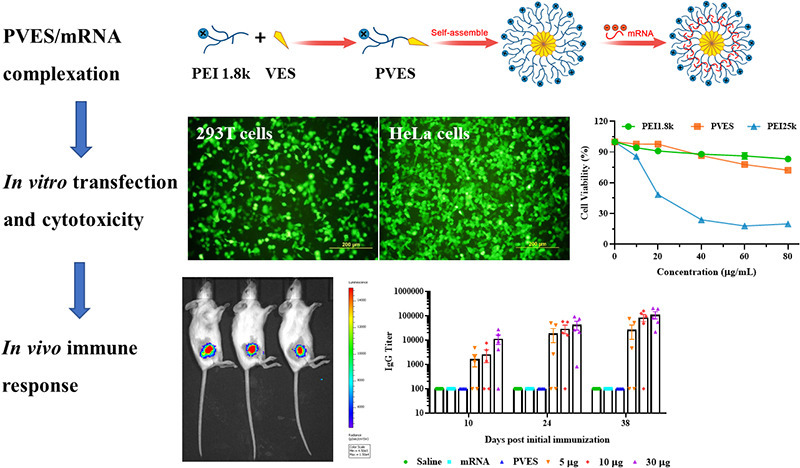
Abstract
mRNA-based therapy has been evaluated in preclinical and clinical studies for the treatment of a wide variety of disease such as cancer immunotherapies and infectious disease vaccines. However, it remains challenging to development safe and efficient delivery system. Here, we have designed a novel self-assembled polymeric micelle based on vitamin E succinate modified polyethyleneimine copolymer (PVES) to delivery mRNA. In vitro, PVES could transfect mRNA into multiple cell lines such as HEK-293T, HeLa and Vero and the transfection efficiencies were much higher than PEI 25 k. In addition, the cytotoxicity of PVES was much lower than PEI 25 k. Furthermore, mice administered intramuscularly with PVES/SARS-CoV-2 mRNA vaccine induced potent antibody response and show no obvious toxicity. These results demonstrated the potential of PVES as a safe and effective delivery carrier for mRNA.
Keywords: mRNA therapy, Polymeric micelle, Immune response
Graphical abstract
1. Introduction
Over the past decade, major technological innovation and research investment have enabled mRNA to become a promising therapeutic tool in the fields of vaccine development and protein replacement therapy [1,2]. mRNA vaccines have several advantages over other vaccine approaches, such as a high safety, a flexibility to encode any protein as antigen, cost-effective and rapid production, which is of great importance in case of pandemic crisis [3,4]. The recent coronavirus disease 2019 (COVID-19) outbreak has demonstrated how quickly emerging infectious disease can spread and underlined the crucial need for a rapid response vaccine. As expected, mRNA vaccines (mRNA-1273 and BNT162b2) were the first kind of vaccines approved by FDA for emergency use. For mRNA-1273, it took only 25 days and 63 days respectively, from sequence selection to vaccine manufacture for the first clinical batch and the first human dosing. Moreover, there are four candidates have already advanced to clinical trials. The bright prospect of mRNA is attracting the attentions of scientists, investors, and even common people [5,6].
However, mRNA therapy is still facing the challenge of lacking safe and effective delivery system, because the large size and dense negative charge make naked mRNA difficult to pass through the cell membrane. In addition, mRNA is also an inherently unstable molecule that is susceptible to degradation [7]. Thus, the development of appropriate delivery systems is urgently required. Numerous strategies have been developed for mRNA delivery, such as lipid nanoparticle, liposome, lipoplex and polyplex [8,9]. At present, lipid nanoparticle (LNP) is the main mRNA vaccine delivery system for clinical application and it can protect mRNA from degradation and aid endosomal escape. Present approved mRNA vaccines developed by Moderna and Pfizer/BioNTech were all encapsulated by LNP and their effectiveness in supporting cell-mediated and humoral immune responses match or surpass other vaccines [10,11]. However, more and more side effects causing by LNP were reported such as pain, redness, fever and flulike symptoms [12]. Recently, a study about inflammatory of LNP showed that LNPs used in many preclinical studies were highly inflammatory in mice. Intradermal injection of these LNPs led to rapid and robust inflammatory responses, characterized by massive neutrophil infiltration, activation of diverse inflammatory pathways, and production of various inflammatory cytokines and chemokines. The same dose of LNP delivered intranasally led to similar inflammatory responses in lungs and a high mortality rate [13]. Thus, the development of more efficient and safe delivery systems is vital and highly necessary.
