Abstract
Background
Drug repurposing is a cost-effective strategy to identify drugs with novel effects. We searched for drugs exhibiting inhibitory activity to Herpes Simplex virus 1 (HSV-1). Our strategy utilized gene expression data generated from HSV-1-infected cell cultures which was paired with drug effects on gene expression. Gene expression data from HSV-1 infected and uninfected neurons were analyzed using BaseSpace Correlation Engine (Illumina®). Based on the general Signature Reversing Principle (SRP), we hypothesized that the effects of candidate antiviral drugs on gene expression would be diametrically opposite (negatively correlated) to those effects induced by HSV-1 infection.
Results
We initially identified compounds capable of inducing changes in gene expression opposite to those which were consequent to HSV-1 infection. The most promising negatively correlated drugs (Valproic acid, Vorinostat) did not significantly inhibit HSV-1 infection further in African green monkey kidney epithelial cells (Vero cells). Next, we tested Sulforaphane and Menadione which showed effects similar to those caused by viral infections (positively correlated). Intriguingly, Sulforaphane caused a modest but significant inhibition of HSV-1 infection in Vero cells (IC50 = 180.4 µM, p = 0.008), but exhibited toxicity when further explored in human neuronal progenitor cells (NPCs) derived from induced pluripotent stem cells.
Conclusions
These results reveal the limits of the commonly used SRP strategy when applied to the identification of novel antiviral drugs and highlight the necessity to refine the SRP strategy to increase its utility.
Keywords: Drug repurposing, sulforaphane, Herpes Simplex Virus type 1, neural progenitor cell
Background
Herpes Simplex Virus 1 (HSV-1) infection remains a massive public health problem. HSV-1 causes common infections as well as more severe morbidities including blindness and encephalitis. Encephalitis can lead to long term neurological sequel and/or death, even if treated with current antivirals. HSV-1 has the ability to cause recurrent infections at the same sites as a result of its reactivation from the neuronal latent state that was established upon the initial infection. Latency is a poorly understood, untreatable process that is a dynamic interplay between the maintenance of virus DNA with a silenced or limited gene expression program and the host cell chromatin remodeling processes. Currently, only the lytic productive infection can be treated with antivirals, despite vigorous efforts to attempt to target the latent state. The most effective drugs are derivatives of Acyclovir (ACV) and Pencyclovir, and include esterified prodrugs, Valacyclovir (VAL) and Famvir, which have higher oral bioavailability. These drugs inhibit lytic infections by acting as nucleoside analogues that are selectively activated in virus-infected cells and then block viral DNA replication. However, breakthrough infections can still occur, particularly among long-term prophylactic antiviral receivers, which may lead to the development of ACV resistant mutants. Mutations arising in the viral thymidine kinase (TK), and/or in the DNA polymerase cause changes in the affinity for the antiviral resulting in resistancy. Importantly, TK is required for the growth of HSV in neurons during reactivation; therefore, TK mutations are selected against. Higher prevalence of HSV infections accompanied with decreased response to ACV is observed in immunocompromised patients, including HIV-positive patients.1 The second line agent for ACV resistant infections is foscarnet, which requires intravenous administration and is considerably more toxic than ACV or VAL. Currently, no antivirals are effective at eradicating latent infection or removing the latent human reservoir that gives rise to recurrent disease. Several strategies targeting the latent genome for cleavage using meganucleases or gRNA directed CRSPR-cas9 are under development but they are far from clinical trial evaluation. The search for new antivirals that act differently from ACV remains a paramount goal and an urgent public health necessity.
Compound or drug repurposing (also known as repositioning or reprofiling) refers to the process of developing an existing compound/drug for new use beyond its predetermined indication. Compared to the traditional process of developing an entirely new drug for a specific purpose, repurposing bears lower risk of failure, especially concerning safety, and has much more rapid return on investment.2 For instance, using the traditional method, DiMasi et al. searched commercial databases to identify 1,914 investigational compounds between 1995 and 2017, intended for treating a wide array of infectious diseases.3 Of those, 1,323 entered clinical testing. After an average of 100 months of testing, only 20.7% were finally approved. While almost half of them are still in active clinical testing, 29.9% of them have since been abandoned.3 If a compound is approved by the drug regulatory authorities for one indication and is found to be effective for another indication by reliable repurposing processes, the prior approval enables investigators to bypass pre-clinical studies, and phase I clinical trials. As such, the time to apply for the approval of a novel clinical use is greatly shortened.4 Even if a compound is not approved by regulatory authorities, the source platform or pharmacological database provides us with copious physicochemical information and correlated experiments that may have already been performed. Pre-clinical cost per approved drug on average is estimated to comprise of 32% of total out-of-pocket costs. Moreover, considering that the average time from synthesis to initial human testing is 31.2 months,5 repurposing investigational compounds is extremely cost-effective.
Drug repurposing can be opportunistic or serendipitous. There are some typical successful examples of off-target effects or newly discovered on-target effects, including the use of sildenafil for erectile dysfunction,6 and thalidomide for multiple myeloma.7 However, with rapidly developing computational approaches to probe large amounts of drug-related research data that is available, systematic methods can be used to identify reliable target compounds in an organized, data-driven way.8 Considering the urgent needs for new broad-spectrum anti-viral agents, incorporative drug repurposing methods can serve as an approach to explore novel, anti-viral drugs in a cost-efficient way. Various successful discoveries of novel, anti-viral drugs have been achieved through drug repurposing approaches.9 However, there are possible pitfalls of this approach in that: i) while failures due to toxicity are less likely to happen with pre-characterization, failures at late stages of clinical trials are not unusual.10 Such late stage failures might result from limited primary screening methods (concentrations, delivery method, etc.) and/or the cell types/model systems employed to determine toxicity. Other concerns include: i) for compounds with poly-pharmacology, different indications may require different dosages; ii) once a compound failed for one desirable purpose, significant resources are going to be likely required for re-profiling the compound, potential lag time risk might also develop during this period;8 iii) repurposing processes could be restricted by legal and regulatory forces in concern of exclusivity and marketing secure because repurposing focuses on existing agents.11
Despite these pitfalls, the obvious benefit from drug-repurposing is well recognized and prompts the need to develop systematic approaches to investigate potential candidates. In this study, we propose a new strategy to generate anti-viral candidates based on the comparison of gene profile datasets by applying a platform called BaseSpace Correlation Engine (Illumina®). Gene profile changes of human induced pluripotent stem cell (hiPSC)-derived neurons after exposure to HSV-1 infection were chosen as inputs for comparison, and a set of compounds that induce opposite or similar gene expression changes were selecte as contrasts. We started from two negatively correlated compounds based on the traditional signature reversion principle (SRP), which assumes that if a drug could reverse the expression pattern of a set of hallmark genes for a particular disease phenotype, the drug might be able to reverse the phenotype itself.10 While this principle has been successfully applied to various therapeutic scenarios,12–16 in the case of virus infections, it could be argued that the gene expression effects in cell culture models of infection represent host-defense systems. Therefore, we also investigated drugs with similar gene expression patterns to those observed in the infection cell culture models. We next investigated the antiviral efficacy of the putative antiviral drugs in two cell culture models. Finally, we investigated mechanisms of action for Sulforaphane (SFN), the most promising candidate by studying its effects on canonical pathways; we also compared the upstream regulators suggested in its most significantly correlated bioset to those responsible for gene profile changes in our original input dataset.
