in five of six donors (Figure 2B; also see supplementary Figure 4). In Beta/B.1.351 variant spike-stimulated T cells, the authors observed reactivity to the variant spike in five of six donors (Figure 2C; also see supplementary Figure 5). These observations strongly suggest that epitope-specific T-cell responses to reference spike cross-react with epitopes in the mutated regions of Alpha/B.1.1.7 and Beta/B.1.351, which is consistent with homologies in these predicted epitopes (see supplementary Figure 1) for the HLA types represented by the seronegative donors the authors evaluated (see supplementary Table 2).
To determine whether CD4+ T cells or CD8+ T cells contribute to the reactivity to spike, the authors performed intracellular cytokine stains and demonstrated that both CD4 and CD8 compartments showed specificity against reference spike protein following vaccination (Figure 2D,E; also see supplementary Figure 6). CD4+ and CD8+ T cells secreted IFN-γ and TNF-α in response to the spike antigen, and in some donors also recognized spike from Alpha/B.1.1.7 or Beta/B.1.351.
These data support the hypothesis that COVID-19 vaccination elicits immunological responses that are cross-reactive with novel strains and may be protective through a combination of humoral and adaptive T-cell immunity. These data also suggest that immunity may be incomplete after the first vaccine dose. Cellular immune responses peaked after the second spike mRNA vaccine dose, emphasizing the importance of completing the validated two-dose vaccine strategy advocated by the manufacturers to elicit both humoral and cell-mediated immunity to SARS-CoV-2 in vivo. The presence of CD8+ T-cell responses in donors post-vaccination is therefore encouraging since cytotoxic T cells directly lyse their targets, and the authors posit that their presence may offset the unwanted excessive cytokine secretion mediated by unregulated T-cell activity.
Conclusions
The authors contend that ongoing surveillance of both antibody and T-cell responses to emerging variants is an important measure to evaluate the immune protection afforded by current mRNA vaccines. T-cell response data can potentially serve as indicators for future vaccination strategies, including the need for booster vaccines as well as designs for more universal vaccines.
Funding
This work was partially supported by the intramural research program of the National Institute of Allergy and Infectious Diseases and the National Institute of Dental and Craniofacial Research.
Author Contributions
Conception and design of the study: HL, MDK, MS, CMB and CRYC. Acquisition of data: HL, MJW, KW, PB, JC, MS, AG, DS, BO, CL and CRYC. Analysis and interpretation of data: MS, MDK, CMB and CRYC. Drafting or revising the manuscript: MS, MDK, CMB and CRYC. All authors have approved the final article.
Declaration of Competing Interest
CL, MDK, CRYC and CMB have intellectual property related to developing T-cell therapies for infectious diseases, including SARS-CoV-2.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.jcyt.2021.07.006.
Appendix. Supplementary materials
References
- 1.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom. October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 3.CDC . 2021. US COVID-19 Cases Caused by Variants.https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html [Google Scholar]
- 4.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 Spike variants bind human ACE2 with increased affinity. The Lancet Infectious Diseases. 2021;21(8):1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen R.E., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jangra S., et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv (Preprint) 2021 doi: 10.1101/2021.01.26.21250543. [DOI] [Google Scholar]
- 8.Gupta S., Su H., Narsai T., Agrawal S. SARS-CoV-2-Associated T-Cell Responses in the Presence of Humoral Immunodeficiency. Int Arch Allergy Immunol. 2021;182:195–209. doi: 10.1159/000514193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifoni A., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rydyznski Moderbacher C., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.038. 996-1012 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collier D.A., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 12.Diamond M., et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res Sq (Preprint) 2021 doi: 10.21203/rs.3.rs-228079/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarke A., et al. Negligible impact of SARS-CoV-2 variants on CD4 (+) and CD8 (+) T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv (Preprint) 2021 doi: 10.1101/2021.02.27.433180. [DOI] [Google Scholar]
- 15.Keller M.D., et al. SARS-CoV-2-specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein. Blood. 2020;136:2905–2917. doi: 10.1182/blood.2020008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tada T., et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv (Preprint) 2021 [Google Scholar]
- 17.Burbelo P.D., et al. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients With Coronavirus Disease 2019. J Infect Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vita R., et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.