Although not as clinically advanced as lipid systems for mRNA delivery, polyethyleneimine (PEI) has been shown considerable potential in protein replacement, vaccine and other applications related to mRNA therapeutics [14]. The cationic charge and buffering capacity of PEI are beneficial for complexation with nucleic acids and endosomal/lysosomal release via “proton-sponge” effect [15]. However, its broad therapeutic application has been limited by toxicity associated with high molecular weight. Thus, Low-molecular-weight PEIs were often modified and used for mRNA delivery to reduce toxicity [16]. For example, a polymer synthesized from stearic acid and branched PEI 2 k was used to deliver HIV gag encoding mRNA. Following subcutaneous injection, immune responses were notably induced [17]. In another study, the cyclodextrin-PEI 2 k conjugate was used for the intranasal administration of HIV gap120 mRNA, which resulted in a strong systemic and mucosal HIV-specific immune response [18].
In this study, we developed a self-assembled polymeric micelle delivery system based on polyethyleneimine (1.8 kDa) modified by vitamin E succinate for mRNA vaccine delivery. PVES micelles and mRNA could form nanoscale complexes via electrostatic interaction. We thoroughly assessed the physicochemical properties of PVES and PVES/mRNA complexes including particle size and zeta-potential, mRNA delivery efficiency and toxicity in vitro. Next, the SARS-CoV-2 RBD mRNA vaccine was selected to evaluated whether PVES can successfully deliver mRNA vaccine to induce immune response in vivo.
2. Materials and methods
2.1. Material and animals
Branched PEI with molecular weight of 1.8 and 25 kDa, vitamin E succinate (VES), 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Reporter Gene mRNA (eGFP mRNA and Luciferase mRNA) were purchased from TriLink Biotechnologies (San Diego, CA, USA).
HEK-293T and HeLa cells (Preserved by our laboratory) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA), Vero cells (Preserved by our laboratory) were maintained in minimum Eagle's medium (MEM) (Gibco, USA) and DC2.4 (BNCC, BNCC340111) cells were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) (Gibco, USA) in a humidified atmosphere with 5% CO2, supplemental with 10% fetal bovine serum (FBS, Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (HyClone, USA).
BALB/c (female) mice 6–8 weeks old were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All animal studies were performed in strict accordance with the guidelines set by the Chinese Regulations of Laboratory Animals and Laboratory Animal-Requirements of Environment and Housing Facilities. All animal procedures were reviewed and approved by the Animal Experiment Committee of Laboratory Animal Center, Academy of Military Medical Science (AMMS), China (Assurance Number: IACUC-DWZX-2020-064).
2.2. Synthesis of vitamin E succinate-polyethylenimine copolymer
The vitamin E succinate-polyethylenimine copolymer (PVES) was synthesized by amide reaction. Briefly, vitamin E succinate (100 mg) was dissolved in 8 mL CHCl3. Then, EDC (2.0 eq.) was added. The mixture was stirred at room temperature for 1 h. PEI 1.8 k (0.5 eq.) was then added in the mixture and stirred at room temperature for 8 h. After the reaction was completed, solvent was removed by vacuum rotary evaporation. Product was dialyzed against distilled water for 24 h and lyophilized overnight.
2.3. Formulation and characterization of PVES/mRNA complexes
PVES/mRNA complexes were freshly prepared by mixing a fixed amount of mRNA stock solution and varying amounts of PVES in sterile distilled RNase-free water. The ratio of PVES/mRNA was calculated as the molar ration of nitrogen in PEI portion of PVES and phosphate in mRNA. After mild vortex, the mixture was incubated at room temperature for 30 min to allow particles form. As the control, PEI (1.8 k or 25 k)/mRNA complexes were similarly prepared.
