Abstract
Objectives
Adipose-derived stem cells are frequently used for bone regeneration both in vitro and in vivo. N6-methyladenosine (m6A) is the most abundant post-transcriptional modification on eukaryotic RNAs and plays multifaceted roles in development and diseases. However, the regulatory mechanisms of m6A in osteogenic differentiation of human adipose-derived stem cells (hASCs) remain elusive. The present study aimed to build the transcriptome-wide m6A methylome during the osteogenic differentiation of hASCs.
Materials and methods
hASCs were harvested after being cultured in a basic or osteogenic medium for 7 days, and the osteogenic differentiation was validated by alkaline phosphatase (ALP) and Alizarin Red S staining, ALP activity assay, and qRT-PCR analysis of ALP, RUNX2, BGLAP, SPP1, SP7, and COL1A1 genes. The m6A level was colorimetrically measured, and the expression of m6A regulators was confirmed by qRT-PCR and western blot. Moreover, m6A MeRIP-seq and RNA-seq were performed to build the transcriptome and m6A methylome. Furthermore, bioinformatic analyses including volcano plots, Venn plots, clustering analysis, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, gene sets enrichment analysis, and protein-protein interaction analysis were conducted.
Results
In total, 1145 differentially methylated peaks, 2261 differentially expressed genes, and 671 differentially methylated and expressed genes (DMEGs) were identified. GO and KEGG pathway analyses conducted for these DMEGs revealed extensive and osteogenic biological functions. The “PI3K-Akt signaling pathway”; “MAPK signaling pathway”; “parathyroid hormone synthesis, secretion, and action”; and “p53 signaling pathway” were significantly enriched, and the DMEGs in these pathways were identified as m6A-specific key genes. A protein-protein interaction network based on DMEGs was built, and VEGFA, CD44, MMP2, HGF, and SPARC were speculated as the hub DMEGs.
Conclusions
The total m6A level was reduced with osteogenic differentiation of hASCs. The transcriptome-wide m6A methylome built in the present study indicated quite a few signaling pathways, and hub genes were influenced by m6A modification. Future studies based on these epigenetic clues could promote understanding of the mechanisms of osteogenic differentiation of hASCs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-021-02508-1.
Keywords: Adipose-derived stem cell, m6A, MeRIP-seq, N6-methyladenosine, Osteogenic differentiation, RNA-seq
Introduction
Large bone defects, caused by trauma, bone loss, and tumors, lead to heavy social and economic burdens. At present, the clinical gold standard for the treatment of skeletal defects is autogenous or allogeneic bone grafts, which suffers disadvantages of a limited amount of harvested bone and donor site morbidity [1, 2]. Bone tissue engineering, built on stem cells or osteoprogenitor cells, osteoconductive biomaterials, and osteoconductive cytokines facilitating cellular proliferation and osteogenic differentiation, offers a promising solution [2, 3]. Human bone marrow-derived mesenchymal stem cells (hBMSCs), naturally resident in the bone marrow, are pluripotent and have been widely applied in bone tissue engineering for years [4, 5]. However, the access to autogenous hBMSCs is invasive and subject to limited cell incidence. What is more, the self-renewal and proliferative ability of hBMSCs weakens with donor aging and diseases such as osteoporosis and arthritis [6–8].
Adipose stromal cells, available from lipoaspirate, are a cellular niche containing differentiated cells, committed progenitors, and mesenchymal stem cells [9]. Among the multiple cell types, adipose-derived stem cells (ADSCs) are pluripotent, with adipogenic, chondrogenic, neurogenic, and osteogenic potentials. In the presence of ascorbate, glycerophosphate, and dexamethasone, ADSCs can differentiate into osteoblast-like cells [10–12]. Meanwhile, ADSCs have high proliferation rates and immunosuppressive properties and can secrete numerous polypeptides, hormones, and effective growth factors to induce osteogenesis. Moreover, ADSCs can also secrete a mass of suitable chemokines to recruit endogenous stem cells into the bone defect site [12–14]. With the increasing incidence of obesity worldwide, subcutaneous adipose tissue is easy to obtain, making human adipose-derived stem cells (hASCs) a first-rate alternative for hBMSCs in bone regenerative medicine [11].
Eukaryotic RNAs can be modified post-transcriptionally by over 170 modifications, among which, N6-methyladenosine (m6A), mostly resident at the stop codon and the 3′ untranslated region (UTR), is the most abundant [15, 16]. The m6A modification can be dynamically regulated by methyltransferases (also known as “writers”, including METTL3, METTL14, etc.) and demethylases (also known as “erasers,” including FTO, ALKBH5) and can further be recognized by specific RNA-binding proteins (also known as “readers,” including YTHDF1, YTHDF2) [17, 18]. As the first known form of reversible mRNA modification, m6A exerts effects on the stability, alternative splicing, export, cellular localization, and translation of RNAs, therefore influencing gene expression [17–20]. Till now, this epigenetic mark on RNAs has been extensively reported in virtually all major biological processes, normal development, and various diseases [21–26].
In the case of bone biology, m6A modification plays critical roles in bone development as well as bone diseases such as osteoporosis, osteoarthritis, and osteosarcoma [27, 28]. Mechanistically, m6A was found executing dual functions in the osteogenic differentiation of MSCs. On the one hand, m6A methylation enhanced osteogenic differentiation by regulating miR-320, RUNX2, Pth1r, PI3K/Akt, VEGF, or SMAD7/SMURF1 signaling, and inhibited adipogenic differentiation by regulating JAK1, Pth1r, SRSF2/ RUNX1T1, or TRAF4/ PKM2/β-catenin signaling [27–30]. On the other hand, the m6A methylation was also reported to decrease osteogenic differentiation by regulation of MYD88/NF-κB, miR-7212-5p/FGFR3, or FTO/p-AMPK loop [27, 28, 31]. However, all these findings above are rooted in BMSCs, and little is known about the scenario in ADSCs at the present stage.
In the present study, we aimed to investigate the roles of m6A in the osteogenic differentiation of hASCs. High-throughput sequencing for RNA (RNA-seq) and methylated RNA immunoprecipitation and sequencing (m6A MeRIP-Seq) were performed to comprehensively identify differentially regulated m6A peaks and messenger RNAs between two groups: undifferentiated hASCs (uhASCs) and osteogenically differentiated hASCs (dhASCs). The expressions of osteogenic markers, m6A regulating genes, and hub genes were validated by quantitative real-time polymerase chain reaction (qRT-PCR). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for the differentially methylated genes (DMGs), differentially expressed genes (DEGs), and differentially methylated and expressed genes (DMEGs) to reveal the biological significance of m6A in osteogenic differentiation of hASCs. Moreover, we also built the protein-protein interaction (PPI) network based on the DMEGs and calculated the hub genes in this process. These findings could provide a new perspective on the molecular mechanisms of osteogenic differentiation in hASCs.
Materials and methods
Cell culture
The primary hASCs from healthy adult donors in this study were obtained from Cyagen Biosciences (Suzhou, China). The cells were cultured with OriCell™ Human ASC Growth Medium (Cyagen) containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% glutamine. The medium was renewed every 3 days until the cells were passaged upon approximately 80% confluence. All the cells were cultured in an incubator with 5% CO2 at 37 °C. As our preliminary study indicated, the hASCs expressed a high level of stem cell surface markers CD29, CD44, CD105, and HLA-ABC, while less than 5% expressed CD31, CD34, CD45, and HLA-DR [32]. The third to the seventh generation of hASCs were used for osteogenic differentiation.
Osteogenic differentiation
The hASCs were seeded at a density of 2.0 × 105 and 1.0 × 105 cells/well into six- and twelve-well plates, respectively. For osteogenic differentiation, hASCs were cultured in OriCell™ hASC Osteogenic Differentiation Medium (Cyagen) containing 10% fetal bovine serum, 1% penicillin-streptomycin, 10 mM β-glycerophosphate, 10 mM glutamine, 50 μM ascorbate, and 0.1 μM dexamethasone. For the control group, hASCs were cultured in the general growth medium as mentioned before. The medium was renewed every 2 days. Both groups were detected by alkaline phosphatase (ALP) and Alizarin Red S (ARS) staining, ALP activity assay, and qRT-PCR analysis of ALP, RUNX2, BGLAP, SPP1, SP7, and COL1A1 genes [33–35].
ALP and ARS staining
ALP staining was conducted after 10 days of induction of osteogenic differentiation. Cells were fixed with 4% polyoxymethylene for 15 min and incubated with 0.1 M Tris buffer (pH 9.3) containing 0.25% naphthol AS-BI phosphate (Sigma) and 0.75% Fast Blue BB (Sigma) at 37 °C for 45 min. The Alkaline Phosphatase Assay Kit (Beyotime, China) was adopted according to the manufacturer’s instruction, and the ALP activity was quantified by a spectrophotometer (Thermo Fisher Scientific) at 405 nm.
ARS staining was conducted after 14 days of induction of osteogenic differentiation. Cells were fixed with 4% polyoxymethylene for 15 min and stained with 1% Alizarin Red S (ARS) Stain Solution (Cyagen) at room temperature for 5 min. And then, the solution was removed, and the samples were rinsed 3–4 times with deionized water. Alizarin Red-positive area was analyzed using the ImageJ software (NIH) and shown as a percentage of Alizarin Red-positive area over the total area.
Quantitative real-time polymerase chain reaction analysis
The expressions of osteogenic markers, m6A regulating genes, and hub DMEGs were validated by qRT-PCR. Total RNA from hASCs was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. The RNA purity and concentration were measured with a NanoDrop 2000 (Thermo Fisher Scientific). Reverse transcription was performed by PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Japan). qRT-PCR reaction was performed using TB Green™ Premix Ex Taq™ II (Takara) in QuantStudio 3 Real-Time PCR Systems (Thermo Fisher Scientific). Relative gene expression was normalized by GAPDH using a 2−ΔΔCt method. The primers used in this study are listed in Supplementary Table S1.
Western blot
Cells after 7 days of osteogenic induction were lysed in RIPA buffer (Beyotime, China) on ice. The samples were heated at 95 °C for 5 min in a sample buffer containing 20% SDS-PAGE sample loading buffer, separated on 10% SDS-polyacrylamide gels, and transferred to the PVDF membranes by a wet transfer apparatus (Bio-Rad). The membranes were blocked with 5% BSA for 1 h and then incubated overnight with rabbit METTL3 polyclonal antibody (1:1000, Cell Signaling Technology, Cat No: 96391), METTL14 polyclonal antibody (1:1000, ABclonal, Cat No: A8530), FTO polyclonal antibody (1:1000, Affinity, Cat No: DF8421), ALKBH5 polyclonal antibody (1:2000, Proteintech, Cat No: 16837-1-AP), and α-tubulin rabbit polyclonal antibody (1:2000, Beyotime, Cat No: AF0001), followed by incubation with a goat anti-rabbit IgG secondary antibody HRP conjugated (1:5000, Signaling antibody, Cat No: L3012-2). The antibody-antigen complexes were visualized with Bio-Rad Quantity One (Bio-Rad Laboratories Inc., Hercules, CA, USA).
m6A level quantification
Total RNA from hASCs was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. The RNA purity and concentration were measured with a NanoDrop 2000 (Thermo Fisher Scientific). The m6A RNA Methylation Quantification Kit (P-9005, EpiQuik) was used to measure the m6A content in the total RNAs following the manufacturer’s instruction. The m6A levels were quantified colorimetrically by reading the absorbance of each well at a wavelength of 450 nm, and the m6A level was then calculated based on the standard curve.
m6A MeRIP-Seq and RNA-seq
The m6A MeRIP-Seq and RNA-Seq were performed after 7 days of induced osteogenic differentiation. Total RNA was extracted using TRIzol Reagent (Invitrogen, CA, USA) and was tested for quality and quantity with Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number > 7.0. Then, poly-T oligo-attached magnetic beads (Invitrogen) were used to isolate poly(A) mRNA from the total RNA. Next, the poly(A) mRNA fractions are fragmented into ~ 100-nt-long oligonucleotides and then incubated for 2 h at 4 °C with m6A-specific antibody (No. 202003, Synaptic Systems, Germany) in IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% Igepal CA-630) supplemented with BSA (0.5 μg μl−1). The mixture was then incubated with protein-A beads and eluted with elution buffer (1 × IP buffer and 6.7 mM m6A). Eluted RNA was precipitated by 75% ethanol. Both eluted m6A-containing fragments (IP) and untreated control fragments (input) are converted to the final cDNA library. The average insert size for the paired-end libraries was ~ 100 ± 50 bp. And then the paired-end 2 × 150 bp sequencing was performed on an Illumina Novaseq™ 6000 platform at the LC-BIO Bio-Tech Ltd. (Hangzhou, China) following the vendor’s recommended protocol.
Data analysis
First, we used Cutadapt and perl scripts inhouse to remove the reads containing adaptor contamination, low-quality bases, and undetermined bases. Then, sequence quality was validated by fastp. Next, HISAT2 was used to map reads to the genome of Homo sapiens (version: v96). The R package exomePeak was adopted to process mapped reads of IP and input libraries and identifies m6A peaks with bed or bam format that can be adapted for visualization on the IGV software (http://www.igv.org/). HOMER and MEME were used for de novo and known motif finding followed by localization of the motif with respect to peak summit by perl. Called peaks were annotated by intersection with gene architecture using ChIPseeker. Then, StringTie was used to perform the expression level for all mRNAs from input libraries by calculating FPKM. The differentially expressed mRNAs were selected with the standard of p-value < 0.05 and log2 (fold change) > 1 or < −1 and by the R package edgeR.
Venn analysis
Venn analysis was performed to characterize overlapped dysregulated m6A peaks and mRNAs between dhASCs and uhASCs detected by MeRIP-Seq and RNA-Seq. The number of m6A peaks and mRNAs with or without overlapping were shown in different colors in a pie diagram. Up- and downregulated m6A peaks and mRNAs were analyzed separately.
GO and KEGG analyses
GO and KEGG pathway analyses were performed on m6A peaks and mRNAs. GO analysis, including molecular function, biologic process, and cellular component, was performed. p < 0.05 denoted the significance of GO term enrichment. Based on the KEGG database, pathway analysis was performed to analyze the potential functions. GO and pathway enrichment analyses were also performed based on the differentially regulated m6A peaks and mRNAs.
Gene set enrichment analysis
For gene set enrichment analysis (GSEA), gene lists “c2.cp.kegg.v7.3.symbols” were obtained from MSigDB, and the analysis was performed with the GSEA software (http://www.broad.mit.edu/GSEA). In short, we imported our gene lists of interest into the software and examined the significance of the given gene sets. p-values were computed using a bootstrap distribution created by resampling gene sets of the same cardinality.
Construction of the protein-protein interaction network
The protein-protein interaction (PPI) network was constructed to assess the interactions between the differentially methylated and expressed genes (DMEGs) based on the correlation analysis. All the DMEGs were selected to construct the network using Cytoscape 3.8 (Institute of Systems Biology in Seattle). Then, we calculated the hub genes by combining scores from all DMEGs.
Statistical analysis
Statistical tests were performed using R version 3.6.0, SPSS 24.0 (IBM, NY, USA) and GraphPad Prism 8.0. Numerical data was presented as means ± standard deviation. Differences between the two groups were evaluated for statistical significance by a t-test. The appropriate analysis of variance was used when more than two groups were evaluated, followed by Dunnett’s multiple comparisons test to compare the difference between the groups. p ≤ .05 was considered statistically significant. The R packages VennDiagram, ComplexHeatmap, clusterProfiler, and ggplot2 were used for graphing and data visualization.
Results
Osteogenic differentiation of hASCs was successfully induced.
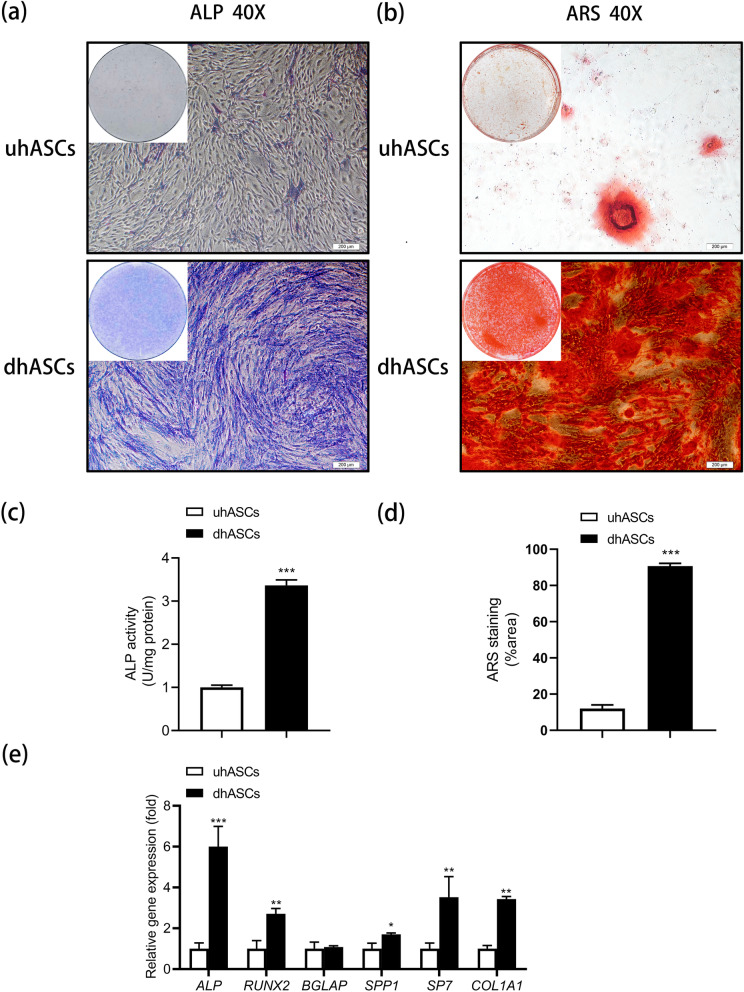
The ALP activity, validated by ALP staining and ALP activity quantification assay, was significantly higher in the dhASCs group than in the uhASCs group. A similar result regarding mineral nodes forming was observed in ARS staining (Fig. 1a–d). Moreover, compared to the control group, the expressions of ALP, RUNX2, SPP1, SP7, and COL1A1 were significantly increased in the dhASCs group (Fig. 1e). These results confirmed the osteogenic differentiation of dhASCs.
Fig. 1.
Osteogenic differentiation of hASCs was successfully induced. a Representative images of ALP staining. b Representative images of ARS staining. c Quantitative analyses of ALP activity. d Quantitative analyses of ARS staining. e qRT-PCR analyses of the expression of ALP, RUNX2, BGLAP, SPP1, SP7, and COL1A1 genes. *p < .05, **p < .01, ***p < .001. ALP, alkaline phosphatase; ARS, Alizarin Red S; hASCs, human adipose-derived stem cells; qRT-PCR, quantitative real-time polymerase chain reaction; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs
m6A level significantly decreased during osteogenic differentiation of hASCs
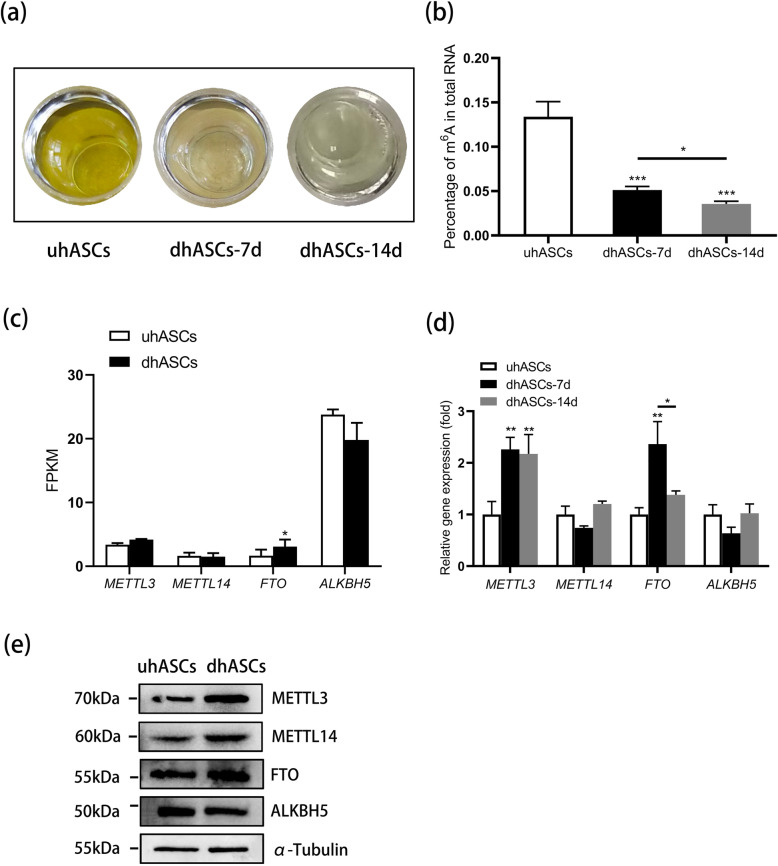
To learn about the m6A methylation in the process of osteogenic differentiation of hASCs, we first detected the relative m6A level between the groups. The m6A quantification assay analysis revealed a significantly reduced m6A level during the osteogenic induction of hASCs from day 0 to day 14, with the most significant and steepest reduction at the early stage (Fig. 2a, b). Then, we investigated the expression of m6A regulators. According to the RNA-seq, compared with the uhASCs group, FTO expression was significantly increased in the dhASCs group at day 7. However, the expression of METTL3, METTL14, and ALKBH5 showed no significant differences between the groups (Fig. 2c). Further, the qRT-PCR revealed the expression of METTL3 was significantly elevated at day 7 and day 14, and the expression of FTO was significantly elevated at day 7 and significantly decreased to baseline at day 14 (Fig. 2d). Moreover, the expressions of METTL3, METTL14, FTO, and ALKBH5 between the groups were confirmed by western blotting (Fig. 2e). In a word, these results demonstrated potential effects of m6A in the osteogenic differentiation of hASCs.
Fig. 2.
m6A level significantly decreased during osteogenic differentiation of hASCs. a Representative image of m6A level quantification assay. b Quantitative analyses of m6A level. c Expression of m6A regulators during the osteogenic differentiation of hASCs by RNA-seq (uhASCs vs dhASCs-7 day). d qRT-PCR analyses of the expression of METTL3, METTL14, FTO, and ALKBH5. e Western blot analysis of METTL3, METTL14, FTO, and ALKBH5 (uhASCs vs dhASCs-7 day). *p < .05, **p < .01, ***p < .001. hASCs, human adipose-derived stem cells; qRT-PCR, quantitative real-time polymerase chain reaction; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs
m6A peaks were significantly dysregulated during osteogenesis of hASCs
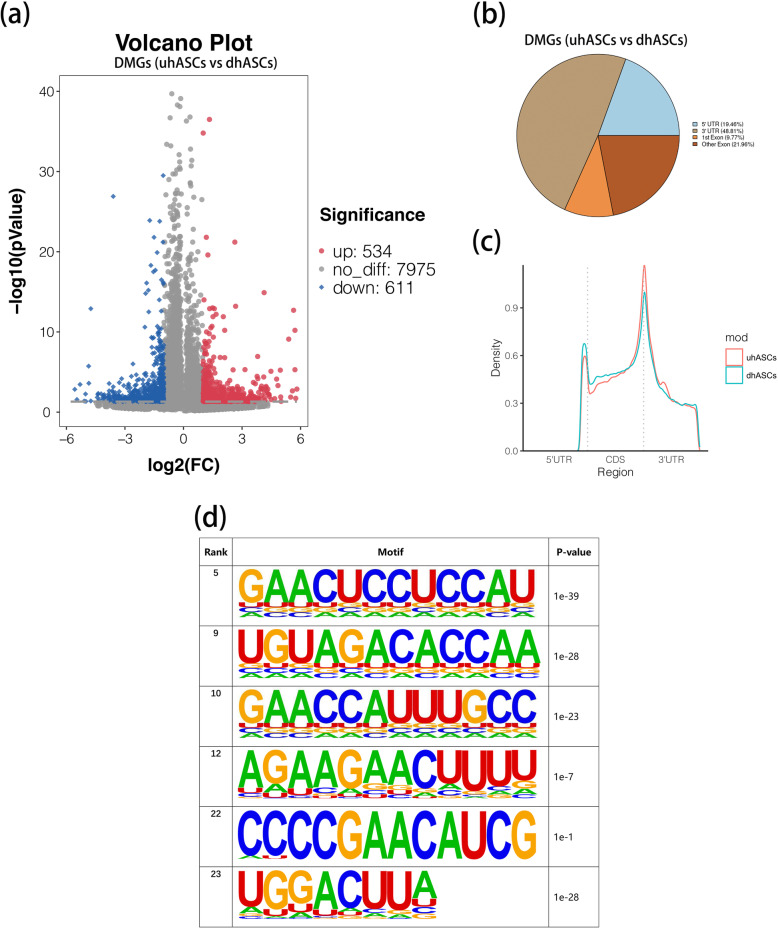
To dissect the underlying roles of m6A in the osteogenesis of hASCs, we performed the m6A MeRIP-Seq to depict the critical picture of m6A methylome. For the whole transcriptome, 39,735 (20431 transcripts) and 44,630 (22789 transcripts) m6A peaks were detected in uhASCs and dhASCs, respectively. For uhASCs, 45.73% of the peaks were detected in the 3′UTR region, 18.39% in the 5′UTR region, 10.6% in the first exon, and 25.28% in the remaining exons (Supplementary Figure S1a-b). For dhASCs, 43.85% of the peaks were detected in the 3′UTR region, 20.48% in the 5′UTR region, 11.06% in the first exon, and 24.61% in the remaining exons (Supplementary Figure S1c-d). According to the MeRIP-Seq, by the standard of p-value < 0.05 and fold change (fc) ≥ 2, 1145 m6A peaks were significantly dysregulated after a 7-day osteogenic induction of hASCs, of which 534 peaks were hypermethylated and 611 hypomethylated (Fig. 3a). In addition, 48.81% of the dysregulated peaks were in the 3′UTR region, 19.46% in the 5′UTR region, 9.77% in the first exon, and 21.96% in the remaining exons (Fig. 3b, c). The top 20 DMGs were listed in Table 1, and the full list of DMGs was shown in Supplementary Table S2. Furthermore, the motif analysis validated m6A RRACH (R was purine, A was m6A, and H was a non-guanine bas) consensus sequence enrichment between uhASCs and dhASCs (Fig. 3d).
Fig. 3.
m6A peaks were significantly dysregulated during osteogenesis of hASCs. a Volcano plot displaying the DMGs (fold change ≥ 2 and p < 0.05). b Pie chart showing distribution of differentially methylated m6A peaks in different gene contexts. c Accumulation of differentially methylated m6A peaks along transcripts. d The m6A motifs significantly enriched across m6A peaks identified between uhASCs and dhASCs. DMGs, differentially methylated genes; hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs; FC, fold change
Table 1.
The top 20 differently methylated m6A peaks (uhASCs/dhASCs)
| Gene | Transcript ID | Chr | Start | End | p-value | FC | Regulation |
|---|---|---|---|---|---|---|---|
| ASCL2 | ENST00000331289 | chr11 | 2268644 | 2268853 | 0.00 | 56.10 | Hyper |
| DTX2P1-UPK3BP1-PMS2P11 | ENST00000636308 | chr7 | 77024167 | 77024406 | 0.02 | 53.82 | Hyper |
| OR2A9P | ENST00000484189 | chr7 | 144299355 | 144299534 | 0.00 | 52.35 | Hyper |
| SLX1B | ENST00000330181 | chr16 | 29457634 | 29457951 | 0.00 | 52.35 | Hyper |
| SLX1B | ENST00000330181 | chr16 | 29457636 | 29457873 | 0.00 | 49.87 | Hyper |
| C4A | ENST00000428956 | chr6 | 31983915 | 31984330 | 0.00 | 47.84 | Hyper |
| FP565260 | ENST00000623998 | chr21 | 5158279 | 5158428 | 0.01 | 44.94 | Hyper |
| MAGED4 | ENST00000599522 | chrX | 52187058 | 52187386 | 0.00 | 41.93 | Hyper |
| AC092296 | ENST00000589470 | chr19 | 36421976 | 36422123 | 0.05 | 32.00 | Hyper |
| AC002066 | ENST00000446355 | chr7 | 116276826 | 116277005 | 0.01 | 31.56 | Hyper |
| SERF1A | ENST00000317633 | chr5 | 70901805 | 70905325 | 0.00 | 31.12 | Hyper |
| NUDT4B | ENST00000322209 | chr1 | 148748953 | 148749247 | 0.00 | 0.02 | Hypo |
| CHST8 | ENST00000650847 | chr19 | 33772791 | 33773000 | 0.00 | 0.02 | Hypo |
| C11orf44 | ENST00000317019 | chr11 | 130715177 | 130715475 | 0.03 | 0.02 | Hypo |
| MYCL | ENST00000397332 | chr1 | 39895965 | 39896534 | 0.00 | 0.03 | Hypo |
| GOSR2 | ENST00000640792 | chr17 | 46964434 | 46964733 | 0.04 | 0.03 | Hypo |
| ATF3 | ENST00000366981 | chr1 | 212618623 | 212618743 | 0.01 | 0.03 | Hypo |
| AADAC | ENST00000232892 | chr3 | 151814103 | 151814251 | 0.05 | 0.03 | Hypo |
| GOLGA8N | ENST00000448387 | chr15 | 32599455 | 32599770 | 0.00 | 0.03 | Hypo |
| PSMC1 | ENST00000553835 | chr14 | 90267920 | 90268160 | 0.00 | 0.03 | Hypo |
| BSN | ENST00000296452 | chr3 | 49655753 | 49655873 | 0.04 | 0.04 | Hypo |
dhASCs, osteogenically differentiated human adipose-derived stem cells; uhASCs, undifferentiated human adipose-derived stem cells; FC, fold change
The DMGs were associated with extensive and osteogenic biological functions
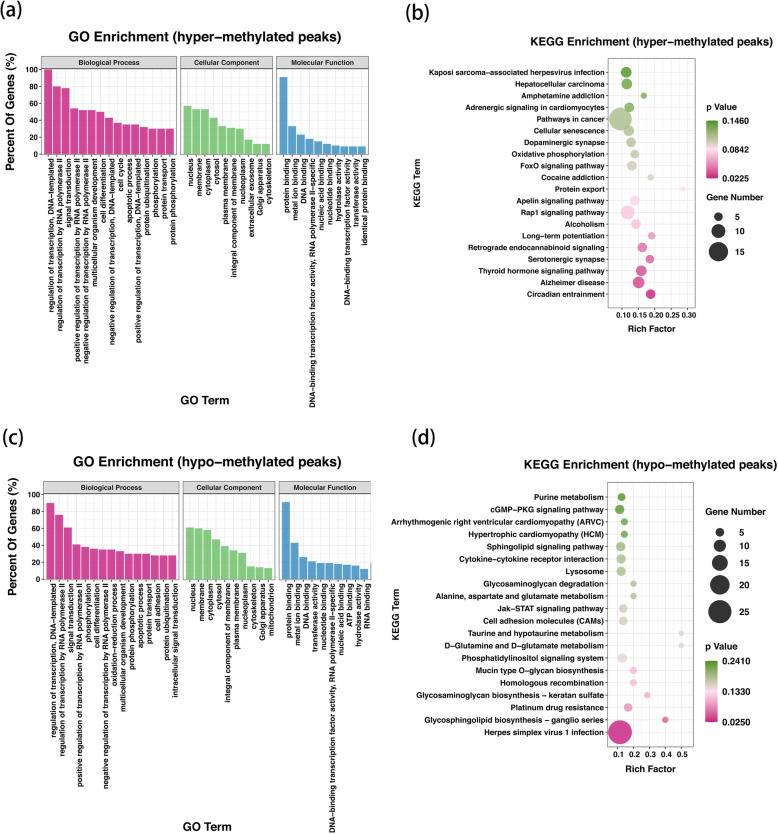
To figure out the biological significance of m6A methylation in the osteogenesis of hASCs, we performed GO and KEGG pathway analyses for DMGs. GO analysis uncovered that both the hypermethylated and hypomethylated genes in dhASCs were significantly involved with “regulation of transcription, DNA-templated,” “regulation of transcription by RNA polymerase II,” “signal transduction,” “cell differentiation,” and “multicellular organism development” (ontology: biological process); “membrane,” “nucleus,” and “cytoplasm” (ontology: cellular component); and “protein binding,” “metal ion binding,” and “DNA binding” (ontology: molecular function) (Fig. 4a, c). Remarkably, KEGG pathway analysis demonstrated that the hypermethylated genes were significantly associated with signaling pathway “thyroid hormone signaling pathway,” “Rap1 signaling pathway,” and “FoxO signaling pathway” (Fig. 4b). However, the hypomethylated genes were not significantly associated with the common osteogenic signaling pathway (Fig. 4d).
Fig. 4.
The DMGs were associated with extensive and osteogenic biological functions. a GO enrichment analysis for hypermethylated peaks. b KEGG pathway analysis for hypermethylated peaks. c GO enrichment analysis for hypomethylated peaks. d KEGG pathway analysis for hypomethylated peaks. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes
Osteogenic pathways and genes were significantly differentially methylated and regulated
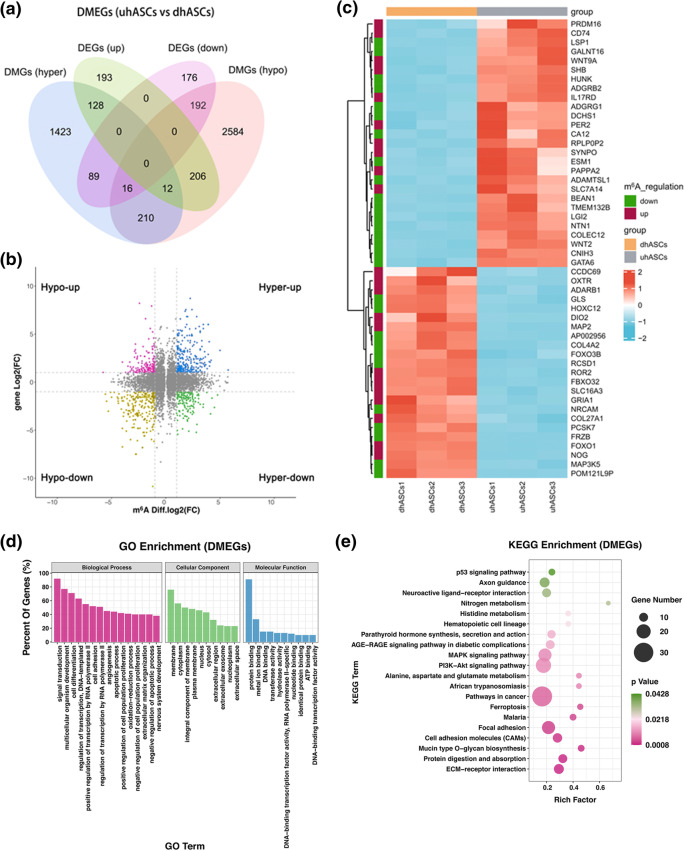
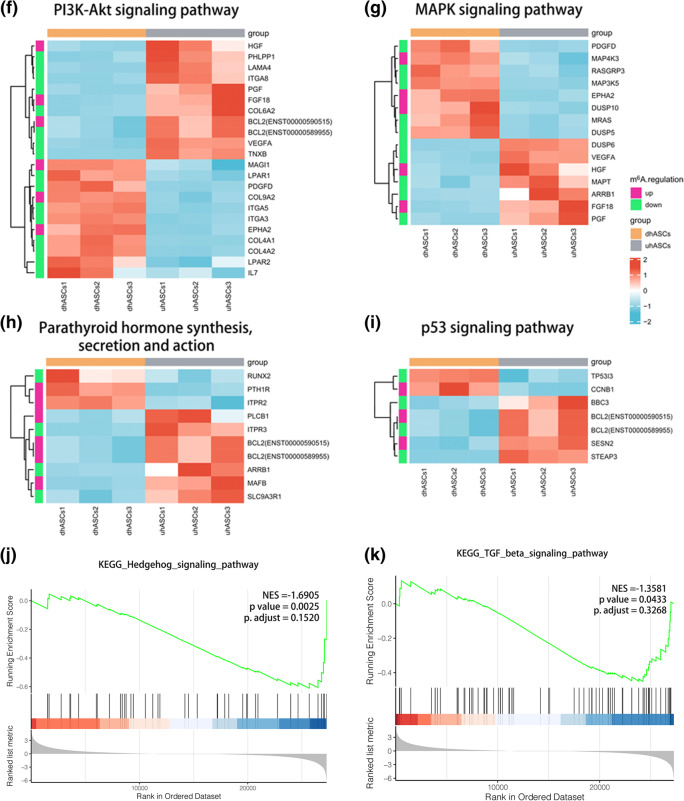
To figure out the alteration of gene expression and signaling pathways in the osteogenesis of hASCs, we performed RNA-seq to figure out the DEGs. By the standard of p-value < 0.05 and fc ≥ 2, compared with uhASCs, 2261 genes were significantly dysregulated in the dhASCs group, of which 1210 were upregulated and 1051 were downregulated (Supplementary Figure S2a). The hierarchical clustering of the top 50 DEGs was shown in Supplementary Figure S2b and the full list of DEGs was shown in Supplementary Table S3. GO analysis and KEGG pathway analysis of DEGs were shown in Supplementary Figure S2b-e. By conjoint analysis of DMGs (by the standard of p-value < 0.05) and DEGs (by the standard of p-value < 0.05 and fold change (fc) ≥ 2), we found 671 differentially methylated and expressed transcripts (598 genes) were mainly divided into four groups, including 140 hypermethylated and upregulated (hyper-up), 208 hypomethylated and downregulated (hypo-down), 105 hypermethylated but downregulated (hyper-down), and 218 hypomethylated but upregulated genes or transcripts (hypo-up) (Fig. 5a, b). The hierarchical clustering of the top 50 DMEGs was shown in Fig. 5c. The top 20 DMEGs were listed in Table 2, and the full list of DMEGs was shown in Supplementary Table S4. GO analysis uncovered that the DMEGs in dhASCs were significantly involved with “signal transduction,” “multicellular organism development,” “regulation of transcription, DNA-templated,” “cell differentiation,” “cell adhesion,” and “regulation of cell population proliferation” (ontology: biological process); “membrane,” “cytoplasm,” and “integral component of membrane” (ontology: cellular component); and “protein binding,” “metal ion binding,” and “DNA binding” (ontology: molecular function) (Fig. 5d). Remarkably, the KEGG pathway analysis demonstrated that the DMEGs were significantly associated with signaling pathway “PI3K-Akt signaling pathway,” “MAPK signaling pathway,” “parathyroid hormone synthesis, secretion and action,” and “p53 signaling pathway” (Fig. 5e). The DMEGs associated with these four signaling pathways were selected and performed a hierarchical cluster analysis (Fig. 5f–i). Furthermore, GSEA analysis of the whole transcriptome revealed these genes were significantly associated with the “Hedgehog signaling pathway” and the “TGF-beta signaling pathway” (Fig. 5j, k).
Fig. 5.
Osteogenic pathways and genes were significantly differentially methylated and regulated. a Venn plot for DMEGs. b Four-quadrant graph for hyper-up, hypo-down, hyper-down, and hypo-up DMEGs. c Hierarchical clustering analysis of the top 50 DMEGs (by the standard fold change ≥ 2 and p < 0.05 for both DEGs and DMGs, ranking by p-value of DEGs). d GO enrichment analysis for DMEGs. e KEGG pathway analysis for DMEGs. f Hierarchical clustering analysis of DMEGs in the PI3K-Akt signaling pathway. g Hierarchical clustering analysis of DMEGs in the MAPK signaling pathway. h Hierarchical clustering analysis of DMEGs in the pathway of parathyroid hormone synthesis, secretion, and action. i Hierarchical clustering analysis of DMEGs in the P53 signaling pathway. j GSEA showed decreased enrichment of the Hedgehog signaling pathway. k GSEA showed decreased enrichment of the TGF-beta signaling pathway. DEGs, differentially expressed genes; DMGs, differentially methylated genes; DMEGs, differentially methylated and expressed genes; hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs; hyper-up, hypermethylated and upregulated genes; hypo-down, hypomethylated and downregulated genes; hyper-down, hypermethylated and downregulated genes; hypo-up, hypomethylated and upregulated genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis
Table 2.
The top 20 differently expressed genes containing differently methylated peaks (uhASCs/dhASCs)
| Gene | Transcript ID | Chr | Start | End | m6A regulation | FC | p-value | Gene regulation |
|---|---|---|---|---|---|---|---|---|
| COLEC12 | ENST00000400256 | chr18 | 318935 | 319136 | hypo | 0.04 | 0.00 | Down |
| FOXO1 | ENST00000636651 | chr13 | 40556102 | 40556341 | hyper | 42.51 | 0.00 | Up |
| LSP1 | ENST00000311604 | chr11 | 1890072 | 1890311 | hypo | 0.07 | 0.00 | Down |
| NRCAM | ENST00000522550 | chr7 | 108149746 | 108156434 | hypo | 37.52 | 0.00 | Up |
| FBXO32 | ENST00000517956 | chr8 | 123541116 | 123541206 | hyper | 9.41 | 0.00 | Up |
| POM121L9P | ENST00000414583 | chr22 | 24252274 | 24252423 | hypo | 8.26 | 0.00 | Up |
| SLC7A14 | ENST00000231706 | chr3 | 170463356 | 170463596 | hyper | 0.05 | 0.00 | Down |
| NOG | ENST00000332822 | chr17 | 56594172 | 56594704 | hyper | 187.37 | 0.00 | Up |
| CNIH3 | ENST00000272133 | chr1 | 224740106 | 224740315 | hypo | 0.04 | 0.00 | Down |
| FRZB | ENST00000295113 | chr2 | 182834653 | 182834922 | hypo | 296.94 | 0.00 | Up |
| TMEM132B | ENST00000299308 | chr12 | 125657702 | 125658152 | hypo | 0.10 | 0.00 | Down |
| MAP2 | ENST00000475600 | chr2 | 209696191 | 209696621 | hyper | 28.70 | 0.00 | Up |
| SHB | ENST00000377707 | chr9 | 37916645 | 37916914 | hyper | 0.16 | 0.00 | Down |
| GALNT16 | ENST00000337827 | chr14 | 69352155 | 69352903 | hypo | 0.09 | 0.00 | Down |
| AP002956 | ENST00000264036 | chr11 | 119308798 | 119309008 | hypo | 12.46 | 0.00 | Up |
| RCSD1 | ENST00000537350 | chr1 | 167697240 | 167705078 | hypo | 242.98 | 0.00 | Up |
| DIO2 | ENST00000422005 | chr14 | 80200075 | 80200314 | hyper | 55.65 | 0.00 | Up |
| OXTR | ENST00000316793 | chr3 | 8750557 | 8750826 | hyper | 11.17 | 0.00 | Up |
| ADARB1 | ENST00000460734 | chr21 | 45145486 | 45146205 | hyper | 7.92 | 0.00 | Up |
| CD74 | ENST00000009530 | chr5 | 150400041 | 150400190 | hyper | 0.13 | 0.00 | Down |
dhASCs, osteogenically differentiated human adipose-derived stem cells; uhASCs, undifferentiated human adipose-derived stem cells; FC, fold change
PPI network and hub genes were discovered among DMEGs
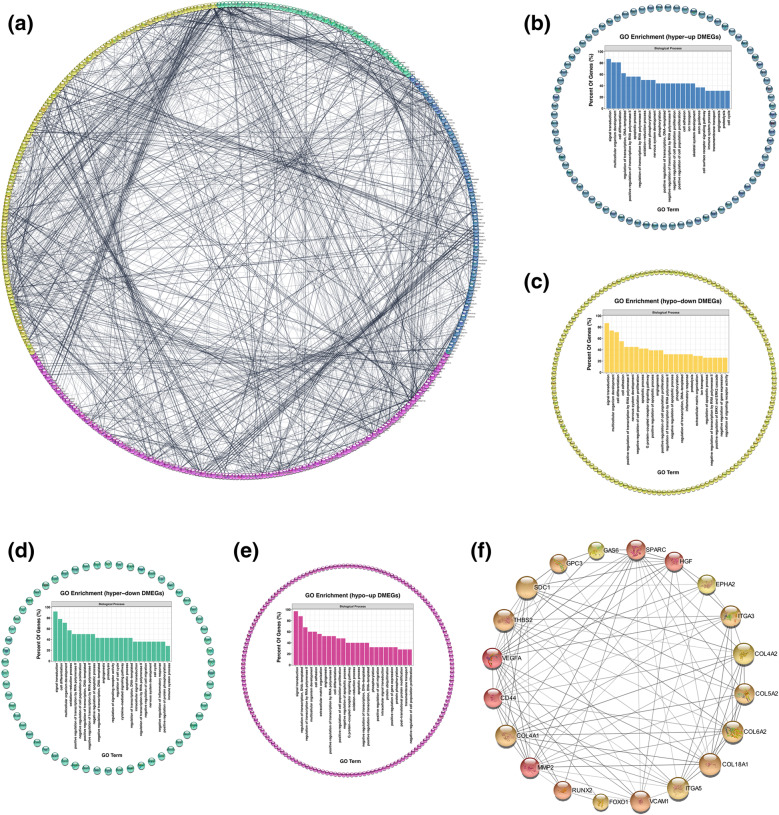
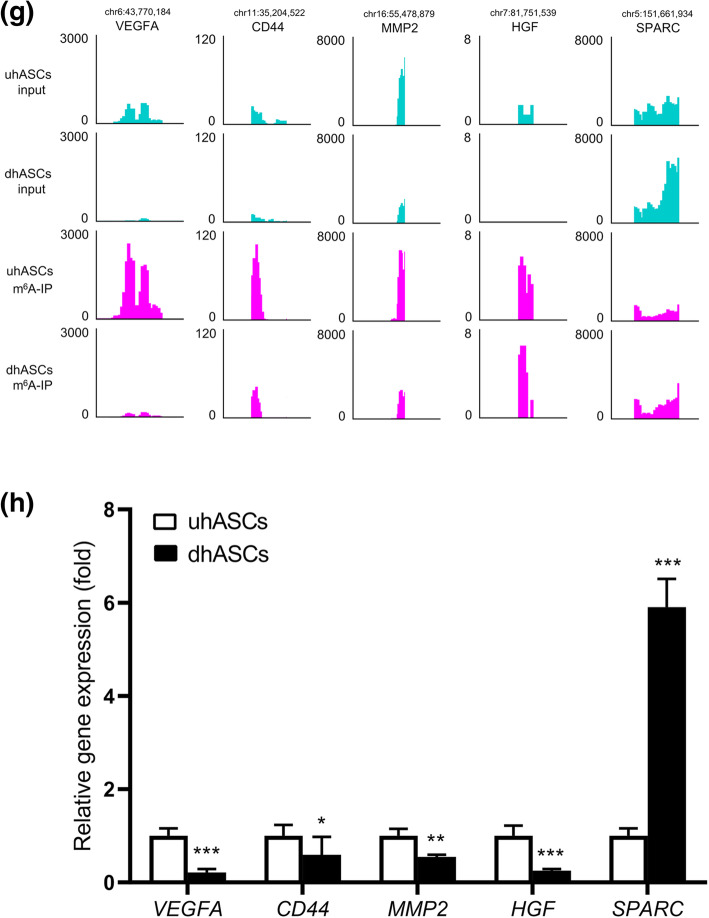
The PPI network of the DMEGs was conducted by STRING database and Cytoscape (Fig. 6a). The network was grouped into four clusters as mentioned before, that is, hyper-up, hypo-down, hyper-down, and hypo-up DMEGs (Fig. 6b–e). GO enrichment analyses were performed for each cluster of DMEGs to elucidate their biological function (Fig. 6b–e). Hub genes in the PPI network were selected using cytoHubba by calculating the protein combining degree (Fig. 6f). And the m6A peak distribution of the top 5 hub genes between the groups was shown in Fig. 6g, and the expressions of the 5 hub DMEGs were shown in Fig. 6h. The PPI network interaction metadata was listed in Supplementary Table S5.
Fig. 6.
PPI network and hub genes were discovered among DMEGs. a PPI network of DMEGs. Blue represents the hypermethylated and upregulated genes, yellow represents the hypomethylated and downregulated genes, green represents the hypermethylated but downregulated genes, and red represents the hypomethylated but upregulated genes. b The hyper-up cluster and the GO enrichment analysis for DMEGs in this cluster. c The hypo-down cluster and the GO enrichment analysis for DMEGs in this cluster. d The hyper-down cluster and the GO enrichment analysis for DMEGs in this cluster. e The hypo-up cluster and the GO enrichment analysis for DMEGs in this cluster. f Hub genes calculated from the PPI network of DMEGs. The size and color of the nodes are proportional to the combining score. g Data visualization of VEGFA, CD44, MMP2, HGF, and SPARC mRNA m6A modification in uhASCs and dhASCs. h qRT-PCR analyses of the expression of hub DMEGs of VEGFA, CD44, MMP2, HGF, and SPARC. *p < .05, **p < .01, ***p < .001. PPI, protein-protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DMEGs, differentially methylated and expressed genes; hyper-up, hypermethylated and upregulated genes; hypo-down, hypomethylated and downregulated genes; hyper-down, hypermethylated and downregulated genes; hypo-up, hypomethylated and upregulated genes; hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs; qRT-PCR, quantitative real-time polymerase chain reaction
Discussion
Adipose-derived stem cells (ADSCs), as one form of mesenchymal stem cells with good accessibility and special biological properties, are frequently used for bone regeneration both in vitro and in vivo [11, 12]. According to the current criteria, the isolation of ADSCs produces cell type heterogeneity, and the purity of ADSCs is essential for their engineering use [9]. In the present study, the hASCs were identified with surface markers and successfully induced into osteoblast-like cells [9, 32]. Identifying factors involved in the osteogenic differentiation of ASCs is important for better understanding the mechanism of osteogenic differentiation. Recent studies have shown that m6A modification plays essential roles in stem cell fate decisions in a stage-, state-, and cell type-specific manner [26, 36–40]. In the case of osteogenic differentiation, reduced m6A level in BMSCs inhibited osteogenic differentiation by affecting the PTH/Pth1r signaling axis, the PI3K-AKT signaling pathways, or the alternative splicing of VEGFA [30, 40, 41]. As for preadipocytes, some studies reported FTO promoted adipogenesis by regulating cell cycle-related genes and the JAK2-STAT3-C/EBPb signaling pathway in an m6A-dependent manner in 3T3-L1 cell lines [42–45]. However, at the present stage, little is known about the roles of m6A modification in the osteogenic differentiation of ADSCs. In this study, using m6A MeRIP-Seq and RNA-seq, we depicted the transcriptome-wide m6A methylome in the process of osteogenic differentiation of hASCs for the first time.
In the present study, we observed that from day 0 to day 14, the level of m6A modification decreased continuously with the most significant and steepest reduction at the early stage. Therefore, we investigated the time point day 7 to explore potential m6A-related signaling pathways. Combined with significant expression changes of m6A regulators METTL3 and FTO, we unveiled the potential effects of m6A modification in osteogenic differentiation of hASCs. The changes of m6A level during osteogenic induction of hASCs appeared stage-specific. This was probably because of the time-dependent expression pattern of m6A regulators. At the early stage, the elevated expression of demethylase FTO predominantly reduced the m6A level. With the osteogenic induction lasting, the expression of FTO decreased while the expression of methyltransferase METTL3 maintained, resulting in a slower reduction of m6A level. Interestingly, this trend was contrary to that of BMSCs where a time-dependent increase of m6A content during osteogenic differentiation was reported [29]. In the cases of BMSCs, from day 0 to day 14, the expression of methyltransferase METTL3 increased while the expression of demethylase FTO and ALKBH5 did not change significantly [41]. The differences between hASCs and BMSCs might reflect the cell type-specific effects of m6A modification in stem cell differentiation [26, 40]. Although both were mesenchymal stem cells, the distinct origins may cause different gene expression pattern and epigenetic modifications, thus producing divergent stemness, proliferation capacity, multilineage differentiation potential, longevity, immunoregulatory properties, and so on [6, 46]. Considering the complex stage- and state-specific roles of m6A modification, the m6A methylome of a prolonged osteogenic induction of hASCs remains to be revealed.
In total, the m6A MeRIP-Seq revealed an abundance of 1.94 and 1.96 peaks per transcript for uhASCs and dhASCs, respectively. Additionally, the m6A peaks of both uhASCs and dhASCs were mostly enriched in the 3′UTR region and near the stop codon; these sites were m6A-specific and consistent with previous studies [15–19, 47]. Furthermore, during the osteogenic differentiation of hASCs, the differentially methylated peaks were significantly enriched in “signal transduction,” “regulation of transcription,” “multicellular organism development,” “cell differentiation,” and so on, indicating the conservative and fundamental roles of m6A in regulating development and cell fate specification [16, 17].
In order to elucidate the mechanism of m6A in affecting osteogenic differentiation of hASCs, we combined the m6A methylome and the transcriptome and found the key signaling pathways that were affected by m6A modification. As with previous studies, several canonical pathways regarding osteogenic differentiation were significantly enriched. The most significant ones were the “PI3K-Akt signaling pathways,” “MAPK signaling pathways,” “parathyroid hormone synthesis, secretion and action,” and “P53 signaling pathways,” in which quite a few key genes were differentially methylated [20, 22, 30, 40, 41, 48]. Moreover, although other common osteogenic pathways were not significantly enriched in the m6A methylome, quite a few key genes in these pathways were significantly methylated. At the same time, we constructed the PPI network of DMEGs and speculated the hub genes based on combining degree, for example, the genes of VEGFA, CD44, MMP2, HGF, and SPARC. All these differentially expressed as well as differentially methylated key genes and hub genes provided the latest and detailed information about how m6A modification affected the osteogenic differentiation of hASCs.
A mass of studies in the past few years have lighted up the biological effects of m6A modification on RNAs [19–22, 40, 47, 49–52]. On the one hand, the m6A methylation process is reversable, and this mark on RNA could be written or erased in case of various stimuli and biological factors [17, 47, 49]. On the other hand, m6A can affect RNA processing and metabolism in multifaceted mechanisms, including alternative splicing, alternative polyadenylation, RNA stability, RNA export, RNA degradation, and translation. Therefore, m6A up- or downregulates the gene expression in a complex and context-depended manner [17–19]. For this reason, we observed four groups of DMEGs in the present study, those were hyper-up, hypo-down, hyper-down, and hypo-up DMEGs. Our function enrichment analyses suggested that these four clusters of DMEGs were associated with the fundamental and different biological processes. Combined with the methylation and expression pattern of the key DMEGs, future studies based on these molecular clues could give us brand-new knowledge on the osteogenic differentiation of hASCs from an m6A-specific epigenetic perspective.
Conclusions
Total m6A level was reduced with osteogenic differentiation of hASCs, and the transcriptome-wide m6A methylome built in the present study indicated quite a few signaling pathways and hub genes were influenced by m6A modification. In total, 1145 DMGs, 2261 DEGs, and 671 DMEGs were detected. GO and KEGG pathway analysis conducted for these differentially methylated and regulated genes revealed extensive and osteogenic biological functions. The “PI3K-Akt signaling pathway,” “MAPK signaling pathway,” “parathyroid hormone synthesis, secretion and action,” and “p53 signaling pathway” were significantly enriched, and the DMEGs in these pathways were identified as m6A-specific key genes. PPI network based on DMEGs was built, and VEGFA, CD44, MMP2, HGF, and SPARC were speculated as the hub DMEGs. Future studies based on these epigenetic clues could promote the understanding of the mechanisms of osteogenic differentiation of hASCs.
Supplementary Information
Additional file 1: Figure S1. m6A peak calling for uhASCs and dhASCs. a Pie charts showing distribution of m6A peaks of uhASCs in different gene context. b Accumulation of m6A peaks of uhASCs along transcripts. c Pie charts showing distribution of m6A peaks of dhASCs in different gene context. d Accumulation of m6A peaks of dhASCs along transcripts. hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs.
Additional file 2: Figure S2. Gene expression profile during osteogenesis of hASCs by RNA-seq. a Volcano plots displaying the DEGs (fold change ≥ 2 and p < 0.05). b Hierarchical clustering analysis of the top 50 DEGs (ranking by p value). c GO enrichment analysis for upregulated genes. d KEGG pathway analysis for upregulated genes. e GO enrichment analysis for downregulated genes. f KEGG pathway analysis for downregulated genes. DEGs, differentially expressed genes; hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FC, fold change.
Additional file 3: Table S1. Primers used for qRT-PCR.
Additional file 4: Table S2. Full list of DMGs (uhASCs vs dhASCs).
Additional file 5: Table S3. Full list of DEGs (uhASCs vs dhASCs).
Additional file 6: Table S4. Full list of DMEG (uhASCs vs dhASCs).
Additional file 7: Table S5. The PPI network metadata.
Acknowledgements
Not applicable.
Abbreviations
- ADSCs
Adipose-derived stem cells
- ALP
Alkaline phosphatase
- ARS
Alizarin Red S
- DEGs
Differentially expressed genes
- dhASCs
Osteogenically differentiated hASCs
- DMEGs
Differentially methylated and expressed genes
- DMGs
Differentially methylated genes
- GO
Gene Ontology
- GSEA
Gene set enrichment analysis
- hASCs
Human adipose-derived stem cells
- hBMSCs
Human bone marrow-derived mesenchymal stem cells
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- m6A
N6-methyladenosine
- m6A MeRIP-Seq
Methylated RNA immunoprecipitation and sequencing
- PPI
Protein-protein interaction
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RNA-seq
High-throughput sequencing for RNA
- uhASCs
Undifferentiated hASCs
- UTR
Untranslated region
Authors’ contributions
J. L and Z. Z. designed this study. W. S., Y. S., and K. X. conducted this study. L. Y. and X. H. analyzed and processed the data. W. S. and J. L. drafted the manuscript. All authors have read and approved the final submitted manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81870743, 81470722, and 81771048) and the Creative Spark Fund of Sichuan University (2018SCUH0007).
Availability of data and materials
All sequencing data have been deposited into the Sequence Read Archive (SRA) database of NCBI with the identifier PRJNA739515 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA739515?reviewer=jqgj6n7reasr8ei9a20riogqok).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sándor GK, Numminen J, Wolff J, Thesleff T, Miettinen A, Tuovinen VJ, Mannerström B, Patrikoski M, Seppänen R, Miettinen S, Rautiainen M, Öhman J. Adipose stem cells used to reconstruct 13 cases with cranio-maxillofacial hard-tissue defects. Stem Cells Transl Med. 2014;3(4):530–540. doi: 10.5966/sctm.2013-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrelli MR, Hu MS, Longaker MT, Lorenz HP. Tissue engineering and regenerative medicine in craniofacial reconstruction and facial aesthetics. J Craniofac Surg. 2020;31(1):15–27. doi: 10.1097/SCS.0000000000005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrovic V, Zivkovic P, Petrovic D, Stefanovic V. Craniofacial bone tissue engineering. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114(3):e1–e9. doi: 10.1016/j.oooo.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Jin YZ, Lee JH. Mesenchymal stem cell therapy for bone regeneration. Clin Orthop Surg. 2018;10(3):271–278. doi: 10.4055/cios.2018.10.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao HT, Chen CT. Osteogenic potential: comparison between bone marrow and adipose-derived mesenchymal stem cells. World J Stem Cells. 2014;6(3):288–295. doi: 10.4252/wjsc.v6.i3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazini L, Rochette L, Amine M, Malka G. Regenerative capacity of adipose derived stem cells (ADSCs), comparison with mesenchymal stem cells (MSCs). Int J Mol Sci. 2019;20(10). [DOI] [PMC free article] [PubMed]
- 7.Ho-Shui-Ling A, Bolander J, Rustom LE, Johnson AW, Luyten FP, Picart C. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi: 10.1016/j.biomaterials.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szpalski C, Barbaro M, Sagebin F, Warren SM. Bone tissue engineering: current strategies and techniques--part II: cell types. Tissue Eng Part B Rev. 2012;18(4):258–269. doi: 10.1089/ten.teb.2011.0440. [DOI] [PubMed] [Google Scholar]
- 9.Squillaro T, Peluso G, Galderisi U. Clinical trials with mesenchymal stem cells: an update. Cell Transplant. 2016;25(5):829–848. doi: 10.3727/096368915X689622. [DOI] [PubMed] [Google Scholar]
- 10.Sheykhhasan M, Wong JKL, Seifalian AM. Human adipose-derived stem cells with great therapeutic potential. Curr Stem Cell Res Ther. 2019;14(7):532–548. doi: 10.2174/1574888X14666190411121528. [DOI] [PubMed] [Google Scholar]
- 11.Tajima S, Tobita M, Mizuno H. Current status of bone regeneration using adipose-derived stem cells. Histol Histopathol. 2018;33(7):619–627. doi: 10.14670/HH-11-942. [DOI] [PubMed] [Google Scholar]
- 12.Shafaei H, Kalarestaghi H. Adipose-derived stem cells: an appropriate selection for osteogenic differentiation. J Cell Physiol. 2020;235(11):8371–8386. doi: 10.1002/jcp.29681. [DOI] [PubMed] [Google Scholar]
- 13.Si Z, Wang X, Sun C, Kang Y, Xu J, Wang X, Hui Y. Adipose-derived stem cells: sources, potency, and implications for regenerative therapies. Biomed Pharmacother. 2019;114:108765. doi: 10.1016/j.biopha.2019.108765. [DOI] [PubMed] [Google Scholar]
- 14.Bacakova L, Zarubova J, Travnickova M, Musilkova J, Pajorova J, Slepicka P, Kasalkova NS, Svorcik V, Kolska Z, Motarjemi H, Molitor M. Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells - a review. Biotechnol Adv. 2018;36(4):1111–1126. doi: 10.1016/j.biotechadv.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Weng H, Chen J. The biogenesis and precise control of RNA m6A methylation. Trends Genet. 2020;36(1):44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 17.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 18.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, He L, Liu Z, Ren X, Qi L, Wan L, Wang W, Tu C, Li Z. Multifaceted functions and novel insight into the regulatory role of RNA N6-methyladenosine modification in musculoskeletal disorders. Front Cell Dev Biol. 2020;8:870. doi: 10.3389/fcell.2020.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, Wu Q, Yuan B, Lu Q, Yang H. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19(1):104. doi: 10.1186/s12943-020-01216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu F, Li W, Yang X, Na L, Chen L, Liu G. The roles of epigenetics regulation in bone metabolism and osteoporosis. Front Cell Dev Biol. 2020;8:619301. doi: 10.3389/fcell.2020.619301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulman Z, Stern-Ginossar N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21(5):501–512. doi: 10.1038/s41590-020-0650-4. [DOI] [PubMed] [Google Scholar]
- 24.McFadden MJ, Horner SM. N6-methyladenosine regulates host responses to viral infection. Trends Biochem Sci. 2020. [DOI] [PMC free article] [PubMed]
- 25.Madugalle SU, Meyer K, Wang DO, Bredy TW. RNA N6-methyladenosine and the regulation of RNA localization and function in the brain. Trends Neurosci. 2020;43(12):1011–1023. doi: 10.1016/j.tins.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu LP, Pickering B, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Tian Y, Zhang Q, Ren D, Zhang Q, Yan X, et al. Novel insights into the role of N6-methyladenosine RNA modification in bone pathophysiology. Stem Cells Dev. 2020. [DOI] [PubMed]
- 28.Chen X, Hua W, Huang X, Chen Y, Zhang J, Li G. Regulatory role of RNA N6-methyladenosine modification in bone biology and osteoporosis. Front Endocrinol. 2019;10:911. doi: 10.3389/fendo.2019.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H, Wang W, du W, Ma T, Liu S, Xu Z, Gao M, Yu M, Bian Y, Pang P, Li X, Yu S, Yang F, Cai B, Yang L. m6A methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids. 2020;19:421–436. doi: 10.1016/j.omtn.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, Li J, Sheng R, Deng P, Wang Y, Zheng R, Jiang Y, Ye L, Chen Q, Zhou X, Lin S, Yuan Q. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9(1):4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi B, Xiong Y, Yan C, Chen L, Xue H, Panayi AC, Hu L, Hu Y, Zhou W, Cao F, Liu G. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020;24(11):6385–6396. doi: 10.1111/jcmm.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo T, Yang X, Sun Y, Huang X, Zou L, Liu J. Effect of microRNA-20a on osteogenic differentiation of human adipose tissue-derived stem cells. Cells Tissues Organs. 2019;208(3-4):148–157. doi: 10.1159/000506304. [DOI] [PubMed] [Google Scholar]
- 33.Di Bernardo G, Messina G, Capasso S, Del Gaudio S, Cipollaro M, Peluso G, et al. Sera of overweight people promote in vitro adipocyte differentiation of bone marrow stromal cells. Stem Cell Res Ther. 2014;5(1):4. doi: 10.1186/scrt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia K, Cen X, Yu L, Huang X, Sun W, Zhao Z, Liu J. Long noncoding RNA expression profiles during the NEL-like 1 protein-induced osteogenic differentiation. J Cell Physiol. 2020;235(9):6010–6022. doi: 10.1002/jcp.29526. [DOI] [PubMed] [Google Scholar]
- 35.Huang XQ, Cen X, Sun WT, Xia K, Yu LY, Liu J, Zhao ZH. CircPOMT1 and circMCM3AP inhibit osteogenic differentiation of human adipose-derived stem cells by targeting miR-6881-3p. Am J Transl Res. 2019;11(8):4776–4788. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, Lu X, Xiao W, Yang YG, Liu F. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 37.Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de los Mozos IR, Sadée C, Lenaerts AS, Nakanoh S, Grandy R, Farnell E, Ule J, Stunnenberg HG, Mendjan S, Vallier L. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature. 2018;555(7695):256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, Xi J, Ye D, Zhu S, Chen W, Jia W, Leng Y, Wan X, Kang J. N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46(8):3906–3920. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, Hanna JH, Ding L. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21(6):700–709. doi: 10.1038/s41556-019-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Zhai Y, Zhang S, Dai X, Li Z. Roles of N6-methyladenosine m6A in stem cell fate decisions and early embryonic development in mammals. Front Cell Dev Biol. 2020;8:782. doi: 10.3389/fcell.2020.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian C, Huang Y, Li Q, Feng Z, Xu Q. Mettl3 regulates osteogenic differentiation and alternative splicing of Vegfa in bone marrow mesenchymal stem cells. Int J Mol Sci. 2019;20(3). [DOI] [PMC free article] [PubMed]
- 42.Wu R, Guo G, Bi Z, Liu Y, Zhao Y, Chen N, Wang F, Wang Y, Wang X. m6A methylation modulates adipogenesis through JAK2-STAT3-C/EBPβ signaling. Biochim Biophys Acta Gene Regul Mech. 2019;1862(8):796–806. doi: 10.1016/j.bbagrm.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, Zhao Y, Wu R, Jiang Q, Cai M, Bi Z, Liu Y, Yao Y, Feng J, Wang Y, Wang X. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biol. 2019;16(12):1785–1793. doi: 10.1080/15476286.2019.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q, Liu Q, Cai M, Wang F, Wang Y, Wang X. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(10):1323–1330. doi: 10.1016/j.bbalip.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Danielsen JMR, Wang XJ, Yang YG. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee RH, Kim B, Choi I, Kim H, Choi HS, Suh K, Bae YC, Jung JS. Characterization and expression analysis of mesenchymal stem cells from human bone marrow and adipose tissue. Cell Physiol Biochem. 2004;14(4-6):311–324. doi: 10.1159/000080341. [DOI] [PubMed] [Google Scholar]
- 47.He PC, He C. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 2021;40(3):e105977. doi: 10.15252/embj.2020105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Gu X, Li D, Cai L, Xu Q. METTL3 regulates osteoblast differentiation and inflammatory response via Smad signaling and MAPK signaling. Int J Mol Sci. 2019;21(1). [DOI] [PMC free article] [PubMed]
- 49.Kim J, Lee G. Metabolic control of m6A RNA modification. Metabolites. 2021;11(2). [DOI] [PMC free article] [PubMed]
- 50.Zhang Y, Xu S, Xu G, Gao Y, Li S, Zhang K, et al. Dynamic expression of m6A regulators during multiple human tissue development and cancers. Front Cell Dev Biol. 2020;8:629030. doi: 10.3389/fcell.2020.629030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N6-methyladenosine m6A modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi YC, Chen XY, Zhang J, Zhu JS. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. m6A peak calling for uhASCs and dhASCs. a Pie charts showing distribution of m6A peaks of uhASCs in different gene context. b Accumulation of m6A peaks of uhASCs along transcripts. c Pie charts showing distribution of m6A peaks of dhASCs in different gene context. d Accumulation of m6A peaks of dhASCs along transcripts. hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs.
Additional file 2: Figure S2. Gene expression profile during osteogenesis of hASCs by RNA-seq. a Volcano plots displaying the DEGs (fold change ≥ 2 and p < 0.05). b Hierarchical clustering analysis of the top 50 DEGs (ranking by p value). c GO enrichment analysis for upregulated genes. d KEGG pathway analysis for upregulated genes. e GO enrichment analysis for downregulated genes. f KEGG pathway analysis for downregulated genes. DEGs, differentially expressed genes; hASCs, human adipose-derived stem cells; uhASCs, undifferentiated hASCs; dhASCs, osteogenically differentiated hASCs; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FC, fold change.
Additional file 3: Table S1. Primers used for qRT-PCR.
Additional file 4: Table S2. Full list of DMGs (uhASCs vs dhASCs).
Additional file 5: Table S3. Full list of DEGs (uhASCs vs dhASCs).
Additional file 6: Table S4. Full list of DMEG (uhASCs vs dhASCs).
Additional file 7: Table S5. The PPI network metadata.
Data Availability Statement
All sequencing data have been deposited into the Sequence Read Archive (SRA) database of NCBI with the identifier PRJNA739515 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA739515?reviewer=jqgj6n7reasr8ei9a20riogqok).