Results
Candidates from negatively and positively correlated compounds
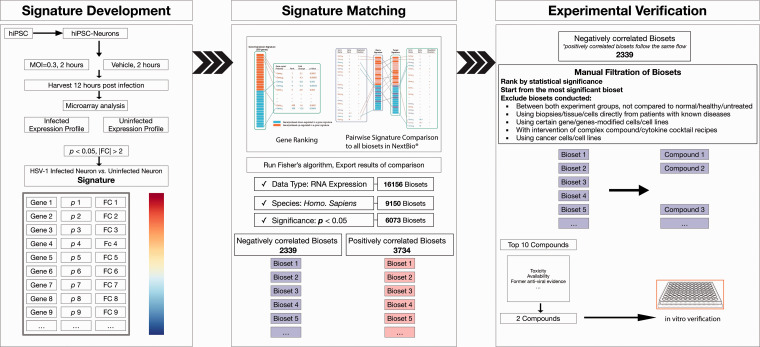
2339 negatively and 3734 positively correlated biosets were ranked based on the significance of correlation (i.e. the rank p value) from the most to least significant. The detail of bioset was manually examined according to the aforementioned exclusion criteria (Figure 1). Ranked by the correlation p value of the No.1 supporting bioset, the top 10 compounds found from the computational process are listed as below (Table 1).
Figure 1.
The process of applying BaseSpace Correlation Engine to find potential compounds. hiPSC: human induced pluripotent stem cell; MOI: Multiplicity of infection; FC: fold change.
Table 1.
Top 10 negatively (left) and positively (right) correlated compounds.
| GSE | Common genes | P | (–) Compound name | GSE | Common genes | P | (+) Compound name |
|---|---|---|---|---|---|---|---|
| 36680 | 4189 | 1.40E-19 | Lipopolysaccharide | 20479 | 4344 | 1.50E-22 | Sulforaphane |
| 20479 | 4836 | 3.30E-12 | Oltipraz | 56308 | 4951 | 3.20E-21 | IL-4 |
| 57051 | 2914 | 3.30E-11 | Dactinomycin | 45251 | 7960 | 4.70E-20 | TNF-alpha |
| 64123 | 2332 | 1.30E-10 | Valproic acid | 10394 | 2049 | 2.90E-18 | PM2.5 |
| 11919 | 2921 | 3.30E-08 | Ascorbate-2-phosphate | NA | 2759 | 1.10E-17 | Ethionine |
| 11919 | 2387 | 9.60E-08 | Ascorbate | 119559 | 6194 | 3.80E-17 | Halofuginone |
| 71127 | 7325 | 5.30E-08 | Vorinostat | 56308 | 2745 | 4.00E-14 | Sphingosine-1-phosphate |
| 74235 | 728 | 1.40E-06 | Hydroxychloroquine | 4301 | 3503 | 1.10E-12 | Dithiothreitol |
| 12198 | 6903 | 3.10E-06 | IL-2 | 4301 | 2477 | 3.60E-12 | Menadione |
| 58395 | 4344 | 7.40E-06 | Hydrogen peroxide | 56308 | 4106 | 5.50E-12 | IL-13 |
GSE: Series number from Gene Expression Omnibus; P: significant level for overlapped genes provided by Correlation Engine; (−)/(+): negative/positive correlations.
The values shown in bold in table 1 are name strings of datasets (GSE datasets) and not data. The names of compounds tested in are in bold. For each GSE dataset, the significant level of overlapping genes were already listed in column 3 and column 7.
To verify the feasibility and reliability of this novel bioinformatic strategy, we started from negatively correlated compounds based aforementioned SRP for subsequent in vitro experiments. Valproic acid and Vorinostat, both ranked high in the list, were selected for further analyses over other candidates, based on the number of studies (only one or two related studies, e.g. Oltipraz, Ascorbate, Ascorbate-2-phosphate, Hydroxychloroquine) and evidence of less significant toxicity (compared to Hydrogen peroxide, Dactinomycin). Lipopolysaccharide was not chosen despite it ranking as the top candidate in the negative list. It has been widely used as an adjuvant to enhance the reactivation of herpes virus.17,18 It also ranked as the 20th compound in the positive list (not shown), which might stand for different set of pathway changes resulted from different experimental conditions.
Negatively correlated valproic acid and vorinostat both failed to produce anti-HSV effect in vero cell culture even at low MOI
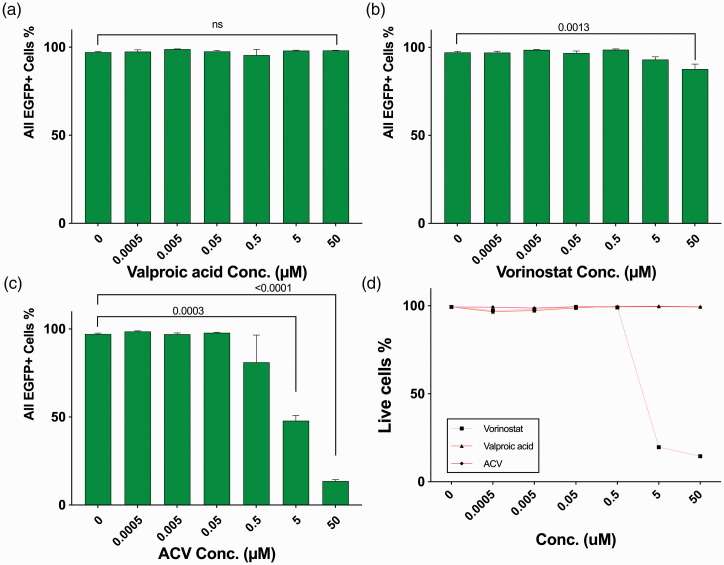
Valproic acid is an anti-convulsant medication, whose mechanism is not fully understood but has traditionally been attributed to the blockade of voltage-gated sodium channels and increased brain levels of gamma-aminobutyric acid (GABA). Vorinostat is a histone deacetylase inhibitor for the treatment of cutaneous T cell lymphoma. These two candidates were applied to treat Vero cell culture acutely infected by HSV-1 at MOI: 0.3, with concentration range from 0.0005 µM to 50 µM. EGFP positive cell percentage (EGFP+%), driven by viral immediate early gene promoter ICP0 was acquired via flow cytometry. Fourty-eight hours of treatment with 5 µM of the anti-HSV-1 drug Acyclovir decreased EGFP+% to 47.87%, compared to infected group without ACV (97.15%) (Figure 2(c)). This was achieved without causing any apparent cell death, indicated by live cell percentage 99.3% of uninfected cells after being cultured with 50 µM ACV for the same time period (Figure 2(d)).
