Abstract
Background
Although lots of quantitative trait loci (QTLs) and genes present roles in litter size of some breeds, the information might not make it clear for the huge diversity of reproductive capability in pig breeds. To elucidate the inherent mechanisms of heterogeneity of reproductive capability in litter size of Xiang pig, we performed transcriptome analysis for the expression profile in ovaries using RNA-seq method.
Results
We identified 1,419 up-regulated and 1,376 down-regulated genes in Xiang pigs with large litter size. Among them, 1,010 differentially expressed genes (DEGs) were differently spliced between two groups with large or small litter sizes. Based on GO and KEGG analysis, numerous members of genes were gathered in ovarian steroidogenesis, steroid biosynthesis, oocyte maturation and reproduction processes.
Conclusions
Combined with gene biological function, twelve genes were found out that might be related with the reproductive capability of Xiang pig, of which, eleven genes were recognized as hub genes. These genes may play a role in promoting litter size by elevating steroid and peptide hormones supply through the ovary and facilitating the processes of ovulation and in vivo fertilization.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40813-021-00226-x.
Keywords: Transcriptome, Alternative splicing, Ovary, Litter size, Xiang pig
Summary
Based on analyzing of the transcriptome and alternative splicing events, twelve candidate genes related with fecundity and litter size trait were found out from the ovary of Xiang pig.
Background
In pig industry, the productive power of sows is one of most concerned economic traits in the world [1]. Reproductive traits are extremely intricate and influenced by multifactors originating from heredity and environment especially in litter size of pigs [2–5]. Lots of genes have functions on the reproduction capability [5, 6]. And number of litter size provides a direct effect on the economic benefits for pig farmer [7]. Ovaries are the main reproductive organs; they perform ovulation and show a direct influence on the efficiency of fecundity. Consequently, the different expression profiles of some important genes in ovary might devote to comprehend the diversity of litter size among breeds [8, 9].
Previous reports focus on the quantitative trait loci (QTL) together with intrinsic genes related with litter size of pigs, and the relationship between genes and the traits [10–12] is explored based on recent technology progress at molecular level [4, 13–15]. Series of genes connected with the fecundity of pig have been found, including FSH-β, ESR, OPN, MTNR1A, PRLR, GDF9 and BMPs members [16, 17]. Many genes and QTLs have been ascertained to have a linkage with the litter size trait of pig [18–20]. Differently expressed genes related with fecundity and litter size have been detected using transcriptome information from gonads in European pigs [4, 13] and Chinese local breeds [15, 21]. Nevertheless, knowledge of previous works aims at several limited pig breeds and they could not make it clear for the huge diversity of reproductive capability in Eurasian pig breeds.
Xiang pig is one of indigenous breeds in China originated from the southeast mountain environment of Guizhou province. It is featured by short stature, early maturity, excellent environmental adaptability as well as with nice meat quality [22, 23]. Furthermore, the populations among Xiang pig herds present great variation in litter size, ranging from 5 to 21 piglets, while most of sows gave 5–8 piglets per litter from the third to seven parities [24]. It was proposed that the cause might due to the specific regulation in the gene expression related with litter size trait. To screen pivotal genes related with litter size, the ovaries were sampled from two groups of Xiang pigs with large and small litter size. The expression profiles and alternative splicing events of transcripts were analyzed by RNA-seq method. The results will benefit to the interpretation of the molecular regulation manner on the diverse reproduction capability and litter size in pig breeds.
Materials and methods
Samples
A total of 40 Congjiang Xiang pigs were prepared from the farm of Dachang pig breeding, Congjiang, Guizhou, China, which born from sows ever giving birth of large litter size as XL group with the total number born (TNB) larger than twelve, or XS group with TNB less than eight. Using the same way from previous reports [15, 25, 26], we randomly chose fourteen pigs that the third estrous time was synchronous from XL group (n = 7) and XS group (n = 7) to sequence the transcriptome using Illumina next sequencing technology. All sampled pigs were 6 to 6.5 months old, weighing 37.50 ± 3.77 kg with five pairs of nipples. The estrous was first detected by B-ultrasound (KX5000V, XuZhou KaiXin Electronic Instrument Company, China) started from the onset of female standing reflex according to the method reported by Lopes et al. [27]. When the numbers of matured follicles were counted to be 4–8 with diameter larger than 6 mm on one ovary [28–30], both ovaries were picked out by standard surgical operation. The follicles numbers and size on two ovaries were directly measured before putting into liquid nitrogen for total RNA isolation. The sampled pigs were alive and kept feeding under routine method together with other pigs.
Library construction and sequencing
Based on protocol of Beijing Genomies Institute (BGI), Shenzhen, China, ovarian RNA was isolated with TRIzol method (Invitrogen) with the values of RNA integrity number (RIN) in the scope from 7.9 to 8.8 to prepare cDNA library. The libraries were sequenced using HiSeqTM 2000 platform (Illumina, USA) and generated 100 bp paired-end reads. The same RNA sample was determined in the RNA-seq and qRT-PCR tests. The cDNA libraries together with RNA sequencing were carried out as described previously [26].
Dataset analysis
The sequencing data from fourteen libraries were taken for analysis of the expression profile and the alternative splicing events of transcripts at estrus stage by RNA-seq method. The raw reads in fastq format were filtered to remove reads under low quality by program Trimmomatic v0.39 using the cutoffs as previous conditions [26], and the clean reads were aligned with the pig reference genomes (Ssc11.1) using HISAT2 software (v2.1.0). The mapped sam files were conversed and sorted into bam format by samtools (v1.1.0). The subread featurecounts software (v2.0.0) was chosen to count the reads amount, which were included in the regions of genes or exons. The expression level of gene was estimated by CPMs values (counts per million mapped reads).
The differently expressed genes with CPM value were calculated by using DESeq2 and edgeR, in which all of CPM values were added 0.001 for logarithm arithmetic. The minimum normalized CPM was 1.0, in which a gene would be eliminated if its CPM value of any sample was not lager than the threshold. The differently expressed genes with CPM values were computed using model featureCounts in subread program (v1.6.3). The threshold for differentially expressed genes (DEGs) was the gene with P < 0.001 (false discovery rate (FDR) < 0.005) and log2 ratio > 1 or < -1. rMATS (v4.0.2) was used to detect the differential AS events with an FDR ≤ 5 % and a |∆PSI | |IncLevelDifference| of ≥ 0.1. The value of PSI or ψ (percentage spliced in) value was calculated by rMATs according to the ratio of the long form on total form present to characterize inclusion of exon, differential splice-site choice, intron retention, etc. The DEGs and differentially spliced genes (DSGs) datasets were uploaded to the platform of KOBAS v3.0 (http://kobas.cbi.pku.edu.cn/kobas3) taking reference Sscrofa11.1 as background based on Ontology Consortium (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for enrichment categories. The ovary DEGs and DSGs from Xiang pig were compared with previous reports [17, 31] via Venn program online. The DEGs and DSGs of Xiang pig related with reproduction were further analyzed. The gene list related with reproduction was first collected from the Gene annotation deposited in NCBI using reproduction as keyword. Then it was converted into the gene names and Ensembl IDs of pig via BioMart online (http://www.ensembl.org/biomart). Based on Venn analysis on the shared genes, the lists of DEGs and DSGs of Xiang pig related with reproduction were selected out.
RT-qPCR verification
To verify the DEGs and DSGs expression patterns deduced by transcriptome analysis, six samples in each group were used as the same aliquot of total RNA for RNA-seq detection. Seven DEGs (LDLR, SCARB1, HSD3B1, CYP11A1, AKR1C2, StAR and LRP8), two highly expressed genes (SERPINE2 and RARRES1) together with five types of AS events were chosen randomly to verify the RNA-seq analysis by quantitative real-time RT-PCR methods. Primers were devised by Primer3 online (http://primer3.ut.ee/). The PCR reaction conditions and proportion were the same as our previous work [32] with each primer concentration of 10 pM/µL, taking GAPDH and β-actin genes as internal controls. Based on dissociation curve analysis for PCR products, the amplification efficiency was controlled within range of 100 ± 10 %. The relative expression level of target gene utilized the method of 2(-ΔΔCt) as reported by Livak et al. [33]. The different level of gene expression between two groups was tested by software SPSS (v21.0) taking the P < 0.05 as threshold of significant difference. The results were presented as mean ± standard deviation. The presence of five types of splicing evens were determined by RT-PCR method using pairs of primers outside of the AS region. And the DSGs levels were further qualitatively detected by qRT-PCR method using one primer span both junction ends of AS event. The nucleotide sequences of primers were listed in Table S1.
Results
Illumina sequencing and assembly
After quality control, the cDNA libraries of each ovary sample generated 49 ~ 66 million clean reads with 100 base pairs (bp) in length. The fourteen samples showed similar matching results, with mapping ranges of 96.17 ~ 99.96 % onto the genome (Ssc11.1) and 69 ~ 75 % being unique matches (Table 1). The results indicated that all of fourteen libraries present high-quality, together with high percentage of coverage throughout pig reference genome. It enabled us to analyze the transcriptomes profiles from ovary at estrus stage of Xiang pigs with large or small numbers of litter size.
Table 1.
Overview of RNA-Seq data
| Sample | Raw Reads | Clean Reads | Raw bases | Clean Pairs (%) | Total_Alignments (bp) | Coverage (Total_Alignments/ Genome_length)%) | Successfully assigned alignments |
|---|---|---|---|---|---|---|---|
| XL1 | 50,011,936 | 49,981,382 | 4,501,074,240 | 24,990,691 (99.94 %) | 25,827,924 | 1.0323273 | 19,158,796 (74.20 %) |
| XL2 | 49,981,708 | 49,963,086 | 4,498,353,720 | 24,981,543 (99.96 %) | 25,815,470 | 1.0318295 | 18,329,302 (71.00 %) |
| XL3 | 50,413,430 | 50,394,894 | 4,537,208,700 | 25,197,447 (99.96 %) | 26,161,129 | 1.0456453 | 18,061,930 (69.00 %) |
| XL4 | 50,375,286 | 50,340,582 | 4,533,775,740 | 25,170,291 (99.93 %) | 25,968,916 | 1.0379626 | 18,816,287 (72.50 %) |
| XL5 | 50,368,172 | 50,349,522 | 4,533,135,480 | 25,174,761 (99.96 %) | 26,071,102 | 1.0420470 | 18,272,816 (70.10 %) |
| XL6 | 52,218,318 | 50,837,574 | 7,832,747,700 | 25,418,787 (97.36 %) | 26,311,441 | 1.0516532 | 19,834,463 (75.40 %) |
| XL7 | 50,004,320 | 48,770,860 | 7,500,648,000 | 24,385,430 (97.53 %) | 25,226,371 | 1.0082835 | 18,583,326 (73.70 %) |
| XS1 | 66,455,060 | 64,122,488 | 6,645,506,000 | 32,061,244 (96.49 %) | 32,672,160 | 1.3058875 | 22,968,529 (70.30 %) |
| XS2 | 66,046,332 | 63,662,060 | 6,604,633,200 | 31,831,030 (96.39 %) | 31,595,025 | 1.2628350 | 22,116,518 (70.00 %) |
| XS3 | 63,770,656 | 63,770,656 | 6,377,065,600 | 30,775,719 (96.52 %) | 29,952,835 | 1.1971976 | 20,607,551 (68.80 %) |
| XS4 | 68,528,104 | 66,122,768 | 6,852,810,400 | 33,061,384 (96.49 %) | 33,824,450 | 1.3519438 | 25,635,551 (75.79 %) |
| XS5 | 66,959,152 | 64,856,636 | 6,695,915,200 | 32,428,318 (96.86 %) | 32,875,650 | 1.3140208 | 23,289,111 (70.84 %) |
| XS6 | 67,665,568 | 65,344,640 | 6,766,556,800 | 32,672,320 (96.57 %) | 31,838,098 | 1.2725505 | 22,703,748 (71.31 %) |
| XS7 | 67,002,792 | 64,436,586 | 6,700,279,200 | 32,218,293 (96.17 %) | 31,845,484 | 1.2728457 | 22,524,311 (70.73 %) |
Differential gene expression analysis between XS and XL groups
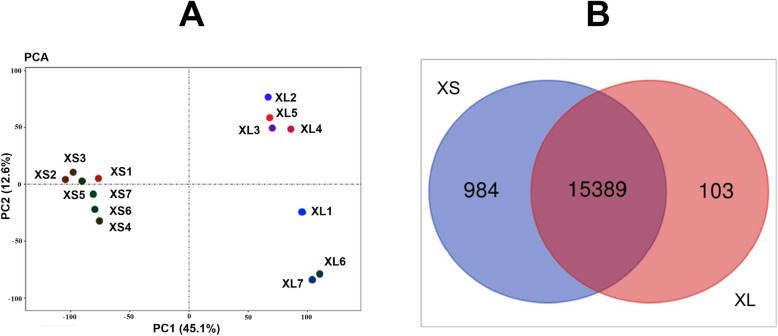
Based on mapping data to the reference of pig genome, we obtained 17,089 and 15,928 genes from XS and XL groups, respectively. Normalized CPM data in Table S2 were inputted for principal component analysis (PCA). It appeared that each point in seven samples of the first group could gather and separate from the points in another group according to PC1, which accounted for percentage of 45.1 % of total variation in the dataset (Fig. 1A). It meant that the distance within samples in the same group was much close to each other than that in another group. After removing the noncoding RNA and pseudogene transcripts and those genes with CPM < 1.0 in each sample, the sequencing data of 16,476 genes could be used for following analysis. Of them, 15,389 genes were expressed in both groups; 984 genes were specifically expressed in the libraries of XS group, while 103 genes were determined only from XL group (Fig. 1B). Most of the genes specially expressed in XS or XL group presented in low or very low CPM values. GO analysis indicated that the especially expressed genes in XS libraries enriched mainly in the cellular process, regulation of biological process and metabolic process. However, the genes only expressed in XL libraries had no statistically significant GO terms (Table S3).
Fig. 1.
Profiles of gene expression in Xiang pig ovaries between XL and XS groups. A: PCA cluster of the gene expression profile of fourteen libraries. B: Venn diagram of expression genes in two groups. Blue color represented genes only expressed in XS group, orange colors showed genes only expressed in XL group, and the intersection is the common genes in both groups
In total, 2,795 genes were differently expressed between two groups after intersection of results from both softwares of edgeR and DESeq2. Compared with the expression in XS group, 1,376 genes were down-regulated and 1,419 genes were up-regulated in XL group (Table S4). The scope of log2FC values was varied from − 8.75 to 9.31 in DEGs. The numbers of genes, with more than four times of difference between two groups, accounted for 26.11 % of the total DEGs. Approximately 18 and 22 % of the DEGs expressed less than 100 normalized CPM in XS and XL respectively, in which 9.26 % of these genes overlapped between two groups. Moreover, a volcano diagram was plotted based on DEGs data (Fig. 2). The expression levels and numbers of DEGs in XL group were more than that in XS group, as displayed in the heat map of Fig. 3. We compared the top ten genes highly expressed in XS group with that in XL groups (Table 2). The expression level in XL group ranged from 3545.875 ~ 15422.571 CPM (logFC from 0.148 to 6.42), which was decreased to 2594.786 ~ 6588.433 CPM (logFC from − 2.238 to 2.307) in XS group. Of those, the expression levels of five genes (StAR, ATP6, COX3, COX1, SELENOP) increased and two genes (MACF1, HSPG2) decreased in XL group.
Fig. 2.
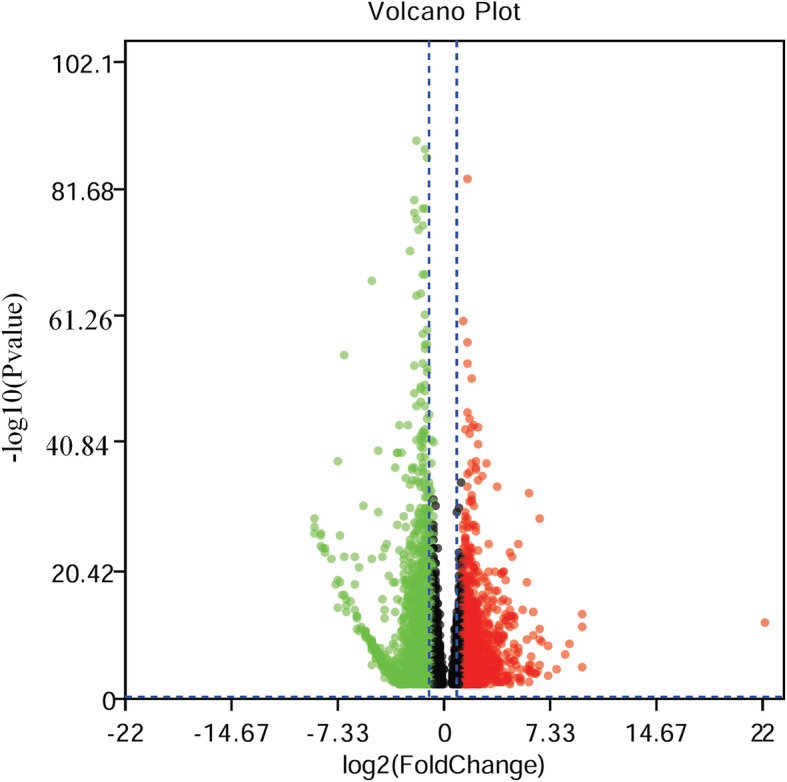
Volcano plots of the differently expressed genes (XL vs. XS). Each point in the figures represented one gene. Red points represented up-regulated genes, green points denoted down-regulated genes. Black points were genes without significant difference
Fig. 3.
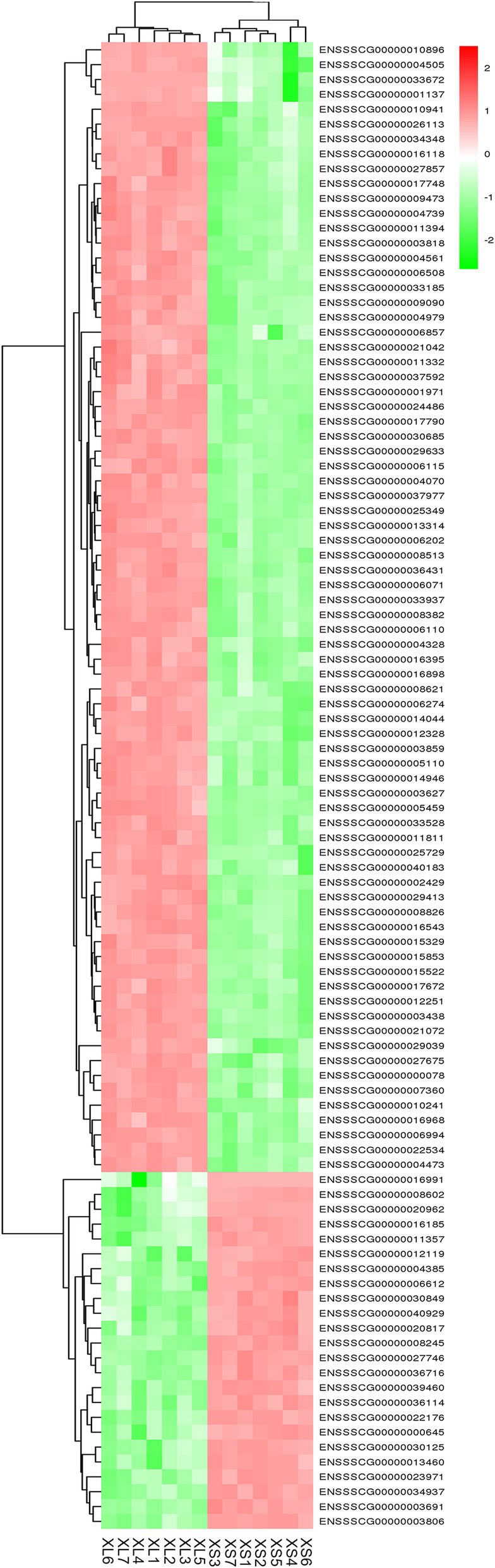
Heatmap showed the most significantly up-regulated and down-regulated genes between XL and XS groups
Table 2.
The top 10 genes at highly expressed level in XS and XL groups
| Gene_ID | Gene symbol | logFC | Pvalue | Padj | Readcount-XL | Readcount-XS | Description |
|---|---|---|---|---|---|---|---|
| ENSSSCG00000018075 | COX1 | 2.307 | 8.16623E-13 | 1.26806E-11 | 15422.571 | 3116.238 | cytochrome c oxidase subunit I |
| ENSSSCG00000018082 | COX3 | 2.607 | 1.74415E-17 | 4.91169E-16 | 6961.009 | 1142.695 | cytochrome c oxidase subunit III |
| ENSSSCG00000036135 | COL1A1 | 0.148 | 0.521083524 | 0.609618558 | 6521.541 | 5887.012 | collagen type I alpha 1 chain |
| ENSSSCG00000004489 | EEF1A1 | 0.531 | 0.000000096 | 0.000000671 | 6455.302 | 4468.317 | eukaryotic translation elongation factor 1 alpha 1 |
| ENSSSCG00000016034 | COL3A1 | 0.295 | 0.169601735 | 0.244697188 | 6382.088 | 5203.422 | collagen type III alpha 1 chain |
| ENSSSCG00000018081 | ATP6 | 4.384 | 8.7737E-25 | 5.28171E-23 | 6356.744 | 304.543 | ATP synthase F0 subunit 6 |
| ENSSSCG00000015326 | COL1A2 | 0.262 | 0.300558594 | 0.392270172 | 5492.596 | 6588.433 | collagen type I alpha 2 chain |
| ENSSSCG00000021208 | SELENOP | 1.372 | 9.40805E-06 | 4.42317E-05 | 4865.024 | 1879.905 | selenoprotein P |
| ENSSSCG00000034942 | StAR | 6.42 | 3.4925E-30 | 3.60421E-28 | 4106.007 | 47.92 | steroidogenic acute regulatory protein |
| ENSSSCG00000011033 | VIM | 0.362 | 0.012616959 | 0.027517822 | 3545.875 | 2758.309 | vimentin |
| ENSSSCG00000025675 | EEF2 | 0.003 | 0.970797113 | 0.977978175 | 2702.43 | 2697.277 | eukaryotic translation elongation factor 2 |
| ENSSSCG00000003514 | HSPG2 | -1.508 | 2.12554E-08 | 1.65259E-07 | 1779.964 | 5061.75 | heparan sulfate proteoglycan 2 |
| ENSSSCG00000000423 | -0.996 | 0.000106900 | 0.000398237 | 1358.776 | 2709.169 | Unknown protein | |
| ENSSSCG00000003654 | MACF1 | -2.238 | 8.38151E-28 | 6.894E-26 | 550.064 | 2594.786 | microtubule actin crosslinking factor 1 |
More strict thresholds were used to screen out the significant DEGs, which included FDR less than 0.001, the value of normalized CPM larger than 100 and the absolute value of log2FC larger than four. We identified 37 significant differently expressed genes from the total DEGs between two groups, such as StAR, TIMP1, NR4A1, PTX3, CYP11A1, PTGFR, OVGP1, SERPINE1, CLDN11, and MSMO1 (Table 3).
Table 3.
The 37 DEGs with great fold changes between XS and XL groups
| Gene_ID | Gene symbol | Readcount -XS | Readcount -XL | log2FC | Pvalue | Padj | Description |
|---|---|---|---|---|---|---|---|
| ENSSSCG00000018081 | ATP6 | 304.543 | 6356.744 | 4.384 | 8.7737E-25 | 5.28171E-23 | ATP synthase F0 subunit 6 |
| ENSSSCG00000034942 | StAR | 47.920 | 4106.007 | 6.420 | 3.4925E-30 | 3.60421E-28 | steroidogenic acute regulatory protein |
| ENSSSCG00000025273 | CYP11A1 | 95.654 | 2902.591 | 4.923 | 6.4979E-26 | 4.61379E-24 | cytochrome P450 family 11 subfamily A member 1 |
| ENSSSCG00000010400 | MSMB | 29.237 | 2419.602 | 6.370 | 1.84815E-12 | 2.71985E-11 | microseminoprotein beta |
| ENSSSCG00000012277 | TIMP1 | 46.398 | 2402.269 | 5.695 | 2.1928E-34 | 2.96206E-32 | TIMP metallopeptidase inhibitor 1 |
| ENSSSCG00000009759 | SCARB1 | 111.493 | 2083.499 | 4.224 | 5.39056E-13 | 8.60367E-12 | scavenger receptor class B member 1 |
| ENSSSCG00000006719 | HSD3B | 124.329 | 2015.189 | 4.019 | 8.923E-14 | 1.56553E-12 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| ENSSSCG00000005216 | RLN2 | 2.618 | 1665.513 | 9.313 | 1.1897E-12 | 1.8052E-11 | relaxin 2 |
| ENSSSCG00000039548 | PTGFR | 46.064 | 1086.103 | 4.559 | 5.30226E-14 | 9.52401E-13 | prostaglandin F receptor |
| ENSSSCG00000026724 | PLBD1 | 45.017 | 939.753 | 4.384 | 4.86394E-07 | 2.93018E-06 | phospholipase B domain containing |
| ENSSSCG00000025698 | SERPINE1 | 46.572 | 859.301 | 4.206 | 1.66233E-14 | 3.20042E-13 | serpin family E member 1 |
| ENSSSCG00000029066 | IDI1 | 40.651 | 690.156 | 4.086 | 8.64835E-15 | 1.73962E-13 | isopentenyl-diphosphate delta isomerase 1 |
| ENSSSCG00000011747 | CLDN11 | 33.833 | 597.989 | 4.142 | 9.07434E-17 | 2.3339E-15 | claudin 11 |
| ENSSSCG00000009645 | ADAMDEC1 | 23.491 | 541.132 | 4.525 | 1.24429E-14 | 2.45241E-13 | ADAM like decysin 1 |
| ENSSSCG00000008857 | MSMO1 | 27.915 | 479.635 | 4.102 | 5.36737E-14 | 9.63054E-13 | methylsterol monooxygenase 1 |
| ENSSSCG00000031321 | NR4A1 | 10.930 | 473.535 | 5.437 | 8.68239E-20 | 3.10901E-18 | nuclear receptor subfamily 4 group A member 1 |
| ENSSSCG00000011727 | PTX3 | 11.869 | 423.841 | 5.158 | 1.30294E-15 | 2.87494E-14 | pentraxin 3 |
| ENSSSCG00000036956 | SOCS3 | 19.526 | 386.909 | 4.308 | 1.15405E-14 | 2.28814E-13 | suppressor of cytokine signaling 3 |
| ENSSSCG00000011683 | PAQR9 | 4.394 | 379.387 | 6.431 | 9.82346E-11 | 1.11946E-09 | progestin and adipoQ receptor family member 9 |
| ENSSSCG00000006359 | ADAMTS4 | 6.917 | 327.492 | 5.565 | 8.02685E-11 | 9.22311E-10 | ADAM metallopeptidase with thrombospondin type 1 motif 4 |
| ENSSSCG00000003753 | PDZK1IP1 | 3.444 | 324.231 | 6.556 | 2.27989E-10 | 2.45339E-09 | PDZK1 interacting protein 1 |
| ENSSSCG00000021576 | CD83 | 15.939 | 270.330 | 4.085 | 3.01992E-20 | 1.15613E-18 | CD83 molecule |
| ENSSSCG00000002013 | DHRS4 | 14.232 | 269.853 | 4.245 | 2.07358E-07 | 1.35533E-06 | dehydrogenase/reductase (SDR family) member 4 |
| ENSSSCG00000015268 | FMO1 | 11.718 | 234.182 | 4.320 | 1.17801E-12 | 1.79072E-11 | flavin containing dimethylaniline monoxygenase 1 |
| ENSSSCG00000023298 | SRXN1 | 10.211 | 218.387 | 4.417 | 4.42571E-24 | 2.48423E-22 | sulfiredoxin 1 |
| ENSSSCG00000009062 | MGARP | 6.837 | 160.851 | 4.559 | 2.91204E-13 | 4.79996E-12 | mitochondria localized glutamic acid rich protein |
| ENSSSCG00000006791 | OVGP1 | 7.036 | 156.771 | 4.478 | 5.01106E-06 | 2.48682E-05 | oviductal glycoprotein 1 |
| ENSSSCG00000008963 | AREG | 0.134 | 149.218 | 9.298 | 9.88649E-15 | 1.97433E-13 | amphiregulin |
| ENSSSCG00000004521 | MRO | 0.401 | 145.096 | 8.434 | 5.4203E-10 | 5.46802E-09 | maestro |
| ENSSSCG00000009219 | IBSP | 0.000 | 111.713 | 9.293 | 2.52194E-06 | 1.32225E-05 | integrin binding sialoprotein |
| ENSSSCG00000028691 | novel gene | 1.780 | 109.434 | 5.936 | 2.32844E-15 | 5.01127E-14 | sulfotransferase 1C1 |
| ENSSSCG00000040843 | MRAP | 5.782 | 106.633 | 4.203 | 1.09225E-08 | 8.92657E-08 | melanocortin 2 receptor accessory protein |
| ENSSSCG00000034167 | SLC5A3 | 476.186 | 26.881 | -4.145 | 4.07214E-26 | 2.91632E-24 | solute carrier family 5 member 3 |
| ENSSSCG00000026113 | ZBTB20 | 197.303 | 5.575 | -5.189 | 2.60958E-68 | 2.16791E-65 | zinc finger and BTB domain containing 20 |
| ENSSSCG00000033672 | HIST1H1E | 215.637 | 1.255 | -7.490 | 2.04215E-39 | 4.03933E-37 | histone cluster 1 H1 family member e |
| ENSSSCG00000034598 | HIST2H2AC | 121.487 | 0.706 | -7.351 | 2.08391E-27 | 1.64877E-25 | histone cluster 2 H2A family member c |
| ENSSSCG00000035473 | novel gene | 110.429 | 0.167 | -8.751 | 1.51499E-27 | 1.21017E-25 | histone H4 |
Compare of DEGs between Xiang and Yorkshire pig
Compared with previous report from Yorkshire pig [17], 71 DEGs were found from ovaries of both pig breeds. Of them, 64 genes were up-regulated and 6 genes were down-regulated in the groups with high litter size (Table 4). And six genes, COX3, STAR, CYTB, CYP11A1, MSMB and SCARB1, were ranked in the highest expression level between two pig breeds.
Table 4.
Compare of DEGs between Xiang and Yorkshire pigs (reported by Zhang et al. [17])
| No. | Gene_ID | Gene name | Xiang pig | Yorkshire sow | Gene description | ||
|---|---|---|---|---|---|---|---|
| Up/Down (XL/XS) | Log2FC | Up/Down (YH/YL) | Log2FC | ||||
| 1 | ENSSSCG00000018082 | COX3 | UP | 2.61 | Up | 2.17 | cytochrome c oxidase subunit III |
| 2 | ENSSSCG00000034942 | STAR | UP | 6.42 | Up | 3.56 | steroidogenic acute regulatory protein |
| 3 | ENSSSCG00000018094 | CYTB | UP | 3.09 | Up | 2.01 | cytochrome b |
| 4 | ENSSSCG00000025273 | CYP11A1 | UP | 4.92 | Up | 3.99 | cytochrome P450 family 11 subfamily A member 1 |
| 5 | ENSSSCG00000010400 | MSMB | UP | 6.37 | Up | 4.58 | microseminoprotein beta |
| 6 | ENSSSCG00000009759 | SCARB1 | UP | 4.22 | Up | 3.59 | scavenger receptor class B member 1 |
| 7 | ENSSSCG00000028512 | LDLR | UP | 3.78 | Up | 2.73 | low density lipoprotein receptor |
| 8 | ENSSSCG00000017164 | TIMP-2 | UP | 1.90 | Up | 1.65 | TIMP metallopeptidase inhibitor 2 |
| 9 | ENSSSCG00000039548 | PTGFR | UP | 4.56 | Up | 3.62 | prostaglandin F receptor |
| 10 | ENSSSCG00000012583 | ACSL4 | UP | 2.87 | Up | 2.33 | acyl-CoA synthetase long chain family member 4 |
| 11 | ENSSSCG00000007435 | PLTP | UP | 3.08 | Up | 3.21 | phospholipid transfer protein |
| 12 | ENSSSCG00000013401 | DKK3 | UP | 2.46 | Up | 2.20 | dickkopf WNT signaling pathway inhibitor 3 |
| 13 | ENSSSCG00000012625 | PGRMC1 | UP | 2.02 | Up | 2.66 | progesterone receptor membrane component 1 |
| 14 | ENSSSCG00000016267 | ITM2C | UP | 2.20 | Up | 1.98 | integral membrane protein 2 C |
| 15 | ENSSSCG00000011747 | CLDN11 | UP | 4.14 | Up | 2.77 | claudin 11 |
| 16 | ENSSSCG00000017024 | CCNG1 | UP | 1.59 | Up | 1.58 | cyclin G1 |
| 17 | ENSSSCG00000018084 | ND3 | UP | 3.87 | Up | 1.87 | NADH dehydrogenase subunit 3 |
| 18 | ENSSSCG00000010554 | SCD | UP | 1.95 | Up | 3.97 | stearoyl-CoA desaturase |
| 19 | ENSSSCG00000014338 | HSPA9 | UP | 1.02 | Up | 1.69 | heat shock protein family A (Hsp70) member 9 |
| 20 | ENSSSCG00000036956 | SOCS3 | UP | 4.31 | Up | 2.90 | suppressor of cytokine signaling 3 |
| 21 | ENSSSCG00000032007 | RTN4 | UP | 1.56 | Up | 2.36 | reticulon 4 |
| 22 | ENSSSCG00000034207 | CEBPB | UP | 3.92 | Up | 2.53 | CCAAT enhancer binding protein beta |
| 23 | ENSSSCG00000025486 | MDH2 | UP | 1.27 | Up | 2.16 | malate dehydrogenase 2 |
| 24 | ENSSSCG00000015709 | SLC35F5 | UP | 2.42 | Up | 2.45 | solute carrier family 35 member F5 |
| 25 | ENSSSCG00000008701 | LRPAP1 | UP | 1.30 | Up | 1.45 | LDL receptor related protein associated protein 1 |
| 26 | ENSSSCG00000003139 | BCAT2 | UP | 1.65 | Up | 2.78 | branched chain amino acid transaminase 2 |
| 27 | ENSSSCG00000001770 | CTSH | UP | 1.47 | Up | 1.77 | cathepsin H |
| 28 | ENSSSCG00000022742 | PRDX6 | UP | 1.53 | Up | 2.52 | peroxiredoxin 6 |
| 29 | ENSSSCG00000027114 | SCP2 | UP | 1.93 | Up | 5.14 | sterol carrier protein 2 |
| 30 | ENSSSCG00000015268 | FMO1 | UP | 4.32 | Up | 3.71 | flavin containing dimethylaniline monoxygenase 1 |
| 31 | ENSSSCG00000007739 | GUSB | UP | 1.29 | Up | 2.39 | glucuronidase beta |
| 32 | ENSSSCG00000013599 | ANGPTL4 | UP | 2.89 | Up | 2.76 | angiopoietin like 4 |
| 33 | ENSSSCG00000009150 | HADH | UP | 1.24 | Up | 1.39 | hydroxyacyl-CoA dehydrogenase |
| 34 | ENSSSCG00000000767 | ATP6V1E1 | UP | 1.04 | Up | 1.63 | ATPase H + transporting V1 subunit E1 |
| 35 | ENSSSCG00000008237 | RETSAT | UP | 1.06 | Up | 1.39 | retinol saturase |
| 36 | ENSSSCG00000005970 | SQLE | UP | 2.40 | Up | 3.42 | squalene epoxidase |
| 37 | ENSSSCG00000009062 | MGARP | UP | 4.56 | Up | 3.66 | mitochondria localized glutamic acid rich protein |
| 38 | ENSSSCG00000008963 | AREG | UP | 9.30 | Up | 5.20 | amphiregulin |
| 39 | ENSSSCG00000006522 | GBA | UP | 1.28 | Up | 1.78 | glucosylceramidase beta |
| 40 | ENSSSCG00000027130 | TNFRSF12A | UP | 3.34 | Up | 1.84 | TNF receptor superfamily member 12 A |
| 41 | ENSSSCG00000030318 | SDHC | UP | 1.43 | Up | 1.83 | succinate dehydrogenase complex subunit C |
| 42 | ENSSSCG00000000757 | ADIPOR2 | UP | 1.13 | Up | 1.86 | adiponectin receptor 2 |
| 43 | ENSSSCG00000011723 | MME | UP | 2.23 | Up | 2.28 | membrane metalloendopeptidase |
| 44 | ENSSSCG00000009219 | IBSP | UP | 9.29 | Up | 3.41 | integrin binding sialoprotein |
| 45 | ENSSSCG00000010853 | EPHX1 | UP | 2.21 | Up | 3.74 | epoxide hydrolase 1 |
| 46 | ENSSSCG00000010537 | GOT1 | UP | 1.48 | Up | 3.39 | glutamic-oxaloacetic transaminase 1 |
| 47 | ENSSSCG00000032213 | DBI | UP | 1.30 | Up | 1.55 | diazepam binding inhibitor, acyl-CoA binding protein |
| 48 | ENSSSCG00000022998 | PKIG | UP | 1.74 | Up | 1.62 | cAMP-dependent protein kinase inhibitor gamma |
| 49 | ENSSSCG00000006296 | ATP1B1 | UP | 2.15 | Up | 2.59 | ATPase Na+/K + transporting subunit beta 1 |
| 50 | ENSSSCG00000001435 | AGPAT1 | UP | 1.91 | Up | 1.42 | 1-acylglycerol-3-phosphate O-acyltransferase 1 |
| 51 | ENSSSCG00000006369 | F11R | UP | 2.76 | Up | 2.34 | F11 receptor |
| 52 | ENSSSCG00000016990 | ATP6V0E1 | UP | 1.76 | Up | 1.81 | ATPase H + transporting V0 subunit e1 |
| 53 | ENSSSCG00000037912 | FITM2 | UP | 1.95 | Up | 2.20 | fat storage inducing transmembrane protein 2 |
| 54 | ENSSSCG00000006337 | HSD17B7 | UP | 1.69 | Up | 3.74 | hydroxysteroid 17-beta dehydrogenase 7 |
| 55 | ENSSSCG00000028943 | ECH1 | UP | 1.57 | Up | 2.16 | enoyl-CoA hydratase 1 |
| 56 | ENSSSCG00000021774 | B3GALNT1 | UP | 1.94 | Up | 2.28 | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| 57 | ENSSSCG00000034896 | HPRT1 | UP | 1.43 | Up | 1.50 | hypoxanthine phosphoribosyltransferase 1 |
| 58 | ENSSSCG00000015299 | STEAP4 | UP | 2.61 | Up | 5.01 | STEAP4 metalloreductase |
| 59 | ENSSSCG00000009245 | SCD5 | UP | 1.13 | Up | 1.89 | stearoyl-CoA desaturase 5 |
| 60 | ENSSSCG00000036893 | PTHLH | UP | 3.24 | Up | 3.37 | parathyroid hormone like hormone |
| 61 | ENSSSCG00000006512 | FDPS | UP | 2.46 | Up | 3.53 | farnesyl diphosphate synthase |
| 62 | ENSSSCG00000038221 | HSD17B2 | UP | 5.76 | Up | 6.95 | hydroxysteroid 17-beta dehydrogenase 2 |
| 63 | ENSSSCG00000007507 | PCK1 | UP | 5.70 | Up | 6.96 | phosphoenolpyruvate carboxykinase 1 |
| 64 | ENSSSCG00000000182 | WNT10B | UP | 3.38 | Up | 2.90 | Wnt family member 10B |
| 65 | ENSSSCG00000016958 | PIK3R1 | Down | -1.05 | Down | -1.50 | phosphoinositide-3-kinase regulatory subunit 1 |
| 66 | ENSSSCG00000025349 | CCDC14 | Down | -2.19 | Down | -1.49 | coiled-coil domain containing 14 |
| 67 | ENSSSCG00000013772 | ASF1B | Down | -1.54 | Down | -1.95 | anti-silencing function 1B histone chaperone |
| 68 | ENSSSCG00000031027 | IRS4 | Down | -2.02 | Down | -3.11 | insulin receptor substrate 4 |
| 69 | ENSSSCG00000005136 | IFNE | Down | -3.02 | Down | -2.10 | interferon epsilon |
| 70 | ENSSSCG00000009490 | DCT | Down | -1.58 | Down | -1.86 | dopachrome tautomerase |
| 71 | ENSSSCG00000006125 | CALB1 | Down | -2.37 | Up | 4.00 | calbindin 1 |
Detection of DSGs and AS events
Five basic types of AS events were classified, including A5SS (alternative 5′splice site), A3SS (alternative 3′splice site), SE (skipped exon), RI (retained intron) and MXE (mutually exclusive exon). The results showed in Table 5. We identified 63,837/64,075 AS events including splice junctions only (JC) and splice junctions and reads on target (JC + ROT) in 11,414/11,468 genes from XS and XL datasets. Thus, approximately 69 % of 16,476 expressed protein-coding genes were subject to alternative splicing. The numbers of AS events were from 1 ~ 28 events (JC or JC + ROT) in a gene. The highest number of alternative splicing event was found out from gene ARHGEF7 (ENSSSCG00000009551) with 28 events. SE was the most prevalent AS event, followed by MXE, and RI. Compared with other AS forms, the high frequency of SE indicated that the manner of skipped exon significantly might impact transcription and resulted in various isoforms during gene transcription. A total of 4,009 / 7,441 (JC / (JC + ROT)) significant differential alternative splicing events were identified from 2,763 / 3,936 genes (Table S5). Of these, 542 differently spliced genes (DSG) also exhibited differently expressing (Table S4). Compared with XL group, the number of up-regulated AS events was significantly more than that of down-regulated events in XS group (Table 5).
Table 5.
The types of AS events in ovaries of Xiang pig with small and large litter size
| EventType | NumEvents.JC.only | SigEvents.JC.only | up | down | NumEvents.JC + ROT | SigEvents. JC + ROT | up | down |
|---|---|---|---|---|---|---|---|---|
| SE | 49,441 | 1353 | 643 | 710 | 49,634 | 2325 | 1723 | 602 |
| MXE | 11,177 | 3386 | 1749 | 1637 | 11,198 | 6160 | 3201 | 2959 |
| A5SS | 622 | 169 | 155 | 14 | 624 | 202 | 191 | 11 |
| A3SS | 837 | 152 | 138 | 14 | 837 | 184 | 173 | 11 |
| RI | 1760 | 1077 | 1076 | 1 | 1782 | 1116 | 1115 | 1 |
| Total | 63,837 | 6137 | 3761 | 2376 | 64,075 | 9987 | 6403 | 3584 |
Notes: NumEvents.JC.only: total number of events detected using Junction Counts only. SigEvents.JC.only: number of significant events detected using Junction Counts only. NumEvents .JC + ROT (ReadsOnTarget): total number of events detected using both Junction Counts and reads on target. SigEvents. JC + ROT (ReadsOnTarget): number of significant events detected using both Junction Counts and reads on target
Compare of DSGs between Xiang and Yorkshire pig
Based on Venn results of the shared genes between Xiang pig and Large White sows [31], 1,597 DSGs from ovaries of Xiang pigs (Table S6) were also detected as much as 2,236 events by using single-molecule long-read sequencing (SMRT) in 39 tissues of Large White sows [31].
Changes in expression and AS of the reproduction genes between XS and XL
To understand the possible effects of the variations in expression and AS types of the reproduction genes on the litter size trait, crowds of major reproductive genes were picked out and explored the difference in expression pattern and AS of these genes in two groups. The RNA-seq data from 162 genes involved in reproduction processes were listed in Table S7. We found that 22 genes, such as ESR1, ESR2, GNRH1, FSHR, AR, GDF5, IRS1, CCND2, and so on, were down-regulated, and 33 genes, such as PTGS2, LIF, ECM1, BMPR1B, GPX3, C4BPA, MMP19, MMP25, STAT3, ect, were up-regulated in the XL group. The alternative splicing analysis indicated that 24 / 86 (JC / JC + ROT) significant differential AS events were presented in 42 reproduction related genes. However, of these, only 11 DEGs were also differential splicing, including AR, G6PD, ESR1, ECM1, BMPR1B, HEXB, STAT3, DNMT1, C4BPA, MMP23B, and LIN9 (Table 6).
Table 6.
The eleven genes related with reproduction harboring differential expression and splicing
| No. | Gene name | Gene ID | Event | ∆PSI | LogFC | Description |
|---|---|---|---|---|---|---|
| 1 | AR | ENSSSCG00000012371 | MXE-1 | -0.253 | -1.56 | androgen receptor |
| MXE-2 | 0.683 | |||||
| 2 | G6PD | ENSSSCG00000025108 | MXE | -0.174 | 1.85 | glucose-6-phosphate dehydrogenase |
| 3 | ESR1 | ENSSSCG00000025777 | MXE-1 | -0.126 | -1.48 | estrogen receptor 1 |
| MXE-2 | 0.139 | |||||
| MXE-3 | -0.174 | |||||
| MXE-4 | -0.112 | |||||
| MXE-5 | 0.364 | |||||
| 4 | ECM1 | ENSSSCG00000029230 | MXE | 0.182 | 3.53 | extracellular matrix protein 1 |
| 5 | BMPR1B | ENSSSCG00000029621 | MXE-1 | 0.152 | 2.27 | bone morphogenetic protein receptor type 1B |
| MXE-2 | 0.441 | |||||
| 6 | HEXB | ENSSSCG00000014073 | RI | 0.209 | 1.70 | hexosaminidase subunit beta |
| MXE | -0.132 | |||||
| 7 | STAT3 | ENSSSCG00000017403 | RI | 0.096 | 1.19 | signal transducer and activator of transcription 3 |
| 8 | DNMT1 | ENSSSCG00000013659 | MXE | 0.325 | -1.29 | DNA methyltransferase 1 |
| 9 | C4BPA | ENSSSCG00000015662 | MXE | -0.275 | 3.45 | complement component 4 binding protein, alpha |
| 10 | MMP23B | ENSSSCG00000003351 | MXE | 0.156 | 1.88 | matrix metallopeptidase 23B |
| 11 | LIN9 | ENSSSCG00000021310 | RI | 0.118 | -0.81 | lin-9 DREAM MuvB core complex component |
Gene ontology and KEGG analysis
To explain the biological effects of DEGs, we carried out GO and KEGG enrichment analysis (Table S3). For the up-regulated genes between XL and XS groups, 59 significantly enriched KEGG pathways were identified (corrected P-value < 0.05), including metabolism pathways (carbon metabolism, citrate cycle (TCA cycle), amino acid metabolism, glycerophospholipid metabolism, cholesterol metabolism etc.), oxidative phosphorylation, illness pathways (rheumatoid arthritis, Parkinson disease, colorectal cancer, type I diabetes mellitus, insulin resistance, hypertrophic cardiomyopathy etc.), physiological process related paths (renin-angiotensin system, complement and coagulation cascades, aldosterone synthesis and secretion, bile secretion, cardiac muscle contraction, adrenergic signaling in cardiomyocytes), immune paths (cell adhesion molecules, antigen processing and presentation), five signaling pathways (MAPK signaling pathway, NOD-like receptor signaling pathway, PI3K-Akt signaling pathway, TNF signaling pathway, PPAR signaling pathway). Notably, three pathways, ovarian steroidogenesis, steroid biosynthesis, FoxO signaling pathway, were included in the regulation of steroid hormone and ovary function. The 904 GO terms were enriched in kinds of physiology process. Significantly, 12 GO terms were gathered in reproductive process, female pregnancy, mammary gland epithelium development and proliferation, placenta development, blastocyst development and embryo development.
Meanwhile, the down-regulated genes enriched in two KEGG pathways, which were oocyte meiosis and progesterone-mediated oocyte maturation (corrected P-value < 0.05). And the GO terms gathered in meiosis I and meiosis I cell cycle processes. It illustrated that the DEGs between XL and XS groups were important in both function and development of reproductive system and hence were probably to contribute to litter size between two Xiang pig groups.
Very few KEGG paths and GO terms in both the up-regulated and down-regulated DSGs were significant at the level of correct P value less than 0.05 (Table S3). It was found that the up-regulated DSGs were enriched in 19 KEGG pathways and 181 GO terms if the threshold was reduced to the P value less than 0.05, including autophagy, endocytosis, and lysosome, phosphatidylinositol signaling system, metabolisms such as protein processing in endoplasmic reticulum, lysine degradation, fructose and mannose metabolism, fatty acid metabolism, ubiquitin mediated proteolysis etc. And the down-regulated DSGs could be enriched in 35 KEGG pathways and 219 GO terms including metabolism, growth cell process and reproduction etc. Of those, the GO term of utero embryonic development, and two KEGG pathways (oocyte meiosis, and progesterone-mediated oocyte maturation) were related with reproduction. These results indicated that the DSGs also participated the regulation processes on oocyte maturation and ovary function in pig.
Candidate genes related with litter size trait in Xiang pig
We performed the Venn analysis on the differently expressed genes, differently spliced genes, the higher expressed genes (CPM ≥ 100), the top expressed genes and the top differently expressed genes between XL and XS groups. Then, we selected these genes that overlapped in three or more datasets for the next GO and KEGG pathway analysis. According to the reported functions in any similar paths that relate to ovarian steroidogenesis, fecundity, pregnant or embryo development, we identified 12 candidate genes (StAR, DHRS4, RLN2, PTX3, HSD3B, MSMO1, SCARB1, COX1, COX3, SELENOP, CYP11A1, and NR4A1) having a linkage with pig reproduction capability and litter size. Of them, eleven candidate genes were identified to be hub genes connected with 12 to 35 genes based on the network relationship of DEGs by using string online platform (Table 7, Fig. S1).
Table 7.
The detection of eleven hub genes and the protein-protein network
| No. | Gene in node1 | Combined gene numbers | Genes in the node 2 |
|---|---|---|---|
| 1 | COX1 | 35 | AICDA,ATP6,ATP8,CAT,COX17,COX18,COX2,COX3,COX5A,COX5B,COX6A1,CYCS,CYTB,EXO1,HFM1,HSPA9,HSPD1,MRPS7,MT-ND2,ND1,ND3,ND4,ND4L,ND5,ND6,NDUFS2,NRF1,PNOC,POLD1,RAG2,REEP5,SDHC,SOD2,TCTP,UBC |
| 2 | COX3 | 23 | AICDA,ATP6,ATP8,COX1,COX2,COX5A,COX5B,COX6A1,CYCS,CYTB,HIGD1A,MT-ND2,ND1,ND3,ND4,ND4L,ND5,ND6,NDUFS2,PNOC,RPS12,SDHC,SOD2 |
| 3 | CYP11A1 | 17 | AR,CEL,CYB5A,CYP21A2,DHCR24,DHRS11,FSHR,GNRH1,HSD11B2,HSD17B1,HSD17B6,HSD17B7,HSD3B1,LIPA,SCARB1,SOAT1,STAR |
| 4 | DHRS4 | 20 | ACOT4,ALDH1A1,ALDH1A2,ARHGAP11A,CAT,CYB5A,CYTH4,DAO1,ECH1,EPHX2,GSTO2,HSD17B2,HSD17B4,IDH1,KIF4A,PECR,RDH12,RDH16,RETSAT,STX12 |
| 5 | HSD3B1 | 20 | CYB5A,CYP11A1,CYP21A2,CYP51,DHCR7,DHRS11,FDFT1,FSHR,HSD11B2,HSD17B1,HSD17B2,HSD17B4,HSD17B6,HSD17B7,LSS,MSMO1,SC5D,SQLE,STAR,TM7SF2 |
| 6 | MSMO1 | 25 | ACAT1,ACAT2,COL6A5,CPOX,CYB5A,CYP51,DHCR24,DHCR7,FDFT1,FDPS,HMGCR,HMGCS1,HSD17B12,HSD17B7,HSD3B1,INSIG1,LSS,MVK,NSDHL,SC5D,SCD,SCD5,SQLE,STARD4,TM7SF2 |
| 7 | NR4A1 | 13 | AR,ATF3,DUSP1,EGR1,EGR2,FOS,GRASP,MAPK3,NOR-1,PCK1,RPS6KA3,RTN4,VEGFA |
| 8 | PTX3 | 20 | ARMC8,ASAH1,C1QA,C1QC,CEP290,CFP,CST3,CSTB,CTSH,CTSZ,F3,GGH,IDH1,NEU1,QPCT,SELP,TIMP2,TNFAIP6,VEGFA,YPEL5 |
| 9 | RLN | 12 | ADCY4,ADCY9,ADM,ADORA2B,ADRB1,FSHR,HTR4,KIAA1109,PTHLH,RAMP2,RLF,RXFP4 |
| 10 | SCARB1 | 16 | ABCA1,ABCG5,APOA1,CD63,CD81,CD82,CYP11A1,EPHA2,HMGCR,LDLR,OLR1,PLTP,PPARA,PPARG,STAR,TSPAN3 |
| 11 | STAR | 17 | ADCY9,AR,CYP11A1,CYP21A2,DHRS11,FSHR,GBA,GBI1,GNRH1,HSD17B1,HSD17B6,HSD3B1,SCARB1,STARD4,TSPO2,UGT8,VDAC1 |
Tests and verification
The trends of expression of nine genes via qRT-PCR detection were positively related to that from RNA-seq data (Fig. 4). And all of five types of AS events were detected out from the transcripts of ovaries (Fig. S2). It indicated that the analysis based on RNA-seq data was precise and effective.
Fig. 4.
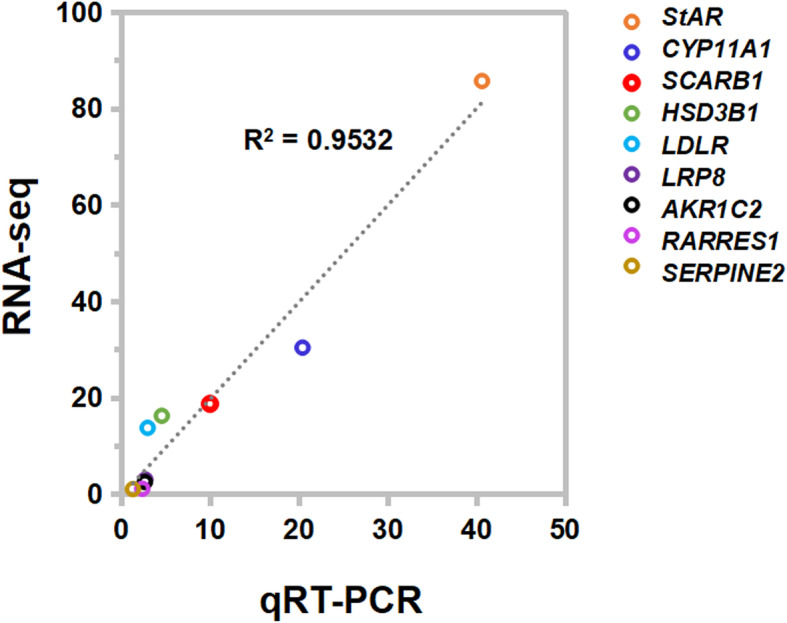
Validation of DEGs by qRT-PCR method. The trend was similar in fold change (XL/XS) from RNA-seq and the ratio of expression levels in groups by qRT-PCR method with the linear correlation coefficient of 0.9532, P = 0.037 based on two-tail T-test
Discussion
In this study, we analyzed the estrus ovarian transcriptome and AS of Xiang pig using Illumina next generation sequencing technology. We detected 16,476 genes that expressed in ovaries from libraries. Of these, there were 15,389 genes expressed in common between two groups (Fig. 1B). The expressed amounts of these genes were much diverse, with CPM values changed from 1 to more than twenty thousand (Table S2). Great amounts of genes expressed specifically in XL or XS group detected to be at low or very low level. Further, the top ten genes highly expressed in XL group were compared with that in XS groups (Table 2), and found that the expression levels of some genes increased in XL group, such as StAR, SELENOP, COX3, COX1, and so on. The genes with expression level ranked top ten occupied 5.04 ~ 6.96 % of the total expression values in XS and XL groups, respectively. It indicated that these high expressed genes were considerable necessary in the function and the development of ovary. For instance, the transport of cholesterol into mitochondria dependents on the effect of steroidogenic acute regulatory protein (StAR), which accelerates the transform of cholesterol into the inner membrane of mitochondrion to trigger steroidogenesis reaction [34]. In mitochondrion, cholesterol is changed into pregnenolone by the cytochrome P450 side-chain cleavage enzyme. And pregnenolone is further transformed into progesterone or dehydroepiandrosterone, two hormones essential for endometrial receptivity, embryo implantation, and the successful establishment of pregnancy [35]. Previous works indicate that selenium (Se) regulates the growth of granulosa cells together with 17-estradiol synthesis in ovary [36]. Another report showed that both of Se and its selenoprotein (SELENOP), as antioxidants, promote the growth and proliferation of granulosa cells [37].
Previous study for goat ovaries suggests that some special differently expressed genes based on RNA-Seq data may improve litter size [9]. In present work, we identified 2,795 DEGs, including 37 most differently expressed genes between XS and XL groups (Table 3). It indicated that these genes might be very important for the litter size of pig. Results from enrichment analysis indicated that the up- and down-regulated DEGs were clustered in many GO terms and pathways, including metabolism, growth, development, and reproduction. And the effects of the top thirty-seven DEGs between XL and XS groups mainly included ovarian steroidogenesis, metabolic pathways, oxidation-reduction process, negative regulation of endopeptidase activity. Compared with Yorkshire sow [17], 71 DEGs (Table 4) and both pathways (steroid biosynthesis and ovarian steroidogenesis) were shared with Xiang pigs (Table S3). Moreover, 15 genes were up-regulated in the group with high litter size of Xiang and Yorkshire ovaries (Table S3). Some of them are reported to have a pivotal role in ovary. For example, Cyp11a1 protein catalyzes the transformation from cholesterol to pregnenolone in mitochondrion in luteal cells. Both of StAR and Cyp11A1 genes are taken as two marker of corpus luteum in mice [38]. And the StAR gene governs the rate-limiting step in steroidogenesis described above [34]. The oxidase HSD3B promotes the oxidation of both delta 5-ene-3-beta-hydroxy steroid and the ketosteroids. The enzyme, 3-beta-HSD, is necessary in the anabolism for all kinds of steroid hormones [39]. As the receptor of HDL, SCARB1 participates in the optional absorption of cholesteryl ether together with the transport outside of HDL-dependent cholesterol, and even accelerates the flow of esterified or free cholesterol on cell surface together with modified lipoproteins [40]. MSMO1 takes part in the first reaction to remove the two C-4 methyl from molecule 4, 4-dimethylzymosterol [41].
Furthermore, the other DEGs listed in Table 3 were reported to have a close connection with ovary function, such as NR4A1, DHRS4, and PTX3. Nuclear receptor subfamily 4 group A member 1 (NR4A1) is an orphan receptor in nucleus, which regulates the transcription of androgen biosynthesis and the expression of paracrine factor insulin-like 3 (INSL3) in thecal cell of ovary. Androgens together with another hormone control the follicle growth in ovary [42]. NR4A1 distributes in many cells of ovary including theca cell, luteal cell, and granulosa cell in human. Furthermore, NR4A1 in Leydig cell is reported to affect the expression of gene StAR in mouse [43]. The dehydrogenase/reductase SDR family member 4 (DHRS4) gene, also known as NADPH-dependent retinol dehydrogenase/reductase (NRDR) gene, is a tetrameric protein that is pivotal to the biosynthesis of steroid hormone. DHRS4 functions as NADPH-dependent 3-ketosteroid reductase to produce the 3β-hydroxysteroids from 3-keto-C19/C21-steroids. Types of 3β-hydroxysteroids are reported to transmit signal and participate various physiology functions, such as binding to estrogen receptor β (ER-β) in nucleus and changing the development of prostate [44]. PTX3 is specifically expressed by cumulus cells around oocyte, and mediates the effect of LH or hCG in preovulatory follicle. PTX3 actively participates in the organization of the hyaluronan-rich provisional matrix required for successful fertilization. And PTX3 is taken as a biomarker of oocyte quality and has a role in oocyte maturity and female fertility based on gene deficiency mice [45].
These genes mentioned above, including Cyp11A1, StAR, HSD3B, SCARB1, MSMO1, NR4A1, PTX3, DHRS4 and so on (Tables 2 and 3), were all up-regulated in the ovaries of Xiang pigs with large litter size. It suggested that these genes might play important roles in promoting litter size by increasing the level of steroid and peptide hormones supply through the ovary and facilitating the oocyte ovulation and in vivo fertilization.
It is interested that many alternative splicing events from DEGs were detected based on comparison between XS and XL groups. About 69 % of all expressed genes contained AS events in both of XL and XS groups, which is much near to the AS rates in human [46]. Total of 1,597 nonredundant genes with differentially splicing in Xiang pig also detected many isoforms from tissues of Large White pigs [31] (Table S6). In DSGs of Xiang pig, skipped exon was the most prevalent AS events. The rates of AS events in XL group were not as high as that in XS group. AS is the main reason leading to change the different transcripts together with proteome varieties [47]. Numbers of reports indicate that alternative splicing interferes the functions of animal genes and alters the receptor structure especially in the processes of development and growth [48]. Lots of hereditary disease appear strong relationship with high frequency of alternative splicing in genes [49]. It was deduced herein that the high percentage of AS in pigs of XS group might cause the decrease of fecundity. Moreover, we found that 542 DEGs were differently spliced at AS levels between two groups (Table S4). The DEGs presented different and special patterns of splicing and events. Many tops differently expressed genes, such as StAR, MSMO1, SCARB1 and PDZK1IP1 showed high percentages of differently alternated splicing events (Table 2, Table S5). However, there were 3,693 genes only undergoing differently AS events between XL and XS groups. Therefore, the expression patterns and AS events of 162 genes related with reproductive processes were explored profoundly from the RNA-seq datasets. And eleven genes were found to be differently expressed and differently spliced in ovarian samples between XS and XL groups (Table S7). In addition, 31 reproductive genes only underwent differential splicing, such as CYP19A1 and FMR1. Estrogens are essential for animal fertility, which are catalyzed by aromatase enzyme coded by gene Cyp19a1 [50]. The gene encoding aromatase of mammals contains two promoters, including gonad specific and brain specific promoters. It exists 10 promoters at tissue-specific manner with the first exon to be chosen differently in diverse tissue cells. In kinds of promoters of CYP19A1 gene, the most vigorous one is promoter II (PII), which drives the transcription of aromatase gene in ovary [50]. The studies from rat find that the expression of aromatase transcription present the diverse and active regulation [51]. Gene FMR1 (FMRP translational regulator 1; FMRP: fragile X mental retardation protein) is composed of seventeen exons occupying about 38 kb in genome [52]. The gene undergoes extensive AS which changes the retain of four exons, 12, 14, 15 and 17, producing various FMR1 transcript isoforms, and some of FMRP isoforms have been reported in several species [52, 53]. In rat follicles, the FMR1 gene was transcribed in granulosa cell, theca cell and germ cell. FMR1 mRNA is much less in pre-ovulatory follicles than that in both preantral and antral follicles. FMRP content raises in the development process of follicles, and could be detected more than four bands by Western blotting method [53]. In present work, the expression of CYP19A1 mRNA isoform with MXE event was significantly down-regulated and FMR1 mRNA isoforms with A3SS event were significantly down-regulated in XL group (Table S5). These results indicated that the changes of gene expression between groups with large or small litter size were moderated at many ways and the splicing variants were highly controlled.
Finally, combined with DEGs, DSGs and the higher expressed genes via Venn analysis, we identified 12 candidate genes related with litter size in Xiang pig, including StAR, DHRS4, RLN2, PTX3, HSD3B, MSMO1, SCARB1, COX1, COX3, SELENOP, CYP11A1, and NR4A1. And eleven of them were identified to be hub genes based on the network relationship of DEGs (Table 7, Fig. S1). It indicated that these candidate genes might play important roles in the regulation of reproduction. The effects of StAR, DHRS4, RLN2, HSD3B, MSMO1, SCARB1, COX1, COX3, CYP11A1 and NR4A1 involve in ovarian hormone biosynthesis and regulation, which could regulate the processes of ovary development, oocytes mature and the quality of embryos. The functions of SELENOP are related to follicular growth and oocyte maturation [37]. Gene PTX3 increases the progress of oocyte ovulation and fertilization in vivo [45]. These genes expressed at a high level in ovaries of XL group and may accelerate ovarian hormone biosynthesis and the quality of oocytes. They might connect to the higher reproduction performance in Xiang pig. There are eleven candidate genes related with the litter size of Yorkshire breed reported from transcriptome analysis [17]. Interestingly, five of them (STAR, COX3, HSD3B, SCARB, CYP11A1) were also found to be related with the litter size trait in Xiang pig in this work. But the other candidates from Yorkshire breed are much different from Xiang pigs. It suggested that trait of litter size in pig breeds shared candidate genes and further controlled by diverse genes because of their various genetic background.
Conclusions
In short, this study showed a transcriptome pattern and AS profiles at estrus stage of ovaries from Xiang pigs. We identified 1,419 genes that showed up-regulated and 1,376 genes that appeared to be down-regulated in the large litter size samples. And it was found that approximately 69 % of expressed genes harbored AS treatment. Of 542 DEGs also exhibited differential alternative splicing. Based on previous finding on those genes, total of 12 candidate genes were found to be corresponding to the reproduction capability and litter size in Xiang pig. These genes play important roles in promoting litter size by increasing steroid and peptide hormones supply by ovary and facilitating the oocyte release and in vivo fertilization.
Supplementary Information
Additional file 1: Table S1. Primers for validation of DEGs and DSGs.
Additional file 2: Table S2. The expressed genes in the ovaries of Xiang pig with large and small litter size.
Additional file 3: Table S3. The KEGG and GO enrichment for DEGs and DSGs of Xiang pig and the compares with Yorkshire pig.
Additional file 4: Table S4. DEGs and DSGs between XS and XL groups.
Additional file 5: Table S5. The DSG analysis by rMATs.
Additional file 6: Table S6. Compare of DSGs between Xiang and Large White pigs.
Additional file 7: Table S7. DEGs and DSGs related with reproduction of pigs.
Additional file 8: Figure S1. Hub gene networks. Figure S2. Confirmation of five types AS events by RT-PCR and qRT-PCR methods.
Acknowledgements
The authors are grateful to Guizhou Dachang pig breeding farm, in Congjiang county, Guizhou province, China, for providing the animal materials. We would like to thank Prof. Franklin Y. and TopEdit LLC (https://www.topeditsci.com) for the linguistic editing and proofreading during the preparation of this manuscript.
Authors' contributions
Conceptualization, X.R., J.W.; data curation, F.H., X.N., L.Y., F.Z., L.T.; investigation, N.M., Y.R., F.Y., S.H.; writing–the draft, X.R.; writing–review and editing, J.W., S.L., J.L. All the authors have read and approved to the submitted and published version of the manuscript and agree to be personally accountable for the author’s own contributions.
Funding
This work are funded by the National Natural Science Foundation of China (31672390, 31960641), the Guizhou Province “Hundred” Innovative Talents Project [2016–4012], the Guizhou Agriculture Research program (QKHZC[2017]2585, QKHZC[2017]2587), the Guizhou Province Science and Technology Innovation Team Building Special (2019–5615), the National High Technology Research and Development Program of China (863 Program) [2013AA102503].
Availability of data and materials
The sequencing data is available from SRA database in NCBI with accession number PRJNA737004.
Declarations
Ethics approval and consent to participate
All the procedures were conducted in adherence with the the guidelines of Guizhou University Subcommittee of Experimental Animal Ethics with no. of EAE-GZU-2020-P002.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xueqin Ran, Email: xqran@gzu.edu.cn.
Jiafu Wang, Email: jfwang@gzu.edu.cn.
References
- 1.Li PH, Ma X, Zhang YQ, Zhang Q, Huang RH. Progress in the physiological and genetic mechanisms underlying the high prolificacy of the Erhualian pig. Yi Chuan. 2017;39(11):1016–24. doi: 10.16288/j.yczz.17-119. [DOI] [PubMed] [Google Scholar]
- 2.Kobek-Kjeldager C, Moustsen VA, Theil PK, Pedersen LJ. Effect of litter size, milk replacer and housing on production results of hyper-prolific sows. Animal. 2020;14(4):824–33. doi: 10.1017/S175173111900260X. [DOI] [PubMed] [Google Scholar]
- 3.Andersson E, Frössling J, Engblom L, Algers B, Gunnarsson S. Impact of litter size on sow stayability in Swedish commercial piglet producing herds. Acta Vet Scand. 2016;58(1):31. doi: 10.1186/s13028-016-0213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon WS, Rahman MS, Ryu DY, Khatun A, Pang MG. Comparison of markers predicting litter size in different pig breeds. Andrology. 2017;5(3):568–77. doi: 10.1111/andr.12332. [DOI] [PubMed] [Google Scholar]
- 5.Niu D, Ma X, Yuan T, Niu Y, Xu Y, Sun Z, et al. Porcine genome engineering for xenotransplantation. Adv Drug Deliv Rev. 2021;168:229–45. doi: 10.1016/j.addr.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Varona L, Legarra A, Herring W, Vitezica ZG. Genomic selection models for directional dominance: an example for litter size in pigs. Genet Sel Evol. 2018;50(1):1. doi: 10.1186/s12711-018-0374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X, Su G, Christensen OF, Janss L, Lund MS. Genome-wide association analyses using a Bayesian approach for litter size and piglet mortality in Danish Landrace and Yorkshire pigs. BMC Genom. 2016;17:468. doi: 10.1186/s12864-016-2806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamberlam G, Lapointe E, Abedini A, Rico C, Godin P, Paquet M, et al. SFRP4 is a negative regulator of ovarian follicle development and female fertility. Endocrinology. 2019;160(7):1561–72. doi: 10.1210/en.2019-00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling YH, Xiang H, Li YS, Liu Y, Zhang YH, Zhang ZJ, et al. Exploring differentially expressed genes in the ovaries of uniparous and multiparous goats using the RNA-Seq (Quantification) method. Gene. 2014;550(1):148–53. doi: 10.1016/j.gene.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Wijesena HR, Kachman SD, Lents CA, Riethoven JJ, Trenhaile-Grannemann MD, Safranski TJ, et al. Fine mapping genetic variants associated with age at puberty and sow fertility using SowPro90 genotyping array. J Anim Sci. 2020;98(10):skaa293. doi: 10.1093/jas/skaa293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sell-Kubiak E, Duijvesteijn N, Lopes MS, Janss LL, Knol EF, Bijma P, et al. Genome-wide association study reveals novel loci for litter size and its variability in a Large White pig population. BMC Genom. 2015;16:1049. doi: 10.1186/s12864-015-2273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai FN, Zhai HL, Cheng M, Ma JY, Cheng SF, Ge W, et al. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus) Sci Rep. 2016;6:38096. doi: 10.1038/srep38096.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer D, Laiho A, Gyenesei A, Sironen A. Identification of reproduction-related gene polymorphisms using whole transcriptome sequencing in the Large White pig population. G3 (Bethesda) 2015;5(7):1351–60. doi: 10.1534/g3.115.018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Fu J, Wang A. Expression of genes involved in progesterone receptor paracrine signaling and their effect on litter size in pigs. J Anim Sci Biotechnol. 2016;7:31. doi: 10.1186/s40104-016-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu Q, Zhou B, Xu F, Chen R, Shen C, Liang T, et al. Genome-wide differential mRNA expression profiles in follicles of two breeds and at two stages of estrus cycle of gilts. Sci Rep. 2017;7(1):5052. doi: 10.1038/s41598-017-04336-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muñoz M, Fernández AI, Ovilo C, Muñoz G, Rodriguez C, Fernández A, et al. Non-additive effects of RBP4, ESR1 and IGF2 polymorphisms on litter size at different parities in a Chinese-European porcine line. Genet Sel Evol. 2010;42(1):23. doi: 10.1186/1297-9686-42-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Huang L, Wu T, Feng Y, Ding Y, Ye P, et al. Transcriptomic analysis of ovaries from pigs with high and low litter size. PLoS One. 2015;10(10):e0139514. doi: 10.1371/journal.pone.0139514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Ye J, Han X, Qiao R, Li X, Lv G, et al. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics. 2020;112(1):199–206. doi: 10.1016/j.ygeno.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Li PH, Zhu MX, He LC, Sui SP, Gao S, et al. Genome-wide association analysis reveals genomic regions on chromosome 13 affecting litter size and candidate genes for uterine horn length in Erhualian pigs. Animal. 2018;12(12):2453–61. doi: 10.1017/S1751731118000332. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Ding X, Tan Z, Xing K, Yang T, Wang Y, et al. Genome-wide association study for reproductive traits in a Large White pig population. Anim Genet. 2018;49(2):127–31. doi: 10.1111/age.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Liu R, Su L, Xiao Q, Yu M. Transcriptome analysis revealed the embryo-induced gene expression patterns in the endometrium from Meishan and Yorkshire pigs. Int J Mol Sci. 2015;16(9):22692–710. doi: 10.3390/ijms160922692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Ran X, Wang J, Li S, Liu J. Detection of genomic structural variations in Guizhou indigenous pigs and the comparison with other breeds. PLoS One. 2018;13(3):e0194282. doi: 10.1371/journal.pone.0194282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo ZY, Dai XL, Ran XQ, Cen YX, Niu X, Li S, et al. Identification and profile of microRNAs in Xiang pig testes in four different ages detected by Solexa sequencing. Theriogenology. 2018;117:61–71. doi: 10.1016/j.theriogenology.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Ran X, Yu C, Xu Q, Niu X, Zhao P, et al. Whole-genome analysis of structural variations between Xiang pigs with larger litter sizes and those with smaller litter sizes. Genomics. 2019;111(3):310–9. doi: 10.1016/j.ygeno.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Knauer MT, Cassady JP, Newcom DW, See MT. Estimates of variance components for genetic correlations among swine estrus traits. J Anim Sci. 2010;88(9):2913–9. doi: 10.2527/jas.2009-2639. [DOI] [PubMed] [Google Scholar]
- 26.Tang LT, Ran XQ, Mao N, Zhang FP, Niu X, Ruan YQ, et al. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology. 2018;119:60–8. doi: 10.1016/j.theriogenology.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Lopes TP, Padilla L, Bolarin A, Rodriguez-Martinez H, Roca J. Ovarian follicle growth during lactation determines the reproductive performance of weaned sows. Animals (Basel) 2020;10:1012. doi: 10.3390/ani10061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui H, Jing R, Zhang M, Zhang Z. Development of reproductive organs and changes of plasma estradial-17β and testoterone in Xiang pigs. Zhongguo Xumu Zazhi. 1989;25(5):12–5. In Chinese ( https://xueshu.baidu.com/usercenter/paper/show?paperid=b5eca108fbec66292ea0e8f43a73d6f6&site=xueshu_se).
- 29.Liu RH, Li YH, Jiao LH, Wang XN, Wang H, Wang WH. Extracellular and intracellular factors affecting nuclear and cytoplasmic maturation of porcine oocytes collected from different sizes of follicles. Zygote. 2002;10(3):253–60. doi: 10.1017/s0967199402002332. [DOI] [PubMed] [Google Scholar]
- 30.Marchal R, Vigneron C, Perreau C, Bali-Papp A, Mermillod P. Effect of follicular size on meiotic and developmental competence of porcine oocytes. Theriogenology. 2002;57(5):1523–32. doi: 10.1016/s0093-691x(02)00655-6. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Fang C, Fu Y, Hu A, Li C, Zou C, et al. A survey of transcriptome complexity in Sus scrofa using single-molecule long-read sequencing. DNA Res. 2018;25(4):421–37. doi: 10.1093/dnares/dsy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran XQ, Pan H, Huang SH, Liu C, Niu X, Li S, et al. Copy number variations of MTHFSD gene across pig breeds and its association with litter size traits in Chinese indigenous Xiang pig. J Anim Physiol Anim Nutr (Berl) 2018;102(5):1320–7. doi: 10.1111/jpn.12922. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Galano M, Li Y, Li L, Sottas C, Papadopoulos V. Role of constitutive STAR in Leydig cells. Int J Mol Sci. 2021;22(4):2021. doi: 10.3390/ijms22042021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlak KJ, Prasad M, McKenzie KA, Wiebe JP, Gairola CG, Whittal RM, et al. Decreased cytochrome c oxidase IV expression reduces steroidogenesis. J Pharmacol Exp Ther. 2011;338(2):598–604. doi: 10.1124/jpet.111.182634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao X, Ei-Samahy MA, Fan L, Zheng L, Jin Y, Pang J, et al. In vitro influence of selenium on the proliferation of and steroidogenesis in goat luteinized granulosa cells. Theriogenology. 2018;114:70–80. doi: 10.1016/j.theriogenology.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Qazi IH, Angel C, Yang H, Pan B, Zoidis E, Zeng CJ, et al. Selenium, selenoproteins, and female reproduction: a review. Molecules. 2018;23(12):3053. doi: 10.3390/molecules23123053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan S, Xie L, Ma T, Lv D, Jing W, Tian X, et al. Effects of melatonin on early pregnancy in mouse: involving the regulation of StAR, Cyp11a1, and Ihh expression. Int J Mol Sci. 2017;18(8):1637. doi: 10.3390/ijms18081637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grindflek E, Berget I, Moe M, Oeth P, Lien S. Transcript profiling of candidate genes in testis of pigs exhibiting large differences in androstenone levels. BMC Genet. 2010;11:4. doi: 10.1186/1471-2156-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanislovaitiene D, Lesauskaite V, Zaliuniene D, Smalinskiene A, Gustiene O, Zaliaduonyte-Peksiene D, et al. SCARB1 single nucleotide polymorphism (rs5888) is associated with serum lipid profile and myocardial infarction in an age- and gender-dependent manner. Lipids Health Dis. 2013;12:24. doi: 10.1186/1476-511X-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalay Yildizhan I, Gökpınar İli E, Onoufriadis A, Kocyigit P, Kesidou E, Simpson MA, et al. New homozygous missense MSMO1 mutation in two siblings with SC4MOL deficiency presenting with psoriasiform dermatitis. Cytogenet Genome Res. 2020;160(9):523–30. doi: 10.1159/000511126. [DOI] [PubMed] [Google Scholar]
- 42.Xue K, Liu JY, Murphy BD, Tsang BK. Orphan nuclear receptor NR4A1 is a negative regulator of DHT-induced rat preantral follicular growth. Mol Endocrinol. 2012;26(12):2004–15. doi: 10.1210/me.2012-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teeli AS, Leszczyński P, Krishnaswamy N, Ogawa H, Tsuchiya M, Śmiech M, et al. Possible mechanisms for maintenance and regression of corpus luteum through the ubiquitin-proteasome and autophagy system regulated by transcriptional factors. Front Endocrinol (Lausanne) 2019;10:748. doi: 10.3389/fendo.2019.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kisiela M, Faust A, Ebert B, Maser E, Scheidig AJ. Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans. FEBS J. 2018;285(2):275–93. doi: 10.1111/febs.14337. [DOI] [PubMed] [Google Scholar]
- 45.Camaioni A, Klinger FG, Campagnolo L, Salustri A. The Influence of pentraxin 3 on the ovarian function and its impact on fertility. Front Immunol. 2018;9:2808. doi: 10.3389/fimmu.2018.02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Codreanu SG, Wen B, Li K, Chambers MC, Liebler DC, et al. Detection of proteome diversity resulted from alternative splicing is limited by trypsin cleavage specificity. Mol Cell Proteomics. 2018;17(3):422–30. doi: 10.1074/mcp.RA117.000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chacko E, Ranganathan S. Genome-wide analysis of alternative splicing in cow: implications in bovine as a model for human diseases. BMC Genom. 2009;10(Suppl 3):11. doi: 10.1186/1471-2164-10-S3-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stocco C. Aromatase expression in the ovary: hormonal and molecular regulation. Steroids. 2008;73(5):473–87. doi: 10.1016/j.steroids.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Domingos JA, Budd AM, Banh QQ, Goldsbury JA, Zenger KR, Jerry DR. Sex-specific dmrt1 and cyp19a1 methylation and alternative splicing in gonads of the protandrous hermaphrodite barramundi. PLoS One. 2018;13(9):e0204182. doi: 10.1371/journal.pone.0204182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Barad DH, Darmon SK, Kushnir VA, Wu YG, Lazzaroni-Tealdi E, et al. Reduced RNA expression of the FMR1 gene in women with low (CGGn < 26) repeats. PLoS One. 2018;13(12):e0209309. doi: 10.1371/journal.pone.0209309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseng E, Tang HT, AlOlaby RR, Hickey L, Tassone F. Altered expression of the FMR1 splicing variants landscape in premutation carriers. Biochim Biophys Acta Gene Regul Mech. 2017;1860(11):1117–26. doi: 10.1016/j.bbagrm.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers for validation of DEGs and DSGs.
Additional file 2: Table S2. The expressed genes in the ovaries of Xiang pig with large and small litter size.
Additional file 3: Table S3. The KEGG and GO enrichment for DEGs and DSGs of Xiang pig and the compares with Yorkshire pig.
Additional file 4: Table S4. DEGs and DSGs between XS and XL groups.
Additional file 5: Table S5. The DSG analysis by rMATs.
Additional file 6: Table S6. Compare of DSGs between Xiang and Large White pigs.
Additional file 7: Table S7. DEGs and DSGs related with reproduction of pigs.
Additional file 8: Figure S1. Hub gene networks. Figure S2. Confirmation of five types AS events by RT-PCR and qRT-PCR methods.
Data Availability Statement
The sequencing data is available from SRA database in NCBI with accession number PRJNA737004.