Abstract
Background
The recent emergence of new SARS CoV-2 variants (variants of concern, VOC) that spread rapidly and may lead to immune escape has emphasized the urgent need to monitor and control their spread.
Methods
We analyzed 2018 SARS-CoV-2 positive specimens collected between February 9 and March 22, 2021 using the Thermofisher® TaqPath™ COVID-19 CE-IVD RT-PCR kit (TaqPath) and the ID solutions® ID™ SARS-CoV-2/UK/SA Variant Triplex RT-PCR (ID triplex) assay to screen for VOCs.
Results
The ID triplex assay identified 62.8% of them as VOCs: 61.8% B.1.1.7 and 0.9% B.1.351/P.1. The agreement between the ID triplex results for B.1.1.7 and the TaqPath S gene target failure (SGTF)/ S gene target late detection (SGTL) profile for this variant agreed very well (k = 0.86). A low virus load was the main cause of discrepancies. Sequencing discordant results with both assays indicated that the TaqPath assay detected the B.1.1.7 lineage slightly better. Both assays suggested that the virus loads of B.1.1.7 variants were significantly higher than those of non-B.1.1.7 strains. Only 10/20 B1.351/P.1 strains detected with the ID triplex assay were confirmed by sequencing.
Conclusions
We conclude that the SGTF/SGTL profiles identified using the TaqPath assay and ID triplex results are suitable for detecting the B.1.1.7 lineage. The ID triplex assay, which rapidly determines all three current VOCs simultaneously, could be a valuable tool for limiting virus spread by supporting contact-tracing and isolation.
Keywords: SARS-CoV-2, COVID-19, RT-PCR, Variants of concern (VOC), TaqPath, ID triplex, Genome sequencing
1. Introduction
Several SARS-CoV-2 variants of concern (VOCs) have emerged in recent months. The alpha variant, also referred as B.1.1.7 lineage, appeared in December 2020, in the United Kingdom [1], [2], [3], [4]. Its increased transmissibility ensured that it quickly became the dominant strain in England and other countries [5], [6], [7], [8], [9]. The emergence of this VOC has been associated with a rapid increase in case numbers and hospitalization rates in several countries [7,10,11] but there is some debate as to its link with disease severity and mortality [12], [13], [14]. The B.1.1.7 variant is defined by multiple mutations in the spike protein including a deletion at position 69–70 that leads to a loss of detection of the S gene target in some SARS-CoV-2 detection assays, including the Thermo Fisher® TaqPath™ COVID-19 CE-IVD RT-PCR kit (TaqPath) [15,16]. This assay was widely used in the United Kingdom and the increased S gene target failure (SGTF) results helped discover the B.1.1.7 strain.
The other two major variants, the B.1.351 (beta) variant first detected in South Africa, and the P1 (gamma) variant first detected in Brazil share with the B.1.1.7 variant the N501Y mutation in the S protein receptor binding domain involved in virus entry. Evidence suggests that other mutations in these variants might confer not only increased transmissibility but might reduce the susceptibility of the virus to neutralizing antibodies [17], [18], [19]. It is essential to monitor these variants because of their increased transmissibility and potential resistance to host immunity and vaccination.
Many countries have consequently increased their sequencing capacity of the whole SARS-CoV-2 genome. Unfortunately, whole genome sequencing is slower than PCR testing and so is not suitable for contact tracing to limit virus spreading. Alternative methods for rapid VOC detection and contact tracing, such as RT-PCR-based screening assays have been developed; these generate results in just a few hours [6,[20], [21], [22]]. The ID™ SARS-CoV-2/UK/SA Variant Triplex (ID triplex) assay is a multiplex RT-PCR assay that detects the B.1.1.7, B.1.351, and P1 variants by targeting the 69–70 deletion and the N501Y mutation [23]. Both targets are detected in the B.1.1.7 variant, while only the N501Y mutation is present in B.1.1351 and P.1 variants.
This study was done to evaluate the capacity of the ID triplex assay to detect VOCs and to determine its correlation with the TaqPath assay used as first line screening RT-PCR assay. We also assessed the virus loads of each variant.
2. Material and methods
All nasopharyngeal specimens sent to the Toulouse university hospital for SARS-CoV-2 detection between February 9 and March 22 were tested within 24 h of collection. Each positive specimen was then screened for the presence of VOCs.