Figure 2.
Effects of negatively correlated compounds towards HSV-1 acute infection compared with Acyclovir. Vero cells were infected with HSV-1 at MOI = 0.3. After inoculated with compounds from 0.0005 to 50 µM for 48 hours, percentages of total EGFP positive (EGFP+) cells were acquired via flowcytometry: (a) Valproic acid; (b) Vorinostat; (c) Acyclovir. (d) Live cell percentages of uninfected cells after cultured with same concentrations of compounds for 48 hours.
Though Valproic acid was not toxic to Vero cell culture at 50 µM (Figure 2(d)), it did not decrease EGFP+% at the highest concentration tested (Figure 2(a)). Vorinostat showed some toxicity on uninfected Vero cells at concentrations higher than 5 µM (live cell percentage 19.63%, Figure 2(d)). No significant anti-viral effects of Vorinostat were observed at lower concentrations (Figure 2(b)).
Positively correlated sulforaphane achieved moderate but significant anti-HSV effect in vero cell culture
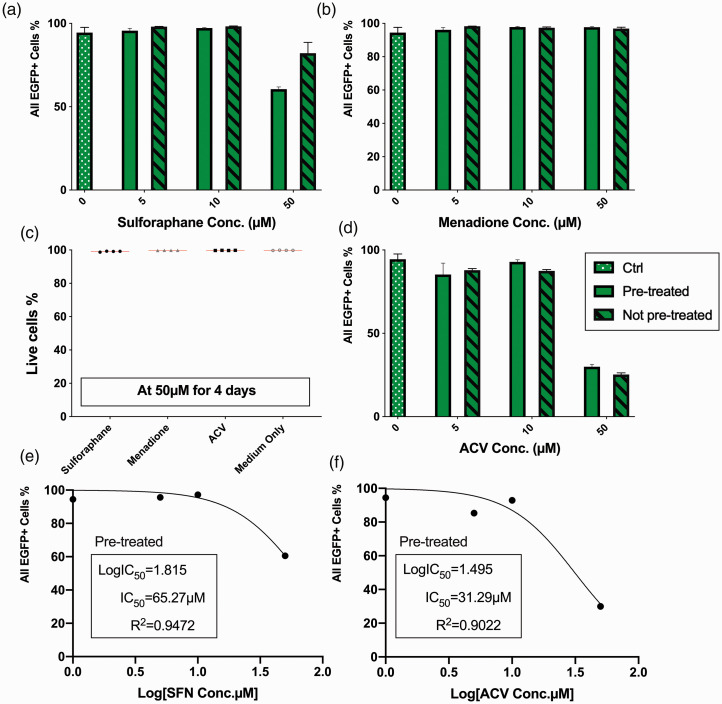
Next, we selected the top significantly positively correlated compound Sulforaphane (SFN, a plant extract widely used as nutraceutical). We also selected Menadione which is also known as vitamin K3, based on aforementioned criteria. In HSV-1 infected Vero cells (MOI of 1), 72 hours treatment with compounds revealed that Menadione failed to show antiviral activity to HSV-1 lytic infection, even at 50 µM with EGFP+ cell percentage 96.8% (Figure 3(b)). In contrast, 50 µM sulforaphane treatment resulted in a moderate but statistically significant reduction of infected cells percentage compared to untreated cultures (from 94.5% to 82.1%, p = 0.0080, Figure 3(a)). A comparable but more significant reduction was achieved with 5 µM Acyclovir. A decrease of EGFP+% to 87.83% (p = 0.0051) with Acyclovir compared to untreated infected (Figure 3(d)). 50 µM ACV was able to decrease the EGFP+ cell percentage to 25.27% (p < 0.0001, Figure 3(d)). Half-maximum inhibitory concentrations were calculated for both SFN (180.4 µM, Figure 3(e)) and ACV (26.69 µM, Figure 3(f)).
Figure 3.
Effects of positively correlated compounds towards HSV-1 acute infection compared with Acyclovir. Vero cells were infected with HSV-1 at MOI = 1. After inoculated with compounds from 5 to 50 µM for 72 hours, percentages of total EGFP positive (EGFP+) cells were acquired via flowcytometry: (a) sulforaphane; (b) Menadione; (c) Live cell percentages of uninfected cells after cultured with 50 µM of compounds for 4 days; (d) Acyclovir; (e) Half-maximal inhibitory concentration of Sulforaphane was calculated using non-linear fit with variable slope; (f) Half-maximal inhibitory concentration of Acyclovir was calculated using non-linear fit with variable slope.
Toxicity overode the anti-viral effect of sulforaphane when applied to human NPC cell culture
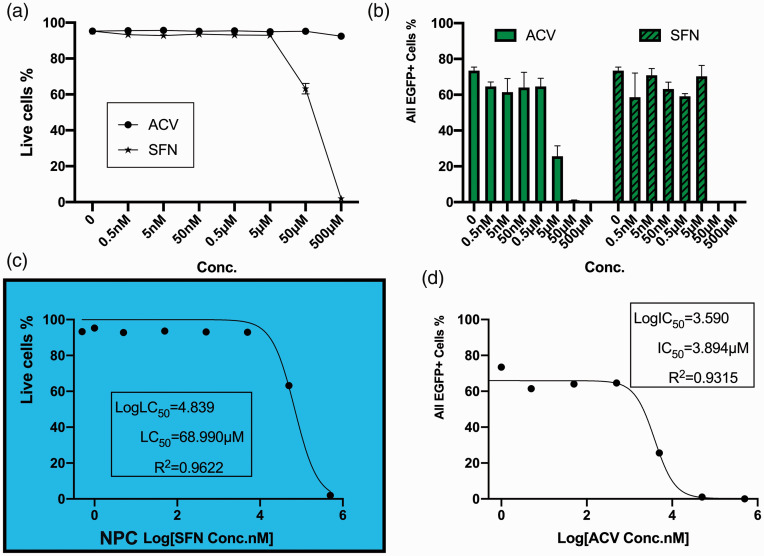
A range of concentrations (0.5 nM ∼ 500 µM) of SFN and ACV were applied to treat HSV-1 acutely infected two-dimensional cultures of neural progenitor cells derived from human pluripotent stem cells. ACV was still protective in NPC cultures with a decrease of EGFP+ percentage to 25.61% at 5 µM, while EGFP+ cells were barely observed under microscope at 50 µM (FC showed 1.02%), (Figure 4(b), left). However, sulforaphane exhibited rather high toxicity to NPCs when concentration was raised to 50 µM with a noticeable reduction in live cell percentage (from 92.9% to 63.20%, Figure 4(a)). 500 µM SFN wiped out uninfected NPCs while the ones incubated with same concentration of ACV remained viable (1.97% compared to 92.47%, Figure 4(a)). Further calculation of half-maximal inhibition concentration (IC50) of ACV from infected and treated cultures revealed an IC50 of 3.894 µM (Figure 4(d)), and since SFN showed toxicity, a half-maximal lethal concentration (LC50) of 68.990 µM was calculated from uninfected but SFN-treated cultures (Figure 4(c)).