Gel retardation assays were performed to confirm the ability of PVES to condense mRNA and provide protection from degradation. First, PVES/mRNA complexes with 1 μg mRNA were prepared as described above at various N/P ratios from 4 to 32 in a final volume of 5 μL. Each of these complexes was mixed with 1 μL 6 × RNA loading buffer (Beyotime Biotechnology, Shanghai, China) and electrophoresed on a 1% (w/v) agarose gel for 45 min at 100 V. Then, mRNA retardation was visualized and photographed by ChemiDoc XRS imaging system (Tanon-5200 Multi, Shanghai, China). In the second study, RNase A (Beyotime Biotechnology, Shanghai, China) was incubated with naked eGFP mRNA (1.0 μg) or the equivalent amount of mRNA complexed with PVES at the N/P ratios of 16 and 32. After 10 min, the mixture was also analyzed by electrophoresis.
Size distribution of PVES and PVES/mRNA complexes of N/P ratios from 4 to 48 were measured using dynamic laser light scattering (DLS) on a particle analyzer (Litesizer 500, Anton Paar, Austria). The zeta potential was analyzed with the same apparatus. The morphology of PVES and PVES/mRNA complexes were analyzed by transmission electron microscopy (TEM, Hitachi H-7650, Tokyo, Japan) using a negative stain technique. PVES (1.0 μg/μL) and PVES/mRNA complexes (N/P = 32) were absorbed to a copper grid for 60 s and stained with phosphotungstic acid (1%) for 20 s before observation.
2.4. In vitro transfection
HEK-293T cells were transfected with PVES/eGFP mRNA complexes. One day before transfection, cells were seeded in a 24-well plate at the density of 2 × 105 cells per well. After 24 h, complete medium was replaced by serum-free medium. PVES/mRNA complexes at various N/P ratios (1.5 μg mRNA/well) were added to cells. 4 h after transfection, the cell culture medium was replaced with fresh complete culture medium and the cells were incubated for another 20 h. Positive controls of transfection were performed with PEI 25 k/mRNA and Lipofectamine 3000/mRNA complexes according to the standard protocol. Negative control was performed with PEI 1.8 k/mRNA complexes at equivalent concentration. The same procedure was also applied to determine eGFP expression in HeLa, Vero and DC2.4 cells after transfected with PVES/mRNA complexes.
To quantify the percentage of eGFP fluorescent-positive cells, the transfected cells were harvested and analyzed by flow cytometry. Briefly, cells were washed with PBS and harvested with 0.25% trypsin/EDTA. The cells were then suspended in PBS and analyzed immediately by flow cytometer (FACSAria II, BD, USA). The percentage of eGFP fluorescent-positive cells were obtained by measuring the number of fluorescent cells verse the control cells. Approximately 10,000 cells were analyzed to obtain the statistical data. The same procedure was also applied to determine eGFP expression in HeLa, Vero cells and DC2.4 cells after being transfected with PVES/mRNA complexes.
2.5. Cytotoxicity of PVES/mRNA complexes and PVES in vitro
The cytotoxicity of PVES/mRNA complexes was measured using a Cell Counting Kit-8 (CCK-8, Dojindo, Japan) according to the instructions. HEK-293T, HeLa, Vero and DC2.4 cells were seeded into a 96-well plate (2 × 104 cells/well). After 24 h, complete medium was replaced by serum-free medium. PVES/mRNA complexes at N/P of 32 and 40 (0.3 μg mRNA/well) were added to cells. 4 h after transfection, the cell culture medium was replaced with fresh complete culture medium and the cells were incubated for another 20 h. Positive and negative controls were performed with PEI 25 k/mRNA and PEI1.8 k/mRNA at the same N/P ratio. After 24 h of incubation, the culture medium was removed and 100 μL fresh medium containing 10% CCK-8 was added to each well. The cells were incubated at 37 °C for 30 min. Absorbance was measured at a wavelength of 450 nm using a microplate reader (Sunrise, TECAN, Switzerland).
The cytotoxicity of PVES was further assessed on HEK-293T cells. Briefly, HEK-293T cells were seeded into a 96-well plate (5 × 103 cells/well). After 24 h of incubation, the culture medium was replaced with fresh medium and PVES at predetermined concentrations (0 to 80 μg/mL) were added to each well. In parallel, PEI 1.8 k and PEI 25 k were used with the same dosage for comparisons. After 48 h of incubation, the culture medium was removed and 100 μL fresh medium containing 10% CCK-8 was added to each well. The cells were incubated at 37 °C for 30 min. Absorbance was measured at a wavelength of 450 nm using a microplate reader (Sunrise, TECAN, Switzerland).