2.1. SARS-CoV-2 detection
RNA was extracted on an MGI SP-960 instrument, a high-throughput fully automated workstation, using the MGIEasy Nucleic Acid Extraction kit (MGI™) and amplified on the QuantStudio™ 5 Real-Time PCR System (Applied Biosystems) using the Thermo Fisher® TaqPath™ COVID-19 CE-IVD RT-PCR kit. The TaqPath assay targets three sequences in the virus ORF1ab, N and S genes. The internal control for nucleic acid extraction was an MS2 phage. Results were interpreted using the COVID-19 Interpretive Software version v.2.5 on QuantStudio™ Design and Analysis Desktop Software v.1.5.1.
Positive results were classified according to cycle threshold (Ct) data obtained for all three targets in S gene target failure (SGTF), S gene target late detection (SGTL) and non SGTF/SGTL as described below.
| SGTF | SGTL | NON SGTF/SGTL | Uninterpretable£ | ||
|---|---|---|---|---|---|
| Targets | N | + (< 33) | + | + | +/- ( ≥ 33) |
| ORF1ab | + | + | + | +/- | |
| S | – | Late +* | +$ | +/- | |
| *At least 5 Ct values higher than N and ORF1ab; $Not meeting the criteria of SGTL; £At least two positive targets | |||||
2.2. SARS-CoV-2 voc screening
All TaqPath positive samples were re-extracted using the MGIEasy Nucleic Acid Extraction kit (MGI™) and amplified using the ID triplex assay. This assay is a multiplex RT-PCR assay that targets several SARS-COV-2 sequences further detected in 3 different channels. The first channel allows the detection of two common SARS-CoV-2 genes, N gene and RNA-dependant RNA polymerase gene (further referred as N), used as an internal control. The two others enable the detection of two single nucleotide polymorphisms (SNP) in the S gene allowing the discrimination between B.1.1.7 and B.1.351/P.1 VOC. The two targeted S gene sequences are the 69–70 deletion and the N501Y mutation. The results for each sample were interpreted as described below.
| SARS-CoV-2 variants | |||||
|---|---|---|---|---|---|
| B.1.1.7 | B.1.351/P1 | NON VOC | Uninterpretable* | ||
| Targets | N501Y | + | + | – | +/- |
| Del69–70 | + | – | +/- | +/- | |
| N | + | + (Ct < 33) | + (Ct < 33) | + (Ct ≥ 33) | |
| *At least one target is missing between N501Y and Del69–70 | |||||
2.3. Next-generation sequencing (NGS), data processing and phylogenetic analysis
SARS-CoV-2 genomes were sequenced according to the Illumina CovidSeq Test instructions (Illumina Inc, USA) [24]. RNA was extracted using the MGIEasy Nucleic Acid extraction kit. Librairies were then prepared with CovidSeq Kit (Illumina Inc, USA) according to the manufacturer instructions. Briefly, cDNA was synthetized with random hexamers. This cDNA was amplified by multiplex PCR (98 amplicons) and the PCR amplified products were then processed for tagmentation and adapter ligation using IDT for Illumina Nextera UD Indexes Set A, B, C and D. Finally, all samples were sequenced with a 100 bp read length on the NovaSeq 6000 platform. Three hundred eighty four samples were multiplexed in a single S4 flow cell.
The sequence data were processed using DRAGEN COVID Lineage (v3.5.1) (Illumina Inc.). Analysis included the SARS CoV-2 consensus sequence, plus generation of Nextclade and Pangolin lineage.
Samples for which genome coverage was at least 75% and spike coverage was at least 85% were further analyzed. The consensus sequences were aligned on the SARS-CoV-2 Wuhan-Hu-1 reference genome (NC_045512.2) and 109 GISAID sequences of different Pangolin lineages detected in France using MAFFT (v.7.475) [25]. We used phylogenetic analysis of this alignment to confirm Nextclade and Pangolin analyses.