Figure 4.
Human iPSC-derived NPCs were cultured with compounds overnight and then infected with HSV-1 at MOI = 0.1 with the presence of compounds. After inoculated with compounds from 0.5 nM to 500 µM for 72 hours, percentages of total EGFP positive (EGFP+) cells and cell viability were acquired via flowcytometry: (a) Live cell percentages of uninfected cells after cultured with different concentrations of compounds for 4 days; (b) Total EGFP+ percentage, left: Acyclovir, right: sulforaphane; (c) Half-maximal lethal concentration of Sulforaphane was calculated using non-linear fit with variable slope; (d) Half-maximal inhibitory concentration of Acyclovir was calculated using non-linear fit with variable slope.
Upstream regulator analysis using ingenuity pathway analysis
To cross-validate the correlation between gene expression changes observed in our input dataset and those resulted from sulforaphane, the most significantly correlated bioset from sulforaphane related studies was chosen (Table 1. right, GSE20479: Primary human hepatocytes + 50uM sulforaphane for 48 hr _vs_ vehicle). Further analyses were conducted with Ingenuity Pathway Analysis software with the threshold for input dataset was set as p < 0.05, |fold change| ≥2.
4508 matching genes from both our input dataset and sulforaphane dataset were identified by Correlation Engine, to demonstrate the strongly correlation between these two datasets, here we only reported the exact up- and down-regulated genes with absolute fold changes higher than or equal to 3. Their original ranks as well as their functions indicated by Correlation Engine are also demonstrated in the table (Table 2).
Table 2.
Up- and down-regulated genes from both our input bioset and sulforaphane bioset with |fold change| ≥ 3.
| Gene | Rank 1 | FC 1 | Rank 2 | FC 2 | Description |
|---|---|---|---|---|---|
| Up-regulated | |||||
| COX11 | 7 | 28 | 2818 | 4.4594 | COX11 cytochrome c oxidase assembly homolog (yeast) |
| NUFIP2 | 52 | 5.45 | 906 | 7.4016 | nuclear fragile X mental retardation protein interacting protein 2 |
| LYAR | 67 | 5.05 | 5555 | 3.1189 | Ly1 antibody reactive homolog (mouse) |
| SBNO1 | 105 | 4.16 | 2018 | 5.2157 | strawberry notch homolog 1 (Drosophila) |
| ABHD5 | 120 | 4.01 | 953 | 7.2345 | abhydrolase domain containing 5 |
| KCTD9 | 122 | 4 | 1785 | 5.5299 | potassium channel tetramerisation domain containing 9 |
| MIR22HG | 159 | 3.68 | 1609 | 5.7696 | MIR22 host gene (non-protein coding) |
| PTGR1 | 168 | 3.63 | 4560 | 3.4829 | prostaglandin reductase 1 |
| CAB39L | 179 | 3.51 | 2710 | 4.5323 | calcium binding protein 39-like |
| SENP5 | 182 | 3.48 | 3763 | 3.8516 | SUMO1/sentrin specific peptidase 5 |
| GDAP2 | 262 | 3.06 | 3378 | 4.0779 | ganglioside induced differentiation associated protein 2 |
| GPATCH8 | 280 | 3.01 | 5096 | 3.2856 | G patch domain containing 8 |
| Down-regulated | |||||
| ADI1 | 1 | –326 | 4748 | –3.4044 | acireductone dioxygenase 1 |
| C1S | 5 | –29 | 5036 | –3.3082 | complement component 1, s subcomponent |
| EAF2 | 18 | –11 | 4077 | –3.707 | ELL associated factor 2 |
| BLVRB | 25 | –7.8 | 2558 | –4.6446 | biliverdin reductase B (flavin reductase (NADPH)) |
| ANXA13 | 33 | –6.7 | 1017 | –7.0587 | annexin A13 |
| CYP4V2 | 45 | –5.6 | 2680 | –4.5541 | cytochrome P450, family 4, subfamily V, polypeptide 2 |
| GPR88 | 53 | –5.4 | 855 | –7.5929 | G protein-coupled receptor 88 |
| KANK2 | 63 | –5.1 | 5874 | –3.0202 | KN motif and ankyrin repeat domains 2 |
| SARDH | 73 | –4.9 | 3764 | –3.851 | sarcosine dehydrogenase |
| SPON2 | 77 | –4.9 | 1272 | –6.4124 | spondin 2, extracellular matrix protein |
| IFIT1 | 78 | –4.9 | 1008 | –7.0904 | interferon-induced protein with tetratricopeptide repeats 1 |
| RCAN2 | 94 | –4.4 | 541 | –9.3165 | regulator of calcineurin 2 |
| ATOH8 | 106 | –4.1 | 918 | –7.3566 | atonal homolog 8 (Drosophila) |
| ST6GAL1 | 115 | –4.1 | 5347 | –3.1994 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| CCDC102A | 124 | –4 | 2606 | –4.6129 | coiled-coil domain containing 102 A |
| ADAMTS5 | 134 | –3.9 | 3381 | –4.077 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| CD24 | 135 | –3.9 | 842 | –7.6378 | CD24 molecule |
| ERBB2 | 136 | –3.9 | 3164 | –4.2119 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 |