2.6. Cellular uptake of PVES/mRNA complexes in vitro
HEK-293T cells were applied to test cellular uptake of PVES/mRNA complexes. Briefly, Luciferase mRNA was labeled by Fluorescein Labeling Kit (Mirus, Madison, USA) and cells were seeded into 24-well plates (2 × 105 cells/well) and allowed to grow for 24 h. Then cells were treated with PVES/MFP488-mRNA complexes at N/P = 32. After transfection for 30 min, 60 min, 120 min and 240 min, cells were washed with PBS, and harvested with 0.25% trypsin/EDTA. The cells were then resuspended in PBS and cellular uptake was analyzed by flow cytometry.
2.7. Bioluminescence imaging in vivo
For detection of in vivo expression of PVES/mRNA complexes, BALB/c mice were administered with 10 μg of luciferase mRNA loaded into PVES via intramuscular route. At indicated times post inoculation (6 h, 24 h and 48 h), mice were injected intraperitoneally with 3 mg luciferase substrate. After reaction for 10min, fluorescence signals were collected by IVIS Spectrum instrument (IVIS Spectrum, PerkinElmer, USA) for 180 s, and the fluorescence signals in regions of interest (ROIs) were quantified using Living Image 3.0.
2.8. In vitro transcription of mRNA vaccine
The SARS-CoV-2 RBD mRNA was prepared by in vitro transcription using the T7 standard mRNA production system (Cellscript, Madison, USA) from a linearized DNA template which encodes codon-optimized RBD region of SARS-CoV-2 (residues 319–541, accession number YP_009724390.1) and a cap capping system (Cellscript, Madison, USA) according to the manufacturer's instructions. The mRNA product was precipitated with phenol/chloroform and resuspended in RNase-free water. The concentration of mRNA was determined by Agilent 2100 bioanalyzer system (Agilent, Palo Alto, USA).
2.9. Western blot analysis of PVES/mRNA vaccine
The expression of PVES/mRNA vaccine was verified by Western blot analysis. Briefly, HEK-293T cells (1 × 106 cells/well) were seeded into a 6-well plate and incubated in a 5% CO2 incubator at 37 °C for 24 h. The PVES/mRNA vaccine complexes at N/P ratio of 32 (5.0 μg mRNA/well) were transfected according to the previous transfection method. After 24 h, the cells were collected, and radio immunoprecipitation assay (RIPA) lysate and proteinase inhibitor were used to lyse the cells. The total protein of the cells was extracted, and a bicinchoninic acid assay kit (Beyotime, Shanghai, China) was used to measure the protein concentration. The same amount of protein was taken for SDS-polyacrylamide gel electrophoresis separation, transferred on to the PVDF membrane, blocked with 5% skimmed milk, detected with SARS-CoV-2 RBD rabbit PAb (1:1000) (Sino Biological, Beijing, China) and secondary antibody (Sino Biological, Beijing, China). The blots were visualized with Clarity Western ECL Substrate (Applygen, Beijing, China) on Chemiluminescence imaging system (Tanon-5200 Multi, Shanghai, China).
2.10. Indirect immunofluorescence assay
Indirect immunofluorescence assay was used to verify the intracellular expression and localization of RBD protein. HeLa cells (2 × 104 cells/dish) were seeded into 35 mm glass-bottom culture dishes (Corning, New York, USA), and cultured for 24 h. Then cells were transfected with PVES/mRNA vaccine (N/P = 32) for 24 h. The cells were washed thrice with PBS and fixed with 4% paraformaldehyde (Beyotime, Shanghai, China) for 20 min. Then, the cells were washed and permeabilized with Triton X-100 (Beyotime, Shanghai, China) for 10 min. After washing with PBS thrice, the cells were blocked with blocking buffer (Beyotime, Shanghai, China) for 15 min. Similarly, the cells were washed thrice and incubated with SARS-CoV-2 RBD rabbit PAb (1:250) (Sino Biological, Beijing, China) overnight at 4 °C. After washing five times, the MFP488-conjugated goat anti-rabbit immunoglobulin G (IgG) antibodies (1:250) were added to the cells and incubated for 1 h at room temperature. Finally, the cells were incubated with DAPI for 10 min and observed by spinning disk confocal with Nikon Ti-E microscope(Ultraview VOX, PerkinElmer, USA).