2.4. Statistical analysis
The ability of the two assays to detect B.1.1.7 variant was evaluated using Cohen's kappa coefficient (κ), the overall agreement, the positive percent agreement, and the negative percent agreement. Confidence intervals (CI: 95%) were calculated by the Clopper and Pearson method using GraphPad Prism. The percentages of uninterpretable results for the two assays were compared using Fisher's exact tests. Statistically significance was defined as p < 0.05. The N gene Ct values for all samples obtained with the two assays were compared using the Wilcoxon matched-pairs signed rank test. We compared the TaqPath Ct values for the N and ORF1ab targets for SGTF/SGTL and non-SGTF/SGTL samples using the Mann-Whitney U test. Only samples in which the N gene Ct was < 33 were included. The ID triplex assay N Ct values (Ct < 33) were compared between B.1.1.7 and non B.1.1.7 samples using the Mann-Whitney U test. The virus loads for each variant detected using the ID triplex were compared using the median Ct value (< 33) for N with the Kruskal-Wallis test and Dunn's multiple comparison test.
3. Results
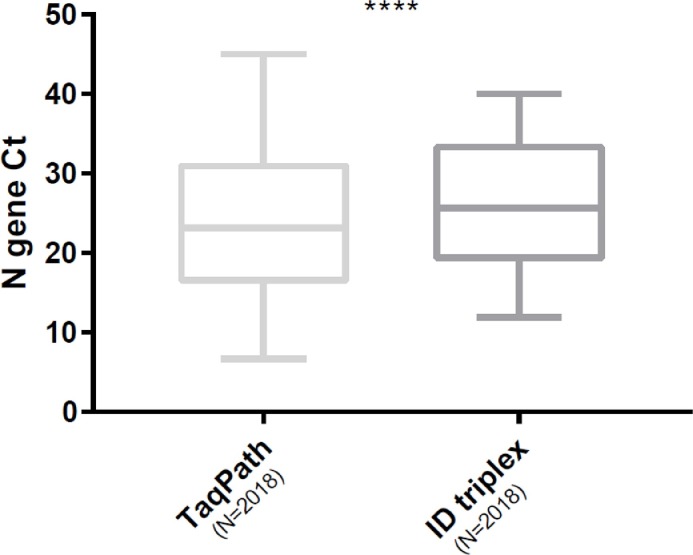
We screened 2018 nasopharyngeal samples that tested positive in the TaqPath assay for VOCs with the ID triplex assay. TheTaqPath assay detected 1339 (66.3%) samples with SGTF profiles, 19 (0.9%) with SGTL profiles, and 19.4% with non-SGTF-SGTL profiles. Analysis using the ID triplex assay identified 1268 (62.8%) VOCs, including 1248 (61.8%) B.1.1.7 variants and 20 (0.9%) B.1.351 or P.1 VOC, but no VOCs were detected in 14.1% (285) of specimens ( Table 1 ). The ID triplex assay gave significantly more uninterpretable results (23%) than the TaqPath assay (13.3%; p < 0.001). Similarly, analysis of N gene Ct values indicated that the ID triplex assay showed significantly higher Ct values (25.6) than the TaqPath assay (23.2; p < 0.001) ( Fig. 1 ).
Table 1.
Screening results of positive samples with the ID triplex and TaqPath assays.
| ID triplex | ||||||
|---|---|---|---|---|---|---|
| B.1.1.7 | B.1.351/P1 | NON VOC | Uninterpretable | ∑ | ||
| TaqPath | SGTF | 1177 | 4 | 38 | 120 | 1339 |
| SGTL | 10 | 0 | 1 | 8 | 19 | |
| NON SGTF/SGTL | 20 | 16 | 236 | 120 | 392 | |
| Uninterpretable | 41 | 0 | 10 | 217 | 268 | |
| ∑ | 1248 | 20 | 285 | 465 | 2018 | |
SGTF: S gene target failure; SGTL: S gene target late detection.
Fig. 1.
Box plot of Ct values for the N gene in all positive specimens. Comparison of both assays. P < 0.0001 (Wilcoxon matched-pairs signed rank test).
3.1. B.1.1.7 detection
Only 1502/2018 specimens (74.4%) gave interpretable results in both the ID triplex and the TaqPath assays for detecting the B.1.1.7 variant. Agreement between the assays was very good (κ = 0.86 [95% CI: 0.83 – 0.90]), with only 63/1502 (4.2%) discordant results(Table 2). Overall agreement was 95.8% [95% CI: 94.7% − 96.8%], positive percent agreement was 98.3% [95% CI: 97.4% − 99.0%], and negative percent agreement was 85.4% [95% CI: 80.9% − 89.2%]. Those samples that gave discordant results in the two assays had significantly higher Ct values than the concordant ones (median difference: 11 Ct; p < 0.001).
Table 2.
Concordance evaluation of B1.1.7 detection between assays.
| ID triplex | ||||
|---|---|---|---|---|
| B.1.1.7 | Non B.1.1.7 | ∑ | ||
| TaqPath | B.1.1.7 | 1187 | 43 | 1230 |
| Non B.1.1.7 | 20 | 252 | 272 | |
| ∑ | 1207 | 275 | 1502 | |
Discordant results:
-
-
The ID triplex assay detected no B.1.1.7 variant in 43 specimens whereas the TaqPath assay found they had an SGTF/SGTL profile. The SARS-CoV-2 genome was successfully sequenced in 12/43 (27.9%) of them. 10 were confirmed to be B.1.1.7 variants (including 4 specimens screened B.1.351/P1) and 2 were sequenced as B.1.525; they had the 69–70 deletion but no N501Y mutation.
-
-
The ID triplex assay detected 20 specimens as B.1.1.7 variant while the TaqPath assay indicated they were non-SGTF-SGTL. Only one of them had a Ct value suitable for sequencing, it was confirmed to be a B.1.1.7 variant.
Thus NGS confirmed the TaqPath results for 10/13 (76.9%) samples and the ID triplex for 3/13 (23.1%).
3.2. B.1.351/P1 variant detection with the id triplex assay
The ID triplex assay detected the B.1.351/P1 variant in 20 specimens and the genomes of 16/20 were sequenced. Of these, NGS confirmed 10 as B.1.351 variants. Two samples were not confirmed as VOC and belonged to a lineage with a N501Y mutation but no associated 69–70 deletion (variant A27 (19B/501Y)). The remaining 4 samples were B.1.1.7 variants with a TaqPath assay SGTF/SGTL profile.
3.3. Comparison of viral load between variant
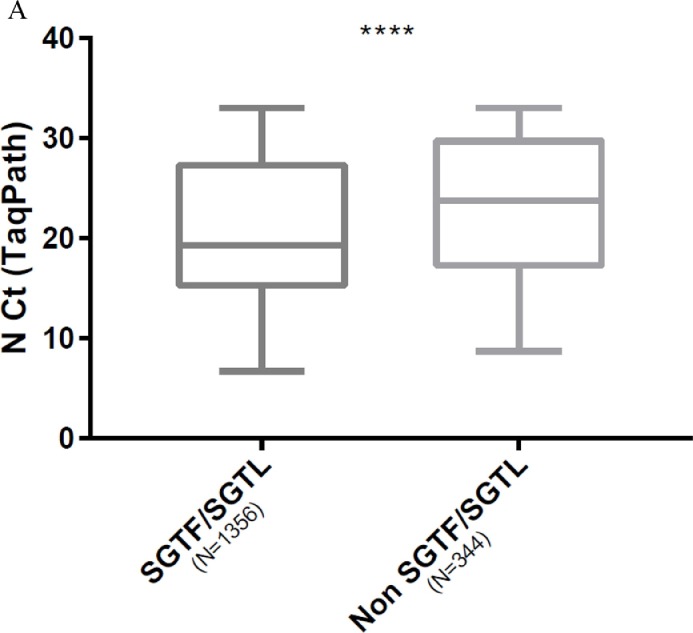
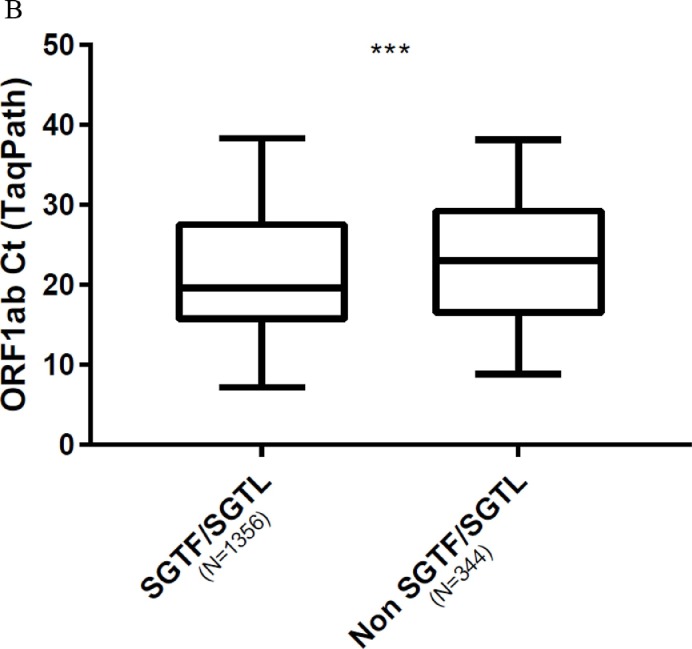
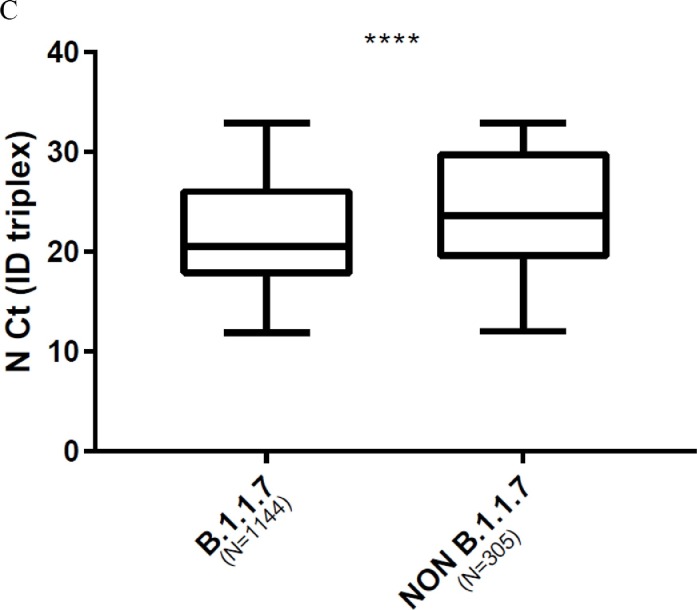
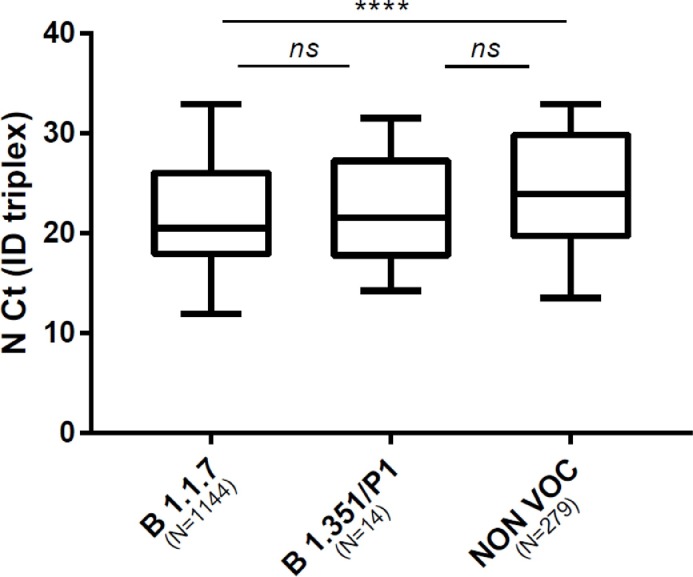
We compared viral loads between variant by using Ct values as surrogates. Comparison of the Ct values for N and ORF1ab targets in SGTF/SGTL and non-SGTF-SGTL samples analysed with the TaqPath assay showed that the Ct values for N and ORF1ab targets in SGTF/SGTL were significantly lower than those for non-SGTF/SGTL; median Ct value difference: 4.5 (p < 0.0001) for the N gene and 3.4 (p = 0.0009) for ORF1ab ( Fig. 2 A, 2 B). Analysis using the ID triplex results to compare N gene Ct values for B.1.1.7 and non-B.1.1.7 variants indicated that the median N gene Ct value for B.1.1.7 was lower than that for non-B.1.1.7 (median N gene Ct value difference: 3.1) ( Fig. 2 C). We also compared the N Ct values obtained for each variant category screened with the ID triplex assay (B.1.1.7, B.1.351/P1 and non-VOC). The B.1.1.7 and non-VOC Ct values (p < 0.0001) differed significantly but the B.1.351/P1 and B.1.1.7 (p > 0.99), or non-VOC (p = 0.46) values did not ( Fig. 3 ).
Fig. 2.
Box plot of virus loads of B.1.1.7 and non-B.1.1.7 variants. (Mann-Whitney U test). A. Using TaqPath N gene Ct values for SGTF/SGTL and non-SGTF/SGTL samples as surrogates (p < 0.0001). B. Using TaqPath ORF1ab Ct values for SGTF/SGTL and non-SGTF/SGTL samples as surrogates (p = 0.0009). C. Using ID triplex N gene Ct values for B.1.1.7 and non-B.1.1.7 variants as surrogates (p < 0.0001).
Fig. 3.
Box plot of virus loads of B.1.1.7, B.1.351/P1 and non-VOC variants using ID triplex N gene Ct values as surrogates. (Kruskal-Wallis test with Dunn's correction).
4. Discussion
From February 2021, the French health surveillance program records all cases of SARS-COV-2 detected and the corresponding VOC screening results to adapt quarantine and contact tracing measures to the specific variant. Several multiplex RT-PCR assays targeting SNP have been developed to screen specific mutations of interest but few data are available on their performance. We compared the results of screening for VOCs by two assays, a first line SARS-CoV-2 diagnosis assay which was subsequently found to detect the B.1.1.7 variant (TaqPath assay) and one developed for VOC screening (ID triplex). The two assays gave similar results for detecting B.1.1.7, with the TaqPath assay performing slightly better according to sequencing results. However, the ID triplex assay, which rapidly detected B.1.1.7 and B.1.351/P1 promises to be a valuable VOC monitoring system. We also found that the loads of the B.1.1.7 variant virus were significantly greater than those of other strains, which is compatible with its greater transmissibility.
Our results highlight how widely the B.1.1.7 variant has spread in the south of France. The B.1.1.7 variant accounted for an estimated 2.9% of infections in our region of Occitanie in early January 2021, and for the majority of them between February and March [8]. The two assays agreed well for detecting the B.1.1.7 variant. This is somewhat surprising as the association of the 69–70 deletion and N501Y mutations detected by the ID triplex assay is specific for B.1.1.7, while the 69–70 deletion detected by the TaqPath COVID-19 assay is shared by other variants, including the B.1.525, and B.1.620 variants. This good correlation could be because B.1.1.7 has rapidly replaced other strains to become the prevalent variant in the South of France. It confirms earlier data showing that the probability for the B.1.1.7 lineage using the S gene dropout depends on the local epidemiology [6], being highly correlated when the circulation of this variant is predominant [7,16]. Moreover, with all SGTL results confirmed B.1.1.7 variants either by the ID triplex assay or by NGS, our data confirm that in addition to the S gene target failure, the S gene late detection can be a useful tool for evaluating the spread of B.1.1.7[7].
Analysis of the sequences of discordant results indicates that the TaqPath assay is better at detecting B.1.1.7 than the ID triplex assay, although the agreement between assays was very good. The sequencing data highlighted the few weaknesses of the ID triplex assay; problems with detecting the 69–70 deletion lead to the misclassification of B.1.1.7 variants as B.1.351/P1 strains. There were also problems with the detection of N501Y leading to the misclassification of B.1.1.7 variants as non VOC. As the majority of discrepancies observed concerned samples with low viral loads, it could be explained by the delayed detection of del 69–70 and N501Y targets compared to N target in ID triplex assay (data not shown). It emphasises the importance of applying a N Ct cut-off above which variants should not be identified when all targets are not detected.
Whereas the TaqPath assay only detects the B.1.1.7 variant, the ID triplex assay allowed the detection of 20 possible B.1.351/P1 infections. However, the positive predictive value of B.1.351/P1 screening is imperfect, as only 10/16 were confirmed by sequencing, perhaps because this was a minor variant at the time and this screening strategy is not very specific. Nevertheless, this strategy is worth considering for effective contact tracing, as long as other strains with the same profile remain minor components.
Despite higher N gene Ct values, the ID triplex assay was not less sensitive than the TaqPath assay. As it detected all 2018 positive samples tested, it could be used as a first line assay despite its commercialization as a second line assay for VOC screening. This would reduce costs and speed up VOC screening.
We investigated the relationship between virus load and the transmissibility of the B.1.1.7 and B.1.351 variant using Ct values as surrogates, as these variants both spread rapidly [11,26]. The Ct values for B.1.1.7 and non-B.1.1.7 samples differed significantly, regardless of the assay used, which corroborates reports of SGTF/SGTL strains having higher virus loads than non-SGTF/SGTL specimens. This may account for the greater transmissibility and rapid spread of B.1.1.7 [7,12,16,27]. However, while the B.1.351 virus loads were greater than those of non-VOC, the difference was not statistically significant. The sample could have been too small; if so, a larger study should be undertaken. The B.1.351 variant has not spread as widely as the B.1.1.7, although it too is said to be highly transmissible [28,29]; it remains a relatively minor variant. Whether the difference in the spread of B.1.1.7 and B.1.351 is due to differences in virus entry, replication, viral fitness or the host immune response is not clear.
Our study has several limitations. First, only viruses’ strains from samples with discordant results or with B.1.351/P1 screening results were sequenced. Therefore, sequence analysis results could not be used as gold standard to evaluate the sensitivity and the specificity of both assay to detect VOCs. Second, these VOC screening assays do not identify new mutations of interest such as the E484K/Q and L452R mutations. Third, the study was realized in a context of high prevalence of B.1.1.7 lineage and relatively low circulation of other VOCs. We can't assume that these results could be generalized to different epidemiological contexts.
We conclude that both the TaqPath and ID triplex assays are useful tools for detecting B.1.1.7 variants. The ID triplex assay, which can rapidly determine all three current VOCs could be invaluable for limiting their spread by enhancing contact-tracing measures.
5. Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The English text was edited by Dr Owen Parkes.
References
- 1.Public Health England Investigation of novel SARS-CoV-2 variants of concern. GOV.UK. 2000 https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- 2.Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- 3.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, october to november 2020. Euro. Surveill. Bull. Eur. Su.r Mal. Transm. Eur. Commun. Dis. Bull. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galloway S.E. Emergence of SARS-CoV-2 B1.1.7 lineage — United States, december 29, 2020–january 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70 doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goncalves Cabecinhas A.R., et al. SARS-CoV-2 N501Y Introductions and transmissions in Switzerland from beginning of october 2020 to february 2021-implementation of swiss-wide diagnostic screening and whole genome sequencing. Microorganisms. 2021;9 doi: 10.3390/microorganisms9040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borges V., et al. Tracking SARS-CoV-2 lineage B.1.1.7 dissemination: insights from nationwide spike gene target failure (SGTF) and spike gene late detection (SGTL) data, Portugal, week 49 2020 to week 3 2021. Euro. Surveill. Bull. Eur. Su.r Mal. Transm. Eur. Commun. Dis. Bull. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.10.2100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaymard A., et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, january to march 2021. Euro. Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.9.2100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpert T., et al. Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell. 2021 doi: 10.1016/j.cell.2021.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Center for Disease prevention and Control (ECDC). SARS-CoV-2 - increased circulation of variants of concern and vaccine rollout in the EU/EEA - 14th update. 29 (2021).
- 11.Davies N.G., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frampton D., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies N.G., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021:1–5. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham M.S., et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021 doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Health C. for D. and R. Genetic Variants of SARS-CoV-2 may lead to false negative results with molecular tests for detection of SARS-CoV-2 - letter to clinical laboratory staff and health care providers. FDA. 2021 [Google Scholar]
- 16.Kidd M., et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021:1–6. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 18.Hu J., et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R.E., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogels C.B.F., et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kami W., et al. Rapid and simultaneous identification of three mutations by the NovaplexTM SARS-CoV-2 variants I assay kit. J. Clin. Virol. Off. Publ. Pan. Am. Soc. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vega-Magaña N., et al. RT-qPCR Assays for Rapid Detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 Mutations: a Screening Strategy to Identify Variants With Clinical Impact. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haim-Boukobza S., et al. Detecting rapid spread of SARS-CoV-2 variants, France, january 26–february 16, 2021. Emerg. Infect. Dis. 2021;27:1496–1499. doi: 10.3201/eid2705.210397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhoyar R.C., et al. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tegally H., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 27.Calistri P., et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell F., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro. Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roquebert B., et al. The SARS-CoV-2 B1.351 lineage (VOC β) is outgrowing the B.1.1.7 lineage (VOC α) in some French regions in April 2021. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.23.2100447. [DOI] [PMC free article] [PubMed] [Google Scholar]