| ADH5 | 140 | –3.9 | 5861 | –3.0243 | alcohol dehydrogenase 5 (class III), chi polypeptide |
| KDELR3 | 145 | –3.8 | 623 | –8.6848 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| NREP | 150 | –3.8 | 3173 | –4.2053 | neuronal regeneration related protein homolog (rat) |
| MX1 | 152 | –3.7 | 5306 | –3.2138 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| LCAT | 155 | –3.7 | 4938 | –3.3429 | lecithin-cholesterol acyltransferase |
| C2orf72 | 171 | –3.6 | 2062 | –5.1553 | chromosome 2 open reading frame 72 |
| RAMP1 | 184 | –3.5 | 863 | –7.5662 | receptor (G protein-coupled) activity modifying protein 1 |
| TBC1D30 | 190 | –3.4 | 1209 | –6.5538 | TBC1 domain family, member 30 |
| IDH2 | 194 | –3.3 | 2661 | –4.5672 | isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| FAM213A | 194 | –3.3 | 2635 | –4.5889 | family with sequence similarity 213, member A |
| MGMT | 204 | –3.3 | 1300 | –6.3455 | O-6-methylguanine-DNA methyltransferase |
| ACADL | 206 | –3.3 | 1552 | –5.8547 | acyl-CoA dehydrogenase, long chain |
| CCL2 | 211 | –3.3 | 5847 | –3.029 | chemokine (C-C motif) ligand 2 |
| ANO1 | 211 | –3.3 | 5037 | –3.3076 | anoctamin 1, calcium activated chloride channel |
| HSD17B7 | 216 | –3.2 | 2252 | –4.9667 | hydroxysteroid (17-beta) dehydrogenase 7 |
| MYO5B | 218 | –3.2 | 1820 | –5.4814 | myosin VB |
| PTGFR | 236 | –3.1 | 4388 | –3.5731 | prostaglandin F receptor (FP) |
| TRIQK | 241 | –3.1 | 2821 | –4.4572 | triple QxxK/R motif containing |
| GDA | 252 | –3.1 | 1554 | –5.854 | guanine deaminase |
| NAT6 | 258 | –3.1 | 3973 | –3.7483 | N-acetyltransferase 6 (GCN5-related) |
| ZNF599 | 262 | –3.1 | 3438 | –4.0387 | zinc finger protein 599 |
| GPX7 | 262 | –3.1 | 4359 | –3.5837 | glutathione peroxidase 7 |
| NDRG2 | 280 | –3 | 3066 | –4.283 | NDRG family member 2 |
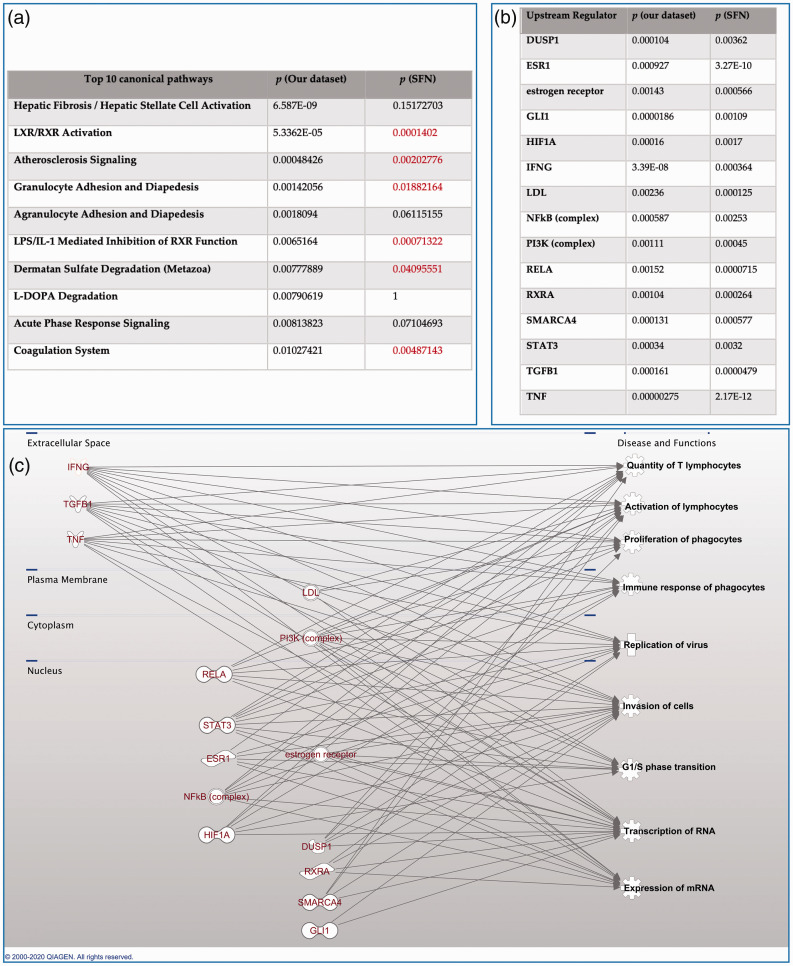
All matching genes identified from both biosets were input into IPA for canonical pathway analysis. Among the top 10 significant canonical pathways identified from our dataset, 6 of them are also significantly (p < 0.05) altered in the Sulforaphan dataset, suggested by IPA (Figure 5(a)). It indicated that SFN resulted in changes of host immune response by influencing several related pathways, like granulocyte adhesion and diapedesis, oxidative stress response, to name a few. Instead of focusing on the huge number of overlapped canonical pathways themselves, we set a much more strict threshold (p < 0.005) to identify common upstream transcriptional regulators (TR) from both datasets to reveal the modulators that might be responsible for transcriptomics changes. For each potential TR, Overlap p value was calculated by the software based on the significant overlap between dataset genes and known targets regulated by a TR.19 We successfully identified a small group of TRs, which were significantly changed in both datasets (Figure 5(b)). They interact with each other at different cellular levels and their common functional pathways are closely related to immunological processes, cell cycle surveillance, as well as directly to replication process of virus (Figure 5(c)).
Figure 5.
Ingenuity pathway analysis. (a) top 10 significantly altered pathways identified from our dataset, compared to their corresponding significant levels when analyzed with SFN dataset; B) list of the 15 upstream transcriptional regulators that are significantly altered (p < 0.001) in both biosets; (c) brief functional analysis of the 15 regulators from (b), suggested by IPA.
Discussion
We tested a simplistic repurposing strategy to identify novel agents with potential antiviral (more specifically anti-HSV-1) characteristics based on BaseSpace® Correlation Engine developed by Illumina®. We identified 10 compounds that induced opposite or similar changes in the gene expression profile compared to what we had observed in HSV-1 infected neurons from previous work. Importantly, we tested the anti-HSV-1 activity of the compounds selected in silico using both Vero cell and human neural progenitor cell cultures; the latter to test for CNS cell relevance. Paradoxically, sulforaphane, which was selected from the positively correlated group exhibited moderate anti-HSV-1 activity in Vero cultures, though it was less efficacious in NPCs, primarily due to toxicity.
Most of the drug-repurposing strategies use an initial data-driven in silico analysis to identify candidate drugs, followed by the experimental validation of their activity with in vitro models.20 The computational methodologies provide information regarding signatures of drugs, such as transcriptomics, metabolomic, chemical structures or even advent event profiles, and so on. Incorporative knowledge databases make the computational analysis more accessible. By incorporating genomic studies and pharmacological knowledge, it is possible to examine the effect of the compound more carefully before moving to experimental validation or clinical trials.21 The Correlation Engine applied in this study incorporates substantial genomic data, proteomic and assay data gathered from different studies and pre-sorted, normalized and re-constructed. These data together with the rank-based enrichment ensure the reliability of subsequent in silico processes.22 The platform enables us to compare our expression profile with biosets from studies that are defined as a ranked list of genes corresponding to a specific treatment of a compound.23 In previous work,24 we observed a series of transcriptomics changes from HSV-1 acutely infected human neurons. The transcriptomic data were imported into the Correlation Engine as the blueprint to conduct a drug-infection matching at transcriptomics level.
Strict filtration processes were applied to the exported list of compounds and their corresponding biosets (Figure 1). We considered it crucial to acquire specific biosets and check its original design and experimental processes in detail. We identified several compounds significantly correlated with the gene profile changes noted in HSV-1 infected neurons (Top 10 listed in Table 1). Further sorting was performed manually (Figure 1, Right Square, Top). Still, our in vitro verification processes revealed that simplistic assumptions of negative correlations may be inadequate, as indicated by the failure of two negatively correlated compounds (Vorinostat and Valproic acid) to protect Vero cultures against acute HSV-1 infection (Figure 2). Both Vorinostat and Valproic acid are well-recognized histone deacetylase inhibitors (HDACs) that have been associated with stimulation of replication of a wide range of viruses such as Human Immunodeficiency Virus (HIV),25–27 Human Herpes Virus 8 (HHV-8),28 and Cytomegalovirus (CMV).29 Besides, valproic acid also interferes with cellular lipid metabolism and affects the composition of cell membranes.30 This makes it a potential antiviral agents towards certain enveloped viruses which acquire their lipid envelopes from host cell membranes.30–32 On the other hand, other HDAC inhibitors can reactivate HSV-1 in neuronal cultures.33 In our study, even at 50 µM concentration, valproic acid did not show anti-HSV-1 activity in Vero cell cultures.
The failure of negatively correlated compounds to show antiviral effects suggests that relying simply on the signature reversion principle might not always lead to the desired novel antiviral compounds, especially when it comes to developing antiviral agents using transcriptomics data. Our partial experimental validation for sulforaphane, a positively correlated drug further support this assumption. Sulforaphane is a phytochemical extracted from cruciferous vegetables like broccoli, which has been shown to have protective effects toward a wide range of viruses such as HIV,34 Respiratory syncytial virus,35 Hepatitis C virus,36 and inhibit Epstein-Barr virus reactivation.37
Due to the complex input dataset, a large number of common genes were identified by the ranking and mapping process with the help of correlation engine. To further link the genes to their functional pathways, we conducted further canonical pathway analysis and compared the common canonical pathways indicated by our bioset and the one acquired from SFN in IPA. Surprisingly, a majoirty of the top 10 canonical pathways identified from our dataset are also significantly altered in SFN dataset (p < 0.05, Figure 5(a)). The functional analysis indicated SFN’s ability to modulate several important host immune response related pathways, such as FXR/RXR, LXR/RXR pathway, granulocyte adhesion and diapedesis, to name a few (Figure 5(a)). To clarify that whether these changes of canonical pathways are resulted from higher level We also sought for the upstream regulators responsible for the trascriptomic changes reflected by our microarray data of HSV-1 infected neurons, with a comparison to upstream regulators identified from the bioset supporting SFN (Figure 5(b)). Those significantly altered upstream regulators were closely related to host immune function and cell cycle surveillance such as lymphocyte proliferation, phagocyte differentiation, antigen-presenting cell differentiation. Some of them are directly related to controlling viral invasion and replication (interferon gamma, SMARCA4, etc., 10 out of 15 TRs) and their close interactions with herpes viridae have been well-established.38,39
Our analysis highlights important limitations of the SRP when utilized to identify anti-herpetic drugs. These limitations may be partially overcome by considering the transcritomic signal from host-defense mechanism. Testing the candidate drugs using disease- or tissue-specific cell types would also be of significant importance.
There are several limitations to this study: i) limited number of compounds from the top 10 list were tested in vitro based on their superiorities over others, but more compounds that are promising were not tested; ii) the range of drug concentrations was limited; iii) oncology medications skews the categories of compounds incorporated into the Correlation Engine platform, and few biosets are reported based on neural cultures; iv) some compounds (e.g. interferon beta, interferon gamma) have been validated as protective agents against HSV infections,39,40 but were not tested. Optimization of the in silico processes should be considered in future studies.
Methods
Data processing with correlation engine
The process of identifying a potential antiviral is based on the comparison of gene expression signature changes. To identify compounds that can induce opposite or similar gene expression changes to the source gene expression signature, the original dataset was compared to a large number of other expression profiles with statistically filtered genes in a comprehensive database. A commercially available genomic knowledgebase called BaseSpace Correlation Engine (Illumina®) fascilitated the screening process (https://www.illumina.com/products/by-type/informatics-products/basespace-correlation -engine.html). The Correlation Engine database contains over 1,69,000 lists of statistically filtered genes from over 22,000 studies carried out in 16 species (as of April, 2019). The expression signature we input into the Correlation Engine are a list of genes with fold changes that reflect expression changes between normal hiPSC-derived neurons and those exposed to HSV-1 and undergoing acute infection (MOI = 0.3, data available at https://www.ncbi.nlm.nih.go-v/geo/query/acc.cgi?acc=GSE111656).24 This list is referred to as ‘bioset’ in the following contexts. One could consider the bioset the primary entity in subsequent analyses, consisting of a ranked list of elements (genes, probes, proteins, compounds, single-nucleotide variants [SNVs], sequence regions, etc.) that corresponds to a given experimental factor or condition in an experiment or an assay, for a gene expression experiment, the biosets will consist of gene lists with associated change values and statistical information for each relevant experimental factor. Significance p values are produced by a t test between treated and control sample intensity values.23
The aforementioned signature is compared to all other biosets in the database using a specialized geneset enrichment algorithm which is a fold-change rank-based statistical test called the Running Fishers test. Its general design is analogous to the Gene Set Enrichment Analysis (GSEA) method. It dynamically detects the most significant enrichment signal in a ranked signature set, allowing the signature set to contain relatively more comprehensive collections of genes at a preselected statistical cutoff. It allows us to assess the overlap in regulated genes and determine if those genes are regulated in a similar or opposite manner.22 However, running Fisher algorithm differs from GSEA in the assessment of the statistical significance, where p-values are computed by a Fisher’s exact test rather than by permutations. Overall, the advantage of this approach is the flexibility of being able to compute correlation scores for data of different sizes and filter thresholds. The directional relationship between 2 signatures from two biosets is capturedby the sign of the correlation score. Upregulated genes and downregulated genes are separated into directional subsets, and correlation scores are computed for each directional subset from one signature against each subset from the other signature. The overall correlation score is the sum of directional subset scores, and the sign of the sum determines whether the 2 signatures are positively or negatively correlated.
Specifically, Pharmaco Atlas, an application in Correlation Engine, was used to assess the bioset and to identify studies of compounds that exhibit similar (positive) or opposite (negative) signatures. This application is designed to find compounds and treatments significantly correlated to the input bioset. A total of 16,383 biosets were identified from all available biosets in the knowledgebase (as of 2019 April 15th). Each bioset was annotated, as well as the corresponding experiment/study using available information derived from the GEO submission and/or original publication. Results were ranked according to the significance of correlation and filtered. Only RNA expression datasets conducted with human tissue or cell lines were included in this exploratory application. Survived biosets were also manually checked for the annotations, starting from highly significantly correlated ones. Comparisons/biosets conducted in following conditions were excluded: i) Between both experiment groups, rather than compared to normal or healthy untreated group; ii) Using samples generated or acquired directly from patients with known diseases; iii) Using cells or cell lines with certain gene (genes) modified; iv) With intervention of a complex compounds or cytokines cocktail recipes; v) Using cancer cells or cell lines (Figure 1).
Cell lines and virus strain
All cells were cultured in standard conditions (37 °C, 5% CO2, and 100% humidity).
African green monkey (Vero) cells (CCL-81; ATCC) were maintained in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone) and 5% anti-biotic/anti-mycotic (Gibco).
Human NPCs were derived from human induced pluripotent stem cell (hiPSC) line 73–56,010–02 using the previously described method.41 hiPSC 73–56,010–02 were established at the National Institute of Mental Health (NIMH) Center for Collaborative Studies of Mental Disorders-funded Rutgers University Cell and DNA Repository (http://www.rucdr.org/mental-health) (RUCDR).
HSV-1 strain applied in this study is a KOS-based (VR-1493; ATCC) recombinant that was previously described, which expresses enhanced green fluorescent protein (EGFP) from the immediately early ICP0 promoter and monomeric red fluorescent protein (RFP) driven by the true late regulated promoter driving expression of Glycoprotein C.42
Cell culture infection
Vero cells were seeded in 96-well flat-bottom cell culture plates at recommended density and incubated in media until approximately 80–85% confluency. Cells were then infected as previously described using the KOS-based HSV-1 at a multiplicity of infection (MOI) of 0.3. Infecting virus was removed 1 hour post infection. Cells were washed and then further incubated in media with or without compounds, so that compounds were applied after absorption. Vero cells were then detached with Trypsin-EDTA solution (0.25%, Cat#T4049) at 48 hours post infection and prepared for flow cytometry.
NPCs were seeded in 96-well cell culture plates coated overnight with Matrigel (Corning, REF#356234) at recommend density and cultured in STEMdiff™ Neural Progenitor medium (NP medium) until cells were approximately 80–85% confluent. The cells were infected with the same virus strain at MOI = 0.1 in the presence or absence of tested compounds supplemented into STEMdiff™ NP medium. In our previous study, this MOI was shown to be effective and high enough to induce lytic infection in NPC culture.43 NPCs infected in the presence of compounds were also pretreated with the antiviral for 24 hours. After one hour, the infection medium was removed, cells were washed with PBS and maintained in STEMdiff™ NP medium with or without antiviral. NPCs were dissociated with Accutase (Biolegend, Cat#423201) 72 hours after infection and prepared for flow cytometry.
Flow cytometry
Flow cytometry was carried out using a BD Fortessa LSR (Beckman Dickenson) for quantitative analysis of fluorescent cells and cell viability was assayed using the Viability Dye 780 (BioGems.Ltd).
Statistical analysis
All experiments were performed in biological triplicates or quadruplicates. When there’s comparison between one experimental group and untreated group, unpaired Student t test was applied and two-tail p value was provided. Error bars in figures represent mean ± standard deviation.
Conclusions
Based on the transcriptomics data from HSV-1 infected neurons, we identified a list of candidate antiviral compounds with opposite or similar signatures. Although the negatively correlated compounds were ineffective against acute HSV-1 infection in Vero cell cultures, a positively correlated compound (Sulforaphane) achieved moderate anti-HSV-1 effect in Vero cell culture, but its toxicity overrides its antiviral effect in NPC culture. More comprehensive methods should be developed to balance the effect of virus and the defense from the host which are reflected in transcriptomics data acquired from infected in vitro models. Our results also indicates that appropriate in vitro cell culture should be chosen to model infection and verify the antiviral effect.
Acknowledgements
We sincerely thank Jacquelynn Jones for her kind edits to improve the readability of the context.
Availability of data and materials: The original dataset analysed with BaseSpace Correlation Engine during the current study are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111656.
Authors’ contributions: WZ performed the experiments with great help from AC (in silico part), LD (viral infection assay) and MD (flow cytometry). WZ, AC, MD, JW and LD were major contributors in analyzing the data. WZ, AC, LD, VM, WM, MD and VN were major contributors in interpreting the results. WZ, WM, LD and VN were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by Stanley Medical Research Foundation 07 R-1712, Veterans Administration Pittsburgh Health System Start-Up Funds (Vishwajit L Nimgaonkar, M.D., PhD.), and by the NINDS R01 NS115082-01A1. We also thank China Scholarship Council (CSC) for providing support to WZ (CSC ID: 201806370301).
ORCID iD: Wenxiao Zheng https://orcid.org/0000-0002-4021-2409
References
- 1.Piret J, Boivin G.Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 2011; 55: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schein CH.Repurposing approved drugs on the pathway to novel therapies. Med Res Rev 2020; 40: 586–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMasi JA, Florez MI, Stergiopoulos S, et al. Development times and approval success rates for drugs to treat infectious diseases. Clin Pharmacol Ther 2020; 107: 324–332. [DOI] [PubMed] [Google Scholar]
- 4.Talevi A, Bellera CL.Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opin Drug Discov 2019; 15: 1–5. [DOI] [PubMed] [Google Scholar]
- 5.DiMasi JA, Grabowski HG, Hansen RW.Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 2016; 47: 20–33. [DOI] [PubMed] [Google Scholar]
- 6.Shah PK.Sildenafil in the treatment of erectile dysfunction. N Engl J Med 1998; 339: 1397–1404. [DOI] [PubMed] [Google Scholar]
- 7.Singhal S, Mehta J, Desikan R, et al. Antitumor activity of thalidomide in refractory multiple myeloma. N Engl J Med 1999; 341: 1565–1571. [DOI] [PubMed] [Google Scholar]
- 8.Parvathaneni V, Kulkarni NS, Muth A, et al. Drug repurposing: a promising tool to accelerate the drug discovery process. Drug Discov Today 2019; 24: 2076–2085. [DOI] [PubMed] [Google Scholar]
- 9.Mercorelli B, Palù G, Loregian A.Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol 2018; 26: 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 2019; 18: 41–58. [DOI] [PubMed] [Google Scholar]
- 11.Breckenridge A, Jacob R.Overcoming the legal and regulatory barriers to drug repurposing. Nat Rev Drug Discov 2019; 18: 1–2. [DOI] [PubMed] [Google Scholar]
- 12.Wagner A, Cohen N, Kelder T, et al. Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol Syst Biol 2015; 11: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh YY, Chou CJ, Lo HL, et al. Repositioning of a cyclin-dependent kinase inhibitor GW8510 as a ribonucleotide reductase M2 inhibitor to treat human colorectal cancer. Cell Death Discov 2016; 2: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel SD, Suneja M, Ebert SM, et al. mRNA expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab 2011; 13: 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirza N, Sills GJ, Pirmohamed M, et al. Identifying new antiepileptic drugs through genomics-based drug repurposing. Hum Mol Genet 2017; 26: 527–537. [DOI] [PubMed] [Google Scholar]
- 16.Shin E, Lee YC, Kim SR, et al. Drug signature-based finding of additional clinical use of LC28-0126 for neutrophilic bronchial asthma. Sci Rep 2015; 5: 17784–17711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domke-Opitz I, Kirchner H.Stimulation of macrophages by endotoxin results in the reactivation of a persistent herpes simplex virus infection. Scand J Immunol 1990; 32: 69–75. [DOI] [PubMed] [Google Scholar]
- 18.Lv X, Wang H, Su A, et al. Herpes simplex virus type 2 infection triggers AP-1 transcription activity through TLR4 signaling in genital epithelial cells 06 biological sciences 0601 biochemistry and cell biology 11 medical and health sciences 1107 immunology. Virol J 2018; 15: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiagen. Ingenuity upstream regulator analysis in IPA®. Qiagen White Pap. Germany: Qiagen, 2014, pp.1–10. [Google Scholar]
- 20.Gns HS, Gr S, Murahari M, et al. An update on drug repurposing: re-written Saga of the drug’s fate. Biomed Pharmacother 2019; 110: 700–716. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, Elemento O, Pathak J, et al. Drug knowledge bases and their applications in biomedical informatics research. Brief Bioinform 2018; 20: 1–14. [DOI] [PubMed] [Google Scholar]
- 22.Illumina. Data analysis details: microarray gene expression. San Diego: Illumina, 2010. [Google Scholar]
- 23.Illumina. Data correlation details: enrichment analysis. San Diego: Illumina, 2010, pp.2–5. [Google Scholar]
- 24.D’Aiuto L, McNulty J, Hartline C, et al. R430: a potent inhibitor of DNA and RNA viruses. Sci Rep 2018; 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Vidal E, Badia R, Pujantell M, et al. Dual effect of the broad spectrum kinase inhibitor midostaurin in acute and latent HIV-1 infection. Antiviral Res 2019; 168: 18–27. [DOI] [PubMed] [Google Scholar]
- 26.Archin NM, Kirchherr JL, Sung JAM, et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest 2017; 127: 3126–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolis DM.Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS 2011; 6: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin HJ, DeCotiis J, Giron M, et al. Histone deacetylase classes I and II regulate Kaposi’s sarcoma-associated herpesvirus reactivation. J Virol 2014; 88: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelis M, Köhler N, Reinisch A, et al. Increased human cytomegalovirus replication in fibroblasts after treatment with therapeutical plasma concentrations of valproic acid. Biochem Pharmacol 2004; 68: 531–538. [DOI] [PubMed] [Google Scholar]
- 30.Vazquez-Calvo A, Saiz J-C, Sobrino F, et al. Inhibition of enveloped virus infection of cultured cells by valproic acid. J Virol 2011; 85: 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vázquez-Calvo Á, Martín-Acebes MA, Sáiz JC, et al. Inhibition of multiplication of the prototypic arenavirus LCMV by valproic acid. Antiviral Res 2013; 99: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crespillo AJ, Praena B, Bello-Morales R, et al. Inhibition of herpes virus infection in oligodendrocyte cultured cells by valproic acid. Virus Res 2016; 214: 71–79. [DOI] [PubMed] [Google Scholar]
- 33.Danaher RJ, Jacob RJ, Steiner MR, et al. Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J Neurovirol 2005; 11: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuya AKM, Sharifi HJ, Jellinger RM, et al. Sulforaphane inhibits HIV infection of macrophages through Nrf2. PLoS Pathog 2016; 12: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho HY, Imani F, Miller-DeGraff L, et al. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med 2009; 179: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu JS, Chen WC, Tseng CK, et al. Sulforaphane suppresses hepatitis C virus replication by up-regulating heme oxygenase-1 expression through PI3K/Nrf2 pathway. PLoS One 2016; 11: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CC, Chuang HY, Lin CY, Chen YJ, et al. Inhibition of Epstein-Barr virus reactivation in nasopharyngeal carcinoma cells by dietary sulforaphane. Mol Carcinog 2013; 52: 946–958. [DOI] [PubMed] [Google Scholar]
- 38.Dudek AH, Pfaff F, Bolte H, et al. Partial inactivation of the chromatin remodelers SMARCA2 and SMARCA4 in virus-Infected cells by caspase-mediated cleavage. J Virol 2018. May 30; 92: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikloska Z, Cunningham AL.Alpha and gamma interferons inhibit herpes simplex virus type 1 infection and spread in epidermal cells after axonal transmission. J Virol 2001; 75: 11821–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low-Calle AM, Prada-Arismendy J, Castellanos JE.Study of interferon-β antiviral activity against herpes simplex virus type 1 in neuron-enriched trigeminal ganglia cultures. Virus Res 2014; 180: 49–58. [DOI] [PubMed] [Google Scholar]
- 41.D’Aiuto L, Zhi Y, Kumar Das D, et al. Large-scale generation of human IPSC-derived neural stem cells/early neural progenitor cells and their neuronal differentiation. Organogenesis 2014; 10: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran S, Knickelbein JE, Ferko C, et al. Development and pathogenic evaluation of recombinant herpes simplex virus type 1 expressing two fluorescent reporter genes from different lytic promoters. Virology 2008; 378: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrahamson EE, Zheng W, Muralidaran V, et al. Patterns of HSV-1 infection in neural progenitor cells. J Virol 2020; 94: e00994–20.32493817 [Google Scholar]