2.11. Animal immunization
Female BALB/c mice aged 6–8 weeks were randomized into 6 groups (5 animals per group) and immunized intramuscularly thrice with PVES/mRNA vaccine complexes (5 μg, 10 μg and 30 μg mRNA/mouse, N/P = 32) at an interval of 14 days. For the negative controls, the mice received the same volume of saline, 30 μg mRNA and PVES at the same time point. Blood was collected from the orbital vein at 10, 24 and 38 days post initial immunization. The collected blood was centrifuged at 4000 rpm to isolate serum (30 min, 4 °C). The serum was stored in aliquots at −20 °C for subsequent detection of SARS-CoV-2 RBD specific IgG.
2.12. Serum antibody evaluation
ELISA was used to measure SARS-CoV-2 RBD specific IgG antibody. SARS-CoV-2 RBD specific IgG titer were determined by a commercial ELISA kit (KIT006, Sino Biological, Beijing, China) according to the manufacturer's instruction. Briefly, serial 10-fold dilutions of serum, starting at 1:100, were added to blocked 96-well plates (100 μL/well) coated with recombinant SARS-CoV-2 RBD protein and plates were incubated for 2 h at room temperature. After five washes with wash buffer, plates were added with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sino Biological, Beijing, China) and incubated for 1 h at room temperature. Plates were then washed five times with wash buffer and added with chromogen solution followed by 20 min of incubation at room temperature. The absorbance at 450 nm was read using a microplate reader (Sunrise, TECAN, Switzerland). The endpoint titers were defined according to the manufacturer's instruction.
2.13. Intracellular cytokine staining assay
An intracellular cytokine staining assay was performed to characterize antigen specific CD4+ and CD8+ immune responses. Briefly, spleens were removed from immunized mice at 4 weeks post immunization and splenocytes were isolated. Mouse splenocytes were added to a 12-well plate (1 × 106 cells/well) and then stimulated with the peptide pool (2 μg/mL of individual peptide) for 2 h. After that, Golgiplug (BD Biosciences) was added to a final concentration of 1 μL/mL and incubated for 4 h. Then, the cells were harvested and stained with anti-CD4 and anti-CD8α surface markers (Biolegend). The cells were subsequently fixed and permeabilized in permeabilizing buffer (BD Biosciences) and stained with anti-IFN-γ and anti-IL-4 (Biolegend). Flow cytometric analysis was performed on a BD FACSAria II flow cytometer and the data were analyzed using FlowJo 10.0.
2.14. In vivo toxicity
To evaluate the in vivo toxicity of PVES and PVES/mRNA vaccine, saline, PVES and PVES/mRNA complexes (N/P = 32) were intramuscularly injected into mice at the doses of 180 μg for PVES. Blood was drawn retro-orbitally and plasma was isolated after 6 h and 24 h injection. Cytokines in plasma were measured with immunoassays based on Bio-Plex MAGPIX system according to the manufacture's protocol.
To investigate whether long-term intramuscular administration of PVES and PVES/mRNA vaccine can cause toxic effect, toxicity was observed throughout the entire immune period. The body weights of all the groups were monitored daily and liver, kidney and spleen were removed and sectioned for histology analysis.
2.15. Statistical analysis
Statistical analysis was performed using GraphPad Prism Version 8.0 software. All of data are presented as the mean ± SM. Statistical difference was analyzed by Student's t-test or one-way ANOVA. All tests are accepted as statistically significant when the p value is less than 0.05.
3. Results
3.1. Synthesis and characterization of PVES polymer
PVES was prepared (Fig. S1) by conjugating vitamin E succinate to branched PEI 1.8 k in CHCl3 using EDC as catalyst. The ratio of vitamin E succinate and PEI 1.8 k was 2:1 and it was achieved by controlling the ingredient proportion. Successful conjugation was confirmed by 1H NMR (Fig. S2). Signals from ethylene protons in PEI (-CH2CH2NH-) appeared at δ 2.8–2.5 ppm. Signals assigned to the methyl protons in vitamin E succinate were observed at δ 0.88–0.84 ppm.
