Abstract
Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA-1) is the essential protein for maintenance of the EBV episome and establishment of latency. The BamHI Q promoter (Qp) is used for the transcription of EBNA-1 mRNA in type I and type II latency, which are EBV infection states exemplified by Burkitt’s lymphoma and nasopharyngeal carcinoma. However, Qp is inactive in type III latency, and other promoters (the BamHI C promoter and/or the BamHI W promoter) are used for EBNA-1. The involvement of interferon regulatory factors (IRFs) in the regulation of Qp is suggested by the presence of an essential interferon-stimulated response element (ISRE) in the promoter. In this work, expression of IRF-2 is shown to be inversely associated with Qp status, i.e., IRF-2 levels are high in type III latency (when Qp is inactive) and low in type I latency (when Qp is active). Also, IRF-2 is identified by electrophoretic mobility shift assay as the major protein binding to the Qp ISRE in type III latency. In transient transfection assays, IRF-2 represses the activity of Qp-reporter constructs. Overexpression of IRF-2 in a type I latency cell line did not activate the endogenous Qp but marginally reduced the EBNA-1 mRNA level. Switching from type III latency (Qp inactive) to type II latency (Qp active), as produced by cell fusion, is directly associated with greatly reduced expression of IRF-2. These data strongly suggest that IRF-2 is a negative regulator of Qp and may contribute to the silencing of Qp in type III latency.
The biologic hallmark of Epstein-Barr virus (EBV) and its usual interaction with B lymphocytes is latency. Three types of latency, each having a distinct pattern of gene expression, have been described. Type I latency is exemplified by Burkitt’s lymphoma (BL) tumors in vivo and earlier passages of cultured cell lines derived from BL biopsy specimens. Only EBV nuclear antigen 1 (EBNA-1) is expressed in this form of latency. Several reports suggest that a type I-like form of latency exists in healthy carriers of EBV (4, 29, 38, 53). Interestingly, cells in type I latency can escape host immune surveillance because EBNA-1 can interfere with its peptide presentation on major histocompatibility complex class I molecules (25), which might explain the lifelong reservoir of virus in immunocompetent, seropositive persons. Type II latency is found in fusions between lymphoblastoid cell lines (LCLs) and epithelial cell lines, in nasopharyngeal carcinoma and in Hodgkin’s disease. EBNA-1, latent membrane protein 1 (LMP-1), LMP-2A, and LMP-2B are expressed in type II latency. Type III latency is represented by LCLs established after EBV infection of adult primary B cells in vitro and by group III BL lines. Nine viral proteins are expressed, including six nuclear proteins (EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, and EBNA-LP) and three integral membrane proteins (LMP-1, LMP-2A, and LMP-2B) (reviewed in references 22 and 40).
EBNA-1 is the sole protein needed for the replication of the EBV episome and maintenance of the latent infection state, events which are essential for cell immortalization (reviewed in references 22 and 40). The promoter usage for expression of EBNA-1 differs in different types of latency. In type I and II latency, the BamHI Q promoter (Qp) is used for the transcription of EBNA-1 mRNA. However, in type III latency Qp is silent, and the BamHI C promoter and/or the BamHI W promoter (C/Wp) are used (see Fig. 1A). The biological consequence of the Qp-to-C/Wp switch and the conversion to type III latency is the expression of the full spectrum of latency genes (reviewed in reference 40), which confer enhanced cell survival, growth, and invasive potential (5, 12, 14, 19, 48, 54, 59).
FIG. 1.
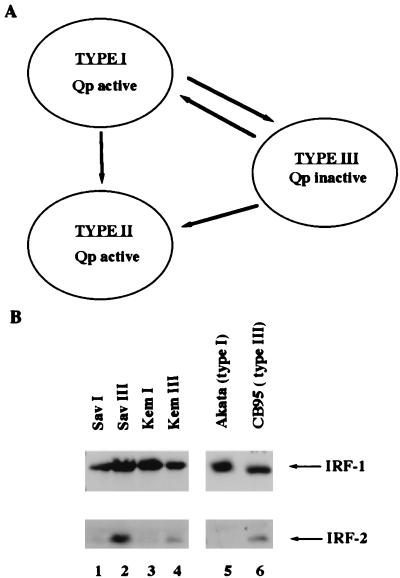
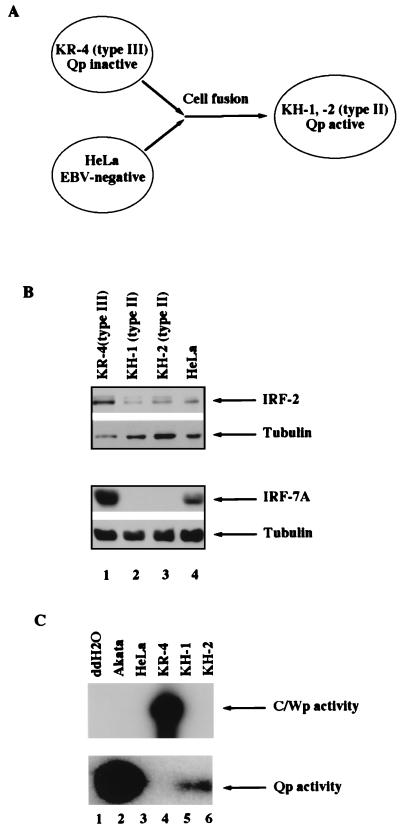
Correlation of Qp status and IRF-2 expression. (A) Schematic diagram of Qp status in EBV latency states. The arrows indicate that conversions can happen naturally or be forced in cell culture. (B) IRF-2 is associated with type III latency. Equal amounts of protein lysates were electrophoresed in sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and stained with Ponceau S Red after the transfer of protein to the membrane. Western blotting with IRF-1 or IRF-2 antibodies was performed. Lanes 1 to 6, cell lysates from Sav I (type I), Sav III (type III), Kem I (type I), Kem III (type III), Akata (type I), and CB95 (type III), respectively.
Since Qp usage not only is essential for the survival of the virus in an immunocompetent host but also is associated with several tumors, understanding the regulation of Qp is essential for understanding the viral program in EBV-associated tumors. Both EBNA-1 and host factors are involved in the transcriptional regulation of Qp. The downstream element of Qp, the Q locus (see Fig. 3A), contains two binding sites for the EBNA-1 protein, which binds to them and acts in an autoregulatory manner to repress Qp transcription (43, 50). However, E2F-1 overcomes EBNA-1-mediated repression of Qp in transient transfection assays, and E2F-1 binds to the Q locus and displaces the binding of EBNA-1 (49), so that the promoter is regulated in a cell cycle-dependent manner (8).
FIG. 3.
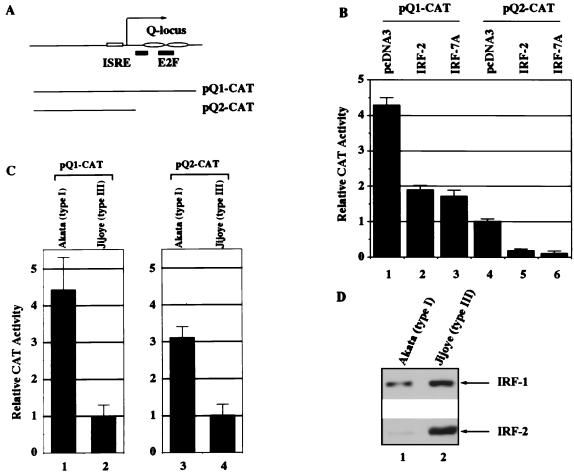
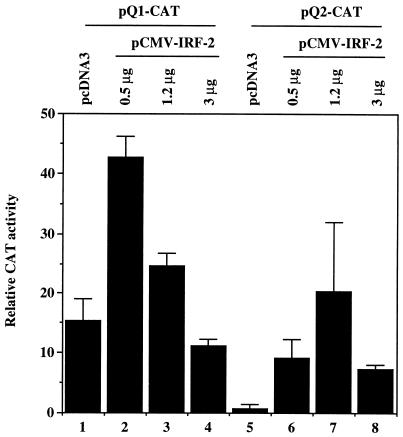
Repression of Qp-reporter constructs by IRF-2. (A) Schematic diagram of Qp and Qp-reporter constructs. Open rectangle, ISRE; solid bars, E2F binding sites; ovals, EBNA-1 binding sites (Q locus). The RNA start site for Qp is indicated by an arrow (46). (B) Repression of Qp by IRF-2. The pQ1-CAT and pQ2-CAT reporter constructs were cotransfected with pCMVβ and either pcDNA3 plasmids, pCMV-IRF-2, or pcDNA-IRF-7A. Transfection efficiency was normalized by β-galactosidase activity. The relative CAT activities from three experiments are shown with standard deviations. (C) Constitutive activities of Qp reporters in type I and type III cell lines. Akata (type I) and Jijoye (type III) cell lines were used for transfection with Qp reporters and pCMVβ. Transfection efficiency was normalized by β-galactosidase activity. The relative CAT activities from four replicates are shown with standard deviations. (D) Endogenous levels of IRF-2 in type I and type III cell lines. Equal amounts of protein lysates were electrophoresed in sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and stained with Ponceau S Red after the transfer of protein to the membrane. Western blotting with IRF-1 or IRF-2 antibodies was performed. Cell lysates from the Akata (type I) and Jijoye (type III) cell lines were used.
An interferon-stimulated response element (ISRE) has been discovered and appears to be essential for the constitutive activity of Qp (see Fig. 3A) (32, 44, 61). Interferon regulatory factors (IRFs), which are a group of transcription factors with multiple functions (reviewed in reference 30), could potentially bind to the ISRE and regulate the activity of Qp. A newly identified IRF-7 has been implicated as a negative regulator of Qp (61). IRF-2, which is usually a repressor of transcription (3, 9, 11, 15, 17, 18, 20, 27, 28, 35), has been reported to be a major positive regulator of Qp based on transient transfection assays in IRF-2-null mouse embryonic fibroblasts (32, 44). The cellular genes activated by IRF-2 that have been identified are the histone H4 gene (55), as well as vascular cell adhesion molecule-1 (VCAM-1), which is highly cell specific (21).
Perhaps the most intriguing aspect of Qp is how it is rendered inactive selectively in type III latency. Since any IRF could potentially bind to the Qp ISRE, the possibility that IRFs other than IRF-7 are involved in the inactivation of Qp was raised. The data presented in this paper suggest that IRF-2 acts as a constitutive repressor of Qp in type III latency.
MATERIALS AND METHODS
Cell culture.
DG75 is an EBV-negative BL line (2). Sav I and Sav III, and Kem I and Kem III, are paired EBV-positive BL lines that differ only in their latent infection types (33, 41). CB95, X50-7, SFC-4, and KR-4 are LCLs (23, 34, 57). Jijoye is an EBV-positive type III BL line, and Akata is a type I BL line (51). FaDuHyg is a head-and-neck squamous carcinoma line (39). All these lines were maintained in RPMI-1640 plus 10% fetal bovine serum (FBS). KH-1 and KH-2 lines were derived from fusion of KR-4 (LCL) and HeLa (cervical carcinoma) cells (6). DKO is a mouse embryonic fibroblast line with targeted disruption of both the IRF-1 and the IRF-2 gene (28). KH-1, KH-2, HeLa, and DKO were maintained in Dulbecco’s modified Eagle medium (DMEM) plus 10% FBS.
Plasmids and antibodies.
IRF-2 and IRF-7A expression plasmids and IRF-7 antibody have been described elsewhere (35, 61). pcDNA/CD4 is a human CD4 expression plasmid (a gift from J. Ting). The β-galactosidase expression plasmid, pCMVβ (6177-1), was purchased from Clontech, pQ1-CAT (−173 to +116 relative to the Qp start site) was obtained by removal of a HindIII-BamHI fragment from pF2-CAT (49), while pQ2-CAT (−173 to +5) was made by cloning the corresponding PCR fragment into pBS-CAT (10). The anti-IRF-1 (C-20) and anti-IRF-2 (C-19) antibodies were purchased from Santa Cruz Biotechnology, Inc. The anti-γ-tubulin antibody (T-6557) was from Sigma. The Western blot analysis with enhanced chemiluminescence (ECL) and protein assay were carried out essentially as described elsewhere (42, 61).
Transient transfection and isolation of transfected cells.
For DG75, Akata, and Jijoye cells, 107 cells in 0.5 ml of medium were used for transfection with a Bio-Rad Gene Pulser (at 320 V and 975 μF). For FaDuHyg cells, Superfect transfection reagents were used according to the manufacturer’s recommendations (Qiagen). For DKO cells in 60-mm culture dishes with 40 to 60% confluence, 24 μg of Lipofectamine reagents (GIBCO BRL) in 400 μl of serum-free Opti-MEM (GIBCO BRL) was mixed gently with another 400 μl of Opti-MEM containing 7.5 μg of DNA. These mixtures were then gently shaken at room temperature for 15 min, and 1 ml of serum-free medium was added before application to DKO cells, which were rinsed once with serum-free medium. After 2 h of incubation in a 37°C incubator, the cells were rinsed again with complete DMEM. Two days after transfection, cells were collected for chloramphenicol acetyltransferase (CAT) assay or for isolation of transfected cells. The CAT and β-galactosidase assays were essentially the same as described elsewhere (24, 42, 61). The CAT assay results were analyzed on a Molecular Dynamics PhosphorImager.
For isolation of transfected cells, cells were collected and CD4-positive cells were enriched by use of anti-CD4-antibody-conjugated magnetic beads according to the manufacturer’s recommendation (Dynal, Inc.). These cells were used for the isolation of total RNA with the RNase kit (Qiagen).
RPA.
RNase protection assays (RPAs) were performed with total RNA by use of the U.S. Biochemicals RPA kit. The hybridization temperature was 37°C. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was provided by U.S. Biochemicals, Inc. The EBNA-1 probe was generated by PCR with the Extend high-fidelity PCR system (Boehringer Mannheim) and specific primers. The sequence of primer A is 5′-GCTCTAGATAATACGACTCACTATAGGG CGACAGACCCAAGCTTGGTACCGAGCTCGGATCCTGTCATAACAAG GTCCTTAATCGCA-3′, and that of primer B is 5′-GCTCTAGAGACTACCGACGAAGGAACTTGG-3′. Primer A contains the T7 RNA polymerase promoter sequence to allow the transcription of the antisense EBNA-1 RNA probe and a spacer to provide separation between the unprotected and protected regions of the probe in the RPA. The protected region of EBNA-1 corresponds to nucleotides (nt) 109437 to 109704 of the EBV B95-8 strain. The purified PCR product was confirmed by enzymatic digestion and used directly for RNA probe synthesis by use of T7 RNA polymerase (Promega) and [α-32P]UTP (Amersham). The H4 histone probe was also generated by PCR with specific primers in a manner similar to that for EBNA-1. The sequences of primers used are 5′-GCTCTAGATTAAGCGGATCTCTGGCCTCAT-3′ and 5′-GCTCTAGAT AATACGACTCACTATAGGGCGACAGACCCAAGCCTTGGTACCGAGC TCGGATCCCTAGCCTCCGAAGCCGTAGAGGGTTCTC-3′. The protected region corresponds to nt 743 to 924 of the published sequence (36).
EMSA.
Cell lysates were made and an electrophoretic mobility shift assay (EMSA) was performed essentially as described elsewhere (26, 37, 61). When antiserum was needed, 1 μl was added to the reaction mixture. The Qp ISRE probe is the same as F7/8, which has a 21-bp region from Qp (61).
RT-PCR for detection of Qp and C/Wp.
C/Wp and Qp activities were detected by reverse transcription-PCR (RT-PCR) with primer pairs which can distinguish the use of C/Wp from that of Qp (53). After RT-PCR, the products were separated on a 2% agarose gel, transferred to a nitrocellulose membrane, and probed with labeled primers for specific detection of either the C/Wp- or the Qp-derived PCR product. C/Wp activity was determined by the detection of both EBNA-2 and EBNA-1 (Y3/U/K spliced form) mRNAs. Qp activity was determined by detection of the Q/U/K spliced form of EBNA-1 mRNA. F promoter (Fp) activity was determined by detection of the F/U/K form of EBNA-1 mRNA (45) by use of induced Akata cDNA as a positive control. The F+9 oligonucleotide (5′-gctctagaGAGAGGAGGGGGATCCGGAG-3′, corresponding to nt 62239 to 62258) was used as a primer for the BamHI F region. (The lowercase letters stand for the non-EBV sequences.) All the other primers and probes were the same as reported elsewhere (53). With reference to the B95-8 genomic sequence, the genome coordinates for these primers and probes are 62440 to 62457, 107986 to 107967, 14802 to 14822, 48583 to 48562, 67544 to 67563, 48397 to 48416, and 47855 to 47904 (53).
RESULTS
Expression of IRF-2 is associated with EBV type III latency.
The association of IRF-7 with EBV type III latency has been established (61), and the levels of expression of other IRFs were investigated. Specifically, the expression of IRF-1 and IRF-2 was examined by Western blot analysis with specific antisera against IRF-1 or IRF-2 proteins. Sav I and Sav III, as well as Kem I and Kem III, are paired, genetically identical EBV-infected cell lines that differ only in their types of latency (33, 41). As shown in Fig. 1B, IRF-2 is expressed at much higher levels in type III latency (Fig. 1B, lanes 2 and 4) than in type I latency (lanes 1 and 3). IRF-2 is also expressed at higher levels in SFC-4, CB95, X50-7, and Jijoye cells (all type III) than in Akata cells (type I) (Fig. 1B, lanes 5 and 6; Fig. 3D, lanes 1 and 2; also data not shown) (34, 51). In contrast, the IRF-1 level is basically unchanged in type I and III cells. The types of latency were confirmed by detection of LMP-1, EBNA-1, and EBNA-2 proteins. In all the cell lines tested, the expression of IRF-2 is higher in type III cells than in type I cells. These data show that a high level of expression of IRF-2 is associated with type III latency and suggest that IRF-2 is a negative regulator of Qp because Qp is inactive in type III latency.
IRF-2 is the major protein binding to the Qp ISRE in type III latency.
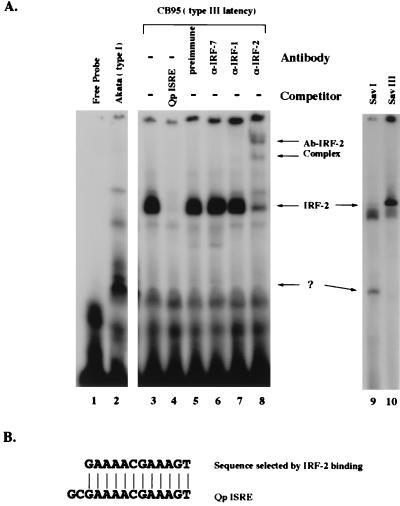
Since the Qp ISRE is essential for Qp activity (50, 61), the binding of various IRFs to Qp is likely to be informative about their relative contributions to the regulation of Qp. An EMSA was used to examine the protein-binding pattern generated with this element in type I and type III cell extracts. Lysates were prepared from Akata (type I) and CB95 (type III) cells, and equal amounts of protein were used with the Qp ISRE as the probe. There are clear differences in the protein-binding patterns with type I and III cell lysates (Fig. 2A). In CB95 lysates, a major band was detected that disappeared when the cold Qp ISRE competitor (100×) was used (Fig. 2A, lane 4) but not by competition with mutated or unrelated probes (data not shown). The protein in that band was identified as IRF-2 by supershifting produced with an IRF-2 antibody and not with IRF-7 or IRF-1 antibodies (Fig. 2A, lanes 5 to 8). Almost identical results could be obtained when SFC-4 (type III) lysate was used for supershift analysis. Under several EMSA conditions tested, IRF-2 was always the major binding protein. As expected, IRF-2 binding was barely detectable with the type I extracts (Fig. 2A, lanes 2 and 9); a large amount of protein from Akata cells was required to detect any binding (data not shown). Similar results were obtained with the paired cell lines, such as Sav I and Sav III (Fig. 2A, lanes 9 and 10). The clear difference in binding of IRF-2 produced with extracts from type I and III cells was consistent with the levels of IRF-2 detected in Western blot analysis (Fig. 1B).
FIG. 2.
IRF-2 is the major protein binding to the Qp ISRE in type III cells. (A) Patterns of binding to the Qp ISRE in type I and type III cells. The probe was Qp ISRE labeled with [α-32P]dCTP. Free probe (lane 1) and lysates from Akata (lane 2), CB95 (lanes 3 to 8), Sav I (lane 9), and Sav III (lane 10) cells were used for EMSA. Cold competitor (the Qp ISRE) was added in lane 4 at a 100-fold molar excess over the hot probe. The following antibodies were added: preimmunization serum (lane 5), IRF-7 antiserum (lane 6), α-IRF-1 antiserum (lane 7), and α-IRF-2 antiserum (lane 8). Arrows with question mark indicate a band found primarily in type I cell lysates. (B) Sequence comparison between a sequence selected randomly by IRF-2 binding and the Qp ISRE.
That IRF-2 is the major protein binding to the Qp ISRE in EMSA suggests that IRF-2 has a high affinity for the Qp ISRE. This point is supported by a previous report that a sequence identical to the core sequence of the Qp ISRE could be selected randomly from an oligonucleotide pool by IRF-2 binding (Fig. 2B) (52).
Interestingly, binding of IRF-1 and IRF-7 to Qp was hardly detectable in this assay (Fig. 2A, lanes 6 and 7). Although IRF-7 is consistently associated with type III latency in all the cell lines tested by Western blot analysis, and in vitro-made IRF-7 binds to Qp, binding of IRF-7 was not detected with cell lysates despite extensive efforts (61). Because IRF-7 itself was cloned based on its binding to the Qp ISRE in a yeast one-hybrid system, it is conceivable that IRF-7 can bind to Qp with low affinity in vivo. Similarly, binding of IRF-1 is hard to detect with cell lysates, although it can bind to Qp in vitro (data not shown). It seems likely that in the presence of IRF-2, other IRFs compete with difficulty for binding to the Qp ISRE. In support of this notion, the IRF-1-Qp ISRE complex could be identified in lysates of an IRF-2-null line (44), which again suggests that IRF-2 has a higher affinity for Qp.
Overexpression of IRF-2 represses activity of Qp-reporter constructs.
The effect of IRF-2 on the activity of Qp was examined by use of Qp-reporter constructs in transient transfection assays. Since EBV can infect epithelial cells, the FaDuHyg epithelial line (39), which has low endogenous expression of both IRF-2 and IRF-7, was chosen for reporter assays. Cotransfection of an IRF-2 expression plasmid with pQ2-CAT, a Qp reporter construct containing the ISRE sequence (Fig. 3A), resulted in a decrease in the constitutive activity of Qp (∼80% [Fig. 3B]). As expected, IRF-7 also repressed pQ2-CAT activity (Fig. 3B). Repression was also observed when pQ1-CAT (Fig. 3A) was used for this experiment (Fig. 3B). IRF-1 has little or no effect on these reporter constructs. No repression by IRF-2 was observed when pFe3M (50), which has point mutations in the ISRE that abolish binding of IRF-2, was used as a reporter. Also, no repression was observed when pBS-CAT was used as a reporter, and IRF-2 could weakly activate a histone H4 reporter construct in this cell line (55) (data not shown). The data here indicate that IRF-2, as well as IRF-7, represses Qp activity.
Repression by IRF-2 of pQ2-CAT, which lacks the Q locus (Fig. 3A), could be detected consistently, although at low levels, in B-cell lines (e.g., DG75 cells); however, repression of the pQ1-CAT reporter construct, which contains the Q locus, was hard to detect in B-cell lines as reported elsewhere (32) (data not shown). These data suggest that the Q locus may contain a positive regulatory element(s) which overcomes the repression of IRF-2. A likely candidate might be an E2F family member(s) (49). In support of this notion, the constitutive activity of pQ1-CAT is about fourfold higher than that of pQ2-CAT (Fig. 3B).
To address whether physiological levels of IRF-2 and IRF-7 might inhibit Qp, Akata (type I) and Jijoye (type III) cell lines were transfected with Qp-reporter constructs along with a β-galactosidase expression plasmid. Akata and Jijoye lines were chosen because they have similar transfection efficiencies, and expression levels of both IRF-2 and IRF-7 are higher in Jijoye than in Akata cells, as expected (Fig. 3D and data not shown). As shown in Fig. 3C, the constitutive activities of Qp-reporter constructs are lower in the Jijoye cell line than in Akata cells. The greater difference in the activity levels of pQ1-CAT between type I and type III latency cells is most likely due to the effect of the additional repressor, EBNA-1 (Fig. 3A) (43, 50). Although other factors may also contribute to the lower activity of Qp in type III cells, the results suggest that physiological levels of IRF-2 and IRF-7 can repress Qp.
IRF-2 did not activate endogenous Qp.
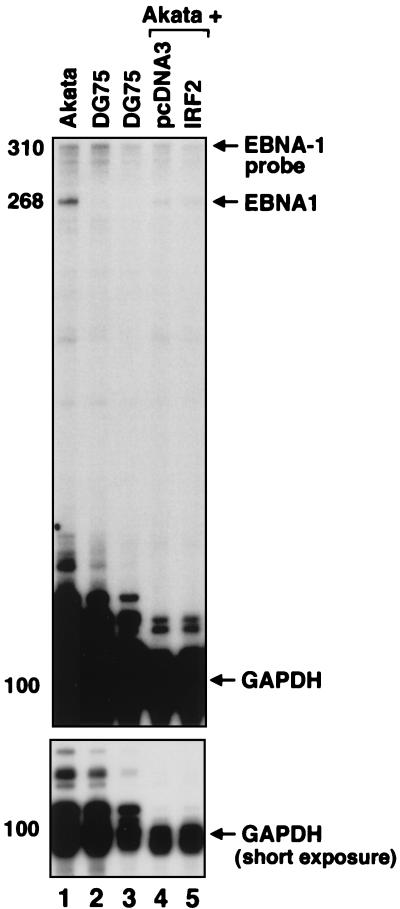
Next, the role of IRF-2 in the regulation of endogenous Qp was addressed by overexpression of IRF-2 in a type I line in an attempt to mimic the high expression of IRF-2 in type III cells. Several attempts to generate a cell line stably expressing IRF-2 failed, and a system that can enrich for transfected cells was used instead. By cotransfection of a CD4 expression plasmid, transfected cells were selected with the use of CD4 antibody-conjugated magnetic beads (see Materials and Methods for details). Akata cells were chosen because of their relatively higher transfection efficiency. Total RNA from vector and IRF-2-transfected cells was extracted, and RPAs were performed with an EBNA-1-specific probe. As shown in Fig. 4, IRF-2 did not activate Qp but marginally reduced the Qp-derived EBNA-1 mRNA (compare lanes 4 and 5). DG75 cells, an EBV-negative BL line, did not show an EBNA-1 RNA band (lane 2 and 3). Only low levels of EBNA-1 mRNA could be detected in type I cells (Fig. 4) (33), which is consistent with the low abundance of Qp-derived EBNA-1 mRNA in type I cells (8).
FIG. 4.
IRF-2 did not activate endogenous Qp. (Top) EBNA-1 and GAPDH probes were labeled with [α-32P]UTP and used for RPA. RNAs from Akata cells (lane 1), DG75 cells (lanes 2 and 3), or Akata cells transfected with pcDNA3 (lane 4) or pCMV-IRF-2 (lane 5) were used. The reduction of EBNA-1 mRNA by IRF-2 was 45%, which was obtained by normalizing EBNA-1 levels to GAPDH by use of a PhosphorImager. Specific protection of EBNA-1, GAPDH mRNA, and undigested probe is indicated. (Bottom) Short exposure time for protected GAPDH mRNA. The sizes, which were confirmed by molecular weight markers, are shown on the left in nucleotides.
Since IRF-2 has been reported to be a positive regulator of the H4 histone promoter (55), the H4 histone mRNA level was examined. IRF-2 could activate the histone H4 gene only slightly, as reported elsewhere (55), and no repression could be observed as determined by RPA with an H4-specific probe. IRF-2 apparently has minimal effect on histone H4 in B-cell lines because no obvious differences in H4 histone mRNA levels were observed in a panel of type I and III lines (data not shown).
The amount of reduction of the EBNA-1 mRNA (∼50%) observed in an RPA was consistent with the degree of repression obtained in the Qp reporter assays (Fig. 3B, bars 1 and 2). These results suggest that IRF-2 is likely to be a repressor, and not an activator, of Qp. The partial reduction of EBNA-1 mRNA detected might be due to the need for more than one repressor to silence Qp (see Discussion), to low enrichment efficiency of transfected cells, or to cotransfection efficiency. Also, IRF-2 may have to compete with a Qp activator(s), which is likely to be specific for type I cells. An additional complex binding to Qp ISRE was consistently detected in type I cell lysates (Fig. 2A, lanes 2 and 9) and might be a candidate for a Qp activator. This complex apparently does not contain IRF-1, because IRF-1 antibody could not supershift and/or block this complex (data not shown).
Qp reactivation is associated with down-regulation of IRF-2 and IRF-7.
It has been clearly shown that fusion of a type III cell line with an epithelial cell line can force a switch from the type III to the type II latency phenotype (6). KR-4 (a type III LCL) and HeLa (EBV-negative cervical carcinoma cells) are parental lines for the KH-1 and KH-2 lines produced by cell fusion (Fig. 5A) (6). That KH-2 and KH-1 are type II latency cells was confirmed by the detection of EBNA-1 and LMP-1 but not EBNA-2 proteins (data not shown). The absence of C/Wp activity in KH-1 and KH-2 lines has previously been reported, but without addressing the Qp status (1). Here we show that Qp is reactivated and C/Wp is inactivated in both fusion cell lines, as expected in cells with a type II phenotype (40), in contrast to the parental KR-4 line, in which C/Wp is active and Qp is inactive (Fig. 5C). The lytic promoter, BamHI F (Fp [45]), is not active in these lines (see Materials and Methods for details). Finally, the percentage of cells containing EBV genomes was examined by using EBER staining followed by fluorescence-activated cell sorter analysis (7). The KH-2 and KR-4 lines have identical amounts of EBER-positive cells, while KH-1 has a relatively lower percentage (data not shown).
FIG. 5.
Reactivation of Qp is associated with down-regulation of IRF-2 and IRF-7 expression. (A) Schematic diagram for generation of KH cell lines (6). (B) Expression of IRF-2 and IRF-7 is down-regulated in type II latency. Cell lysates from KR-4, KH-1, KH-2, and HeLa cells were used for Western blot analysis. γ-Tubulin was used as a loading control. (C) Qp and C/Wp activities in the cell lines. The promoter activities were determined by RT-PCR with Qp- or C/Wp-specific primers followed by Southern blot analysis. Double-distilled water (ddH2O) or cDNAs from the cell lines shown were used.
Since Qp was reactivated in the KH-1 and KH-2 lines, the expression of IRF-2 and IRF-7 was examined by Western blot analysis. As shown in Fig. 5B, IRF-2 is expressed at much higher levels in the parental KR-4 cells than in the KH-1 and KH-2 lines. As expected, IRF-7 is also greatly reduced (61).
These data indicate that switching from type III latency, where Qp is inactive, to type II latency, where Qp is reactivated, is associated with striking reductions in the levels of IRF-2 and IRF-7, supporting the roles of IRF-2 and IRF-7 as negative regulators of Qp.
DISCUSSION
The switch from Qp to C/Wp, which allows transcription of all the other EBNA proteins (EBNA-2, -3A to -3C, and -LP), produces a phenotypic conversion to type III latency. We previously identified an ISRE in Qp and a novel protein, IRF-7, as a putative negative regulator for this promoter (61). In this work, IRF-2 was also found to be strongly associated with type III latency and to act as a negative regulator of Qp.
It is becoming clear that IRFs are involved in the regulation of Qp. Although IRF-2 is reported to be a transactivator that can activate the histone H4 gene and VCAM-1 (21, 55), IRF-2 is usually a transcriptional repressor and an antagonist to IRF-1 (3, 9, 11, 15, 17, 18, 20, 27, 28, 35). It is interesting that IRF-2 has been reported to be the major positive regulator of Qp in mouse fibroblasts (32, 44). However, the data presented here suggest strongly that IRF-2 is a negative regulator of Qp. First, IRF-2 expression is associated with EBV type III latency, where Qp is inactive (Fig. 1, 2, and 3D); second, overexpression of IRF-2 in a type I line did not activate Qp but marginally reduced the level of EBNA-1 mRNA derived from Qp (Fig. 4); third, IRF-2 is the major protein binding to Qp and appears to have a high affinity for its ISRE (Fig. 2); fourth, IRF-2 represses the activity of Qp-reporter constructs in transient transfection assays (Fig. 3); and fifth, Qp reactivation in type II cells converted by cell fusion from type III cells, in which Qp is inactive, is associated with a sharp reduction in the expression of IRF-2 (Fig. 5).
Previous reports that IRF-2 activates Qp may be due to the mouse fibroblast lines that were used (32, 44). Indeed, IRF-2 could activate our Qp-reporter constructs in a mouse IRF-2-null fibroblast line from the same source (Fig. 6). However, these results must be interpreted in relation to their biological relevance. Selection of an appropriate line is especially important for IRF-2 research because IRF-2 possesses both a transcriptional repression domain and a latent activation domain (58). If the repressor domain of IRF-2 is not active in a given cell line, the latent activation domain may be activated, and IRF-2 becomes an activator. This scenario is documented in muscle cells, where IRF-2 can activate the VCAM-1 promoter (21). Since EBV infects neither fibroblasts nor rodent cells, the conclusion based solely on reporter assays conducted in mouse fibroblasts raises questions about biological relevance. In contrast, our results were obtained with human cell lines latently infected or infectible with EBV.
FIG. 6.
Activation of Qp-reporter constructs by IRF-2 in a mouse IRF-2-null cell line. DKO, a mouse embryonic fibroblast line in which both IRF-1 and IRF-2 genes are disrupted (28), was used for transfection. The pQ1-CAT and pQ2-CAT reporter constructs were cotransfected with pcDNA3 plasmids or pCMV-IRF-2. The amounts of plasmids transfected are shown at the top. pCMVβ was also cotransfected, and β-galactosidase activity was used to normalize transfection efficiency. The relative CAT activities and standard deviations are shown.
Another explanation for the different results is that IRF-2 might have a dual function in the regulation of Qp, i.e., it might positively regulate Qp in type I cells when the IRF-2 level is low and negatively regulate Qp in type III cells when expression of IRF-2 is high. Data from transfections of Qp into an IRF-2-null line with high doses of IRF-2 may suggest such a tendency (Fig. 6). This possibility needs to be rigorously addressed. However, a dose-dependent repression of Qp by IRF-2 could be observed in biologically relevant lines, such as FaDuHyg and DG75 (data not shown).
Other than being a regulator of Qp, IRF-2 may also contribute to the transforming properties of EBV in type III latency. EBV can immortalize primary B cells and, at the same time, establish type III latency. IRF-2 has oncogenic potential based on transformation assays (16, 31). It would be interesting to examine EBV-associated immunoblastic lymphomas (type III latency) directly to see if IRF-2 levels are elevated in these malignancies.
The inducer(s) of IRF-2 in type III cells is still unclear, although LMP-1 can stimulate the expression of IRF-7 (62). Further work needs to be done to identify the inducer(s) of IRF-2 in type III latency. Other possibilities are that the type I latency cells, in which IRF-2 and IRF-7 levels are low, are selected by EBV for infection; or that establishment of type I latency itself involves the down-regulation of IRF-2 and IRF-7.
IRF-7 was cloned as a Qp-binding protein and subsequently inferred to be a negative regulator of Qp (61). This function is further supported by the fact that reactivation of Qp by fusion of type III cells with epithelial cells to produce cells with a type II phenotype is associated with a striking reduction in the expression of IRF-7 (Fig. 5). Other than being a Qp repressor, an additional role of IRF-7, namely, virus-induced activation of the interferon-β gene, has just been reported (56).
It is interesting that both IRF-2 and IRF-7 are negative regulators of Qp. There was no apparent synergistic effect between these two factors in terms of Qp repression, and an additive effect was hard to detect when both were overexpressed (data not shown). Why would Qp use two repressors for a single ISRE site? One possible explanation is that IRF-2 and IRF-7 are apparently expressed at different times during the cell cycle (60). Therefore, IRF-2 and IRF-7 may functionally repress Qp in different phases of the cell cycle to keep Qp silenced throughout the cycle.
Apart from IRF-2 and IRF-7, viral proteins definitely play an important role in the silencing of Qp. EBNA-1 can directly repress Qp activity (43, 50), and the higher levels of the protein in type III latency may enhance its repressor effect (references 8, 13, and 47 and our unpublished results). EBNA-2, which is responsible for the increased expression of EBNA-1 in type III latency, could be considered an indirect regulator of Qp.
It is clear that Qp regulation is complicated and that multiple factors, both viral and cellular, are involved. With IRF-2, IRF-7, and EBNA-1 all expressed at higher levels in type III latency, it is reasonable to infer that Qp may be turned off by a combination of IRF-2, IRF-7, EBNA-1, and perhaps another repressor(s), i.e., IRF-2 and IRF-7 may repress Qp through its ISRE while EBNA-1 acts through the Q locus (Fig. 3A). In contrast, in type I latency, when Qp is active, all these factors are expressed at much lower levels. The major positive regulator(s) of Qp operating through the ISRE is unidentified but apparently is associated with type I latency. A band which seems to be specific for type I cells was detected by EMSA (Fig. 2) and was also noticed by other investigators (32); it might be a candidate for such an activator.
Thus, we show that IRF-2, probably acting with IRF-7, and perhaps EBNA-1 as well, silences the promoter used in the most-restricted form of EBV latency, type I, with the indirect consequence that the promoter for the least-restricted form of EBV latency, type III, is used. It is likely that several IRFs, both positive and negative regulators, that have different affinities for the type I promoter and are supplied at different times in the cell cycle, govern the activity of this key, tightly regulated EBV promoter.
ACKNOWLEDGMENTS
We thank Jenny Ting, Maria Masucci, Wanla Kulwichit, Bernard Weissman, Patricia Vaughan, and Gary Stein for providing valuable reagents and/or help in this work. We thank T. Taniguchi for permission to use the IRF knockout cell line. We also thank Shannon Kenney and Nancy Raab-Traub for critical reading of the manuscript, Matt Davenport and Val Zacny for editorial help, and Cyd Johnson for technical help.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (AI 42372-01) and the National Cancer Institute (CA 19014). L.Z. was supported by an NIH Individual National Research Service Award (5F 32 CA67433).
REFERENCES
- 1.Altiok E, Minarovits J, Hu L F, Contreras-Brodin B, Klein G, Ernberg I. Host-cell-phenotype-dependent control of the BCR2/BWR1 promoter complex regulates the expression of Epstein-Barr virus nuclear antigens 2–6. Proc Natl Acad Sci USA. 1992;89:905–909. doi: 10.1073/pnas.89.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwith Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt-like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 3.Cha Y, Deisseroth A B. Human interferon regulatory factor 2 gene. Intron-exon organization and functional analysis of 5′-flanking region. J Biol Chem. 1994;269:5279–5287. [PubMed] [Google Scholar]
- 4.Chen F, Zou J-Z, di Renzo L, Winberg G, Hu L-F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1992;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contreras-Brodin B A, Anvret M, Imreh S, Altiok E, Klein G, Masucci M G. B cell phenotype-dependent expression of the Epstein-Barr virus nuclear antigens EBNA-2 to EBNA-6: studies with somatic cell hybrids. J Gen Virol. 1991;72:3025–3033. doi: 10.1099/0022-1317-72-12-3025. [DOI] [PubMed] [Google Scholar]
- 7.Crouch J, Leitenberg D, Smith B R, Howe J G. Epstein-Barr virus suspension cell assay using in situ hybridization and flow cytometry. Cytometry. 1997;29:50–57. [PubMed] [Google Scholar]
- 8.Davenport, M., and J. S. Pagano. Expression of EBNA-1 mRNA is regulated by cell-cycle during Epstein-Barr virus type I latency. J. Virol, in press. [DOI] [PMC free article] [PubMed]
- 9.Drew P D, Franzoso G, Carlson L M, Biddison W E, Siebenlist U, Ozato K. Interferon regulatory factor-2 physically interacts with NF-kappa B in vitro and inhibits NF-kappa B induction of major histocompatibility class I and beta 2 microglobulin gene expression in transfected human neuroblastoma cells. J Neuroimmunol. 1995;63:157–162. doi: 10.1016/0165-5728(95)00140-9. [DOI] [PubMed] [Google Scholar]
- 10.Furnari F B, Adams M D, Pagano J S. Regulation of the Epstein-Barr virus DNA polymerase gene. J Virol. 1992;66:2837–2845. doi: 10.1128/jvi.66.5.2837-2845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lacoste J, Hiscott J. Viral induction of the human beta interferon promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory C D, Dive C, Henderson S, Smith C A, Williams G T, Gordon J, Rickinson A B. Activation of Epstein-Barr virus latent genes protects human B cells from death by apoptosis. Nature. 1991;349:612–614. doi: 10.1038/349612a0. [DOI] [PubMed] [Google Scholar]
- 13.Gregory C D, Rowe M, Rickinson A B. Different Epstein-Barr virus (EBV)-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J Gen Virol. 1990;71:1481–1495. doi: 10.1099/0022-1317-71-7-1481. [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 15.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 16.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 17.Harada H, Takahashi E, Itoh S, Harada K, Hori T A, Taniguchi T. Structure and regulation of the human interferon regulatory factor 1 (IRF-1) and IRF-2 genes: implications for a gene network in the interferon system. Mol Cell Biol. 1994;14:1500–1509. doi: 10.1128/mcb.14.2.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 19.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 20.Horiuchi M, Koike G, Yamada T, Mukoyama M, Nakajima M, Dzau V J. The growth-dependent expression of angiotensin II type 2 receptor is regulated by transcription factors interferon regulatory factor-1 and -2. J Biol Chem. 1995;270:20225–20230. doi: 10.1074/jbc.270.34.20225. [DOI] [PubMed] [Google Scholar]
- 21.Jesse T L, LaChance R, Iademarco M F, Dean D C. Interferon regulatory factor-2 is a transcriptional activator in muscle where it regulates expression of vascular cell adhesion molecule-1. J Cell Biol. 1998;140:1265–1276. doi: 10.1083/jcb.140.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1995. pp. 2343–2396. [Google Scholar]
- 23.Kozbor D, Lagarde A E, Roder J C. Human hybridomas constructed with antigen-specific Epstein-Barr virus-transformed cell lines. Proc Natl Acad Sci USA. 1982;79:6651–6655. doi: 10.1073/pnas.79.21.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laimins L A, Gruss P, Pozzatti R, Khoury G G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984;49:183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 26.Levy D E, Kessler D S, Pine R, Reich N, Darnell J E J. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 27.Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J Biol Chem. 1994;269:17542–17549. [PubMed] [Google Scholar]
- 28.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig T M, Amakawa R, Kishihara K, Wakeham A, Mak T. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 29.Miyashita E M, Yang B, Lam K M C, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen H, Mustafa A, Hiscott J, Lin R. Transcription factor IRF-2 exerts its oncogenic phenotype through the DNA binding/transcription repression domain. Oncogene. 1995;11:537–544. [PubMed] [Google Scholar]
- 32.Nonkwelo C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagano J S, Jimenez G, Sung N S, Raab-Traub N, Lin J C. Epstein-Barr viral latency and cell immortalization as targets for antisense oligomers. Ann N Y Acad Sci. 1992;28:107–116. doi: 10.1111/j.1749-6632.1992.tb21063.x. [DOI] [PubMed] [Google Scholar]
- 35.Palombella V J, Maniatis T. Inducible processing of interferon regulatory factor-2. Mol Cell Biol. 1992;12:3325–3336. doi: 10.1128/mcb.12.8.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- 37.Pine R. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiss M, Munoz-Antonia T, Cowan J M, Wilkins P C, Zhou Z L, Vellucci V F. Resistance of human squamous carcinoma cells to transforming growth factor beta 1 is a recessive trait. Proc Natl Acad Sci USA. 1993;90:6280–6284. doi: 10.1073/pnas.90.13.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1995. pp. 2397–2446. [Google Scholar]
- 41.Rowe M, Khanna R, Jacob C A, Argaet V, Kelly A, Powis S, Belich M, Croom-Carter D, Lee S, Burrows S R, Trowsdale J, Moss D J, Rickinson A B. Restoration of endogenous antigen processing in Burkitt’s lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur J Immunol. 1995;25:1374–1384. doi: 10.1002/eji.1830250536. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Sample J, Henson E, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer B C, Paulson E, Strominger J L, Speck S H. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol Cell Biol. 1997;17:873–886. doi: 10.1128/mcb.17.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer B C, Strominger J L, Speck S H. The Epstein-Barr virus BamHI F promoter is an early lytic promoter: lack of correlation with EBNA 1 gene transcription in group 1 Burkitt’s lymphoma cell lines. J Virol. 1995;69:5039–5047. doi: 10.1128/jvi.69.8.5039-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer B C, Strominger J L, Speck S H. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer B C, Strominger J L, Speck S H. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol Cell Biol. 1997;17:364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinclair A J, Palmero I, Peters G, Farrell P J. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung N S, Wilson J, Davenport M, Sista N D, Pagano J S. Reciprocal regulation of the Epstein-Barr virus BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol Cell Biol. 1994;14:7144–7152. doi: 10.1128/mcb.14.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung N S, Wilson J, Pagano J S. Characterization of cis-acting elements of the BamHI-F promoter of EBV. In: Tursz T, et al., editors. The Epstein-Barr virus and associated diseases. London, United Kingdom: INSERM/John Libbey Eurotext Limited; 1993. pp. 239–242. [Google Scholar]
- 51.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt’s lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka N, Kawakami T, Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tierney R, Steven N, Young L, Rickinson A. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7378. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan P S, Aziz F, van Wijnen A J, Wu S, Harada H, Taniguchi T, Soprano K J, Stein J L, Stein G S. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 56.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 57.Wilson G, Miller G. Recovery of Epstein-Barr virus from nonproducer neonatal human lymphoid cell transformants. Virology. 1979;95:351–358. doi: 10.1016/0042-6822(79)90490-2. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto H, Lamphier M S, Fujita T, Taniguchi T, Harada H. The oncogenic transcription factor IRF-2 possesses a transcriptional repression and a latent activation domain. Oncogene. 1994;9:1423–1428. [PubMed] [Google Scholar]
- 59.Yoshizaki T, Sato H, Furukawa M, Pagano J S. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc Natl Acad Sci USA. 1998;95:3621–3626. doi: 10.1073/pnas.95.7.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, L., M. Davenport, and J. S. Pagano. Unpublished data.
- 61.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, L., and J. S. Pagano. Stimulation of the expression of interferon regulatory factor 7 by Epstein-Barr virus latent membrane protein 1. Submitted for publication.