3.2. Formulation and characterization of the PVES/mRNA complexes
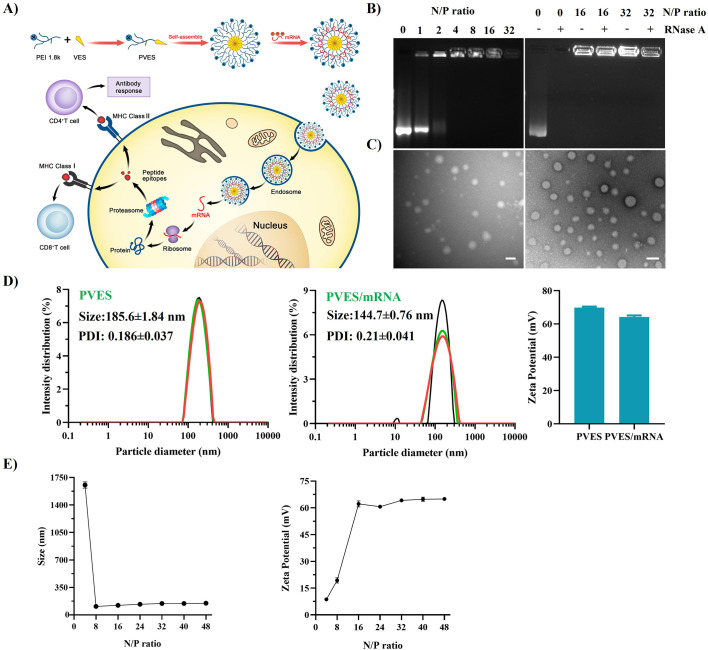
PVES and mRNA were complexed by electrostatic interaction. The ability of PVES to bind mRNA was verified by gel retardation assay. The PVES/mRNA complexes at different N/P ratios (1, 2, 4, 8, 16, 32) were applied and mRNA was efficiently retarded by PVES at a N/P ratio of 4 (Fig. 1B). Agarose gel retardation assay was also performed to study the protection from nuclease degradation of PVES/mRNA complexes. The naked mRNA was completely degraded when treated with Ribonuclease A (RNase A), whereas the mRNA in the complexes with PVES at N/P ratios of 16 and 32 remained intact (Fig. 1B). These results indicate that PVES can efficiently encapsulate mRNA and protect the mRNA from degradation.
Fig. 1.
Characterization of PVES and PVES/mRNA complexes. (A) Schematic illustrating the synthesis of PVES and the process of PVES/mRNA vaccine inducing immune response. (B) Gel retardation assays to detect condensation of mRNA into PVES at different N/P ratios (left) and protection efficiency from RNase A degradation at N/P ratios of 16 and 32 (right). (C) Morphology of PVES (left) and PVES/mRNA at N/P = 32 (right) by transmission electron microscopy. Scale bar, 200 nm. (D) Size distribution and zeta potential of PVES and PVES/mRNA complexes (N/P = 32) determined by DLS. (E) Size distribution (left) and zeta potential (right) of PVES/mRNA complexes at different N/P ratios determined by DLS.
The hydrodynamic diameter and zeta potential of PVES and PVES/mRNA complexes were analyzed. The average size of PVES was 185.6 ± 1.84 nm and zeta potential of 69.8 ± 0.6 mV (Fig. 1D). The average size of PVES/mRNA at N/P ratio of 32 was 144.7 ± 0.76 nm and zeta potential of 64.2 ± 0.9 mV (Fig. 1D). The reduced size after packing mRNA suggested negative charged mRNA facilitate the formation of compact nanoparticles. The particle sizes and zeta potential of PVES/mRNA at different N/P ratios (4, 8, 16, 24, 32, 40 and 48) revealed that PVES could effectively condense mRNA to particle complexes at N/P ≥ 8 (Fig. 1E). Further increase of N/P ratios did not obviously change their particle sizes and zeta potentials.
The morphology of PVES and PVES/mRNA complexes (N/P = 32) determined by TEM (Fig. 1C) illustrated that they were spherical in shape and the size were approximately 200 nm of PVES, 100 nm of PVES/mRNA complexes, which were consistent with the results of DLS.
Altogether, these results confirmed the ability to form stable nanoparticle of PVES and mRNA.
3.3. In vitro transfection of PVES/mRNA complexes
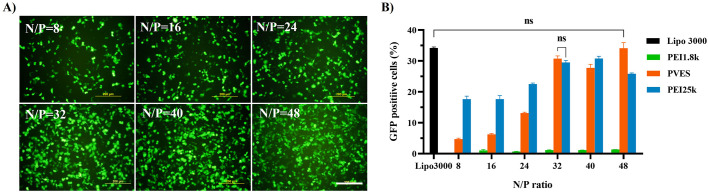
To evaluate the transfection efficiency of PVES/mRNA complexes, eGFP mRNA was used as the reporter gene. HEK-293T cells were transfected with PVES/mRNA complexes at different N/P ratios (Fig. 2 ). The results indicated that the transfection efficiency greatly enhanced with the increase of N/P ratios and reached the plateau at N/P = 32. Further increase of N/P ratios to 40 and 48 did not enhance eGFP expression obviously. At lower N/P ratios, the transfection efficiency of PVES/mRNA complexes was slightly lower than that of positive control PEI 25 k/mRNA complexes. However, When N/P ratio was up to 32, the transfection efficiency of PVES/mRNA complexes was equivalent to PEI 25 k/mRNA complexes. Thus, N/P = 32 was used for subsequent evaluation experiments. As expected, almost no eGFP expression was observed for cells transfected with PEI 1.8 k/mRNA complexes. We also compared the transfection efficiency of PVES with that of a classic nucleic acid transfection reagent lipofectamine 3000 to fully evaluate the potential of PVES as an mRNA delivery vector. The result indicated that the eGFP expression level of the cells transfected with PVES complexes was comparable to lipofectamine 3000.
Fig. 2.
In vitro PVES/mRNA complexes transfection and expression in HEK-293T cells. After 24 h transfection, expression of eGFP mRNA were analyzed by fluorescence microscope and flow cytometry. (A) PVES/mRNA transfection efficiencies in 293T cells at different N/P ratios imaged by fluorescence microscope. Scale bar, 200 μm. (B) eGFP expression efficiency was evaluated by the number of GFP positive cells measured by flow cytometry. Lipofectamine 3000, PEI 25 k and PEI 1.8 k were used as controls. Data were shown as mean ± SEM. Significance was calculated using unpaired t-test (ns, not significant).
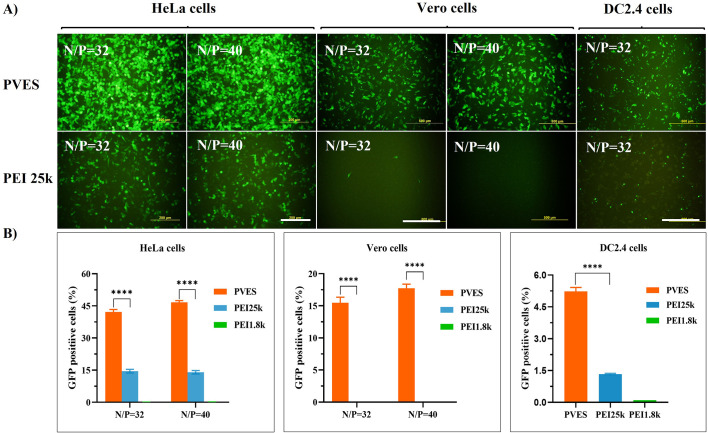
The transfection efficiency of PVES/mRNA complexes on different cell types, including HeLa, Vero and DC2.4 cells were also evaluated (Fig. 3 ). The results showed that in HeLa and Vero cells, the eGFP expression levels of PVES/mRNA were significantly higher than that of PEI 25 k/mRNA complexes. The transfection efficiency of PVES/mRNA complexes was approximate 3.0 times higher than that of PEI 25 k/mRNA complexes in HeLa cells. Besides, there was almost no eGFP expression observed for Vero cells transfected with PEI 25 k/mRNA complexes. As dendritic cells are the most potent professional antigen-presenting cells and play a key role in the immune response, we also studied the efficiency of PVES/mRNA complexes entering to DC2.4 cells. The result indicated that PVES/mRNA complexes (N/P = 32) could successfully enter DC2.4 cells and express GFP protein.
Fig. 3.
In vitro PVES/mRNA complexes transfection and expression in HeLa, Vero and DC2.4 cells. After 24 h transfection, expression of eGFP mRNA were analyzed by fluorescence microscope and flow cytometry. (A) PVES/mRNA transfection efficiencies in HeLa and Vero cells at N/P ratios of 32 and 40; PVES/mRNA transfection efficiency in DC2.4 cells at N/P ratios of 32. Scale bar 200 μm in HeLa cells and 500 μm in Vero and DC2.4 cells. (B) eGFP expression efficiency was evaluated by the number of GFP positive cells measured by flow cytometry. PEI 25 k and PEI 1.8 k were used as controls. Data were shown as mean ± SEM. Significance was calculated using unpaired t-test (****, P < 0.0001).
3.4. Cytotoxicity assay
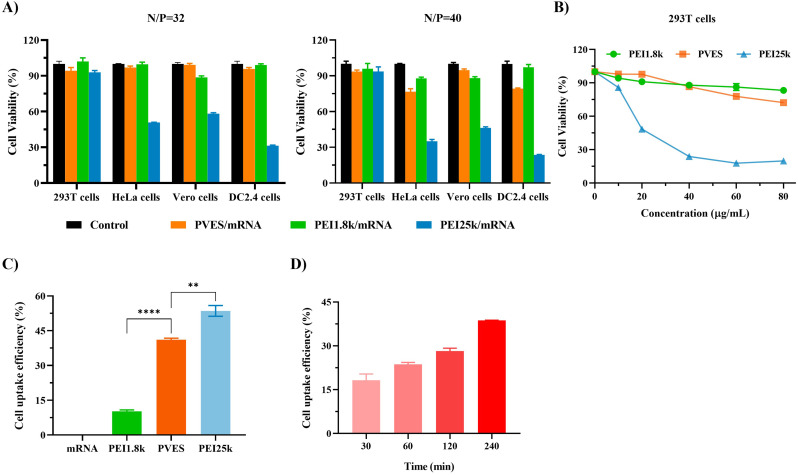
The cytotoxicity of PVES/mRNA complexes at N/P of 32 and 40 were evaluated in HEK-293T, HeLa, Vero and DC2.4 cells (Fig. 4A). The result showed that the cell viabilities of four cell lines after being transfected with PVES/mRNA at N/P of 32 were all close to 100%. When N/P was up to 40, the cell viabilities of HeLa and DC2.4 cells decreased slightly. By comparison, PEI 25 k showed higher cytotoxicity on HeLa, Vero and DC2.4 cells and lower cytotoxicity on HEK 293T cells at N/P of 32 and 40.
Fig. 4.
In vitro cytotoxicity and cell uptake of PVES/mRNA complexes. (A) Cell viability of HEK-293T, HeLa, Vero and DC2.4 cells after being transfected with PVES/mRNA, PEI1.8 k/mRNA and PEI25k/mRNA at N/P of 32 and 40 for 24 h. (B) HEK-293T cell viability at the presence of PVES, PEI 25 k and PEI 1.8 k at different concentrations for 48 h, respectively. (C) Cell uptake of PVES/MFP488-labeled luciferase mRNA by HEK-293T cells after 4 h transfection. (D) Time-dependent uptake of PVES/MFP488-labeled luciferase mRNA by HEK-293T cells. Data were shown as mean ± SEM. Significance was calculated using unpaired t-test (**, p < 0.01, ****, P < 0.0001).
In order to further compare the cytotoxicity of PVES and PEI 25 k on HEK-293T cells, different concentrations (0 to 80 μg/mL) were assessed. The result (Fig. 4B) showed that the cell viabilities of PVES were significantly higher than PEI 25 k. The cell viabilities of PVES were still up to 80% at the concentration of 60 μg/mL and it was only 20% for PEI 25 k at the same concentration.
3.5. Cell uptake of PVES/mRNA complexes
In the cellular uptake experiments, luciferase mRNA was labeled with green fluorescence (MFP488) by nucleic acid labeling kit. The HEK-293T cells were transfected with PVES/MFP488-mRNA complexes (N/P = 32). The results showed that cell uptake of PVES/mRNA complexes were close to positive control PEI 25 k and was higher than PEI 1.8 k after 4 h transfection (Fig. 4C). Time-dependent monitoring of cells treated with PVES/mRNA complexes showed a gradually increase in fluorescent intensity (Fig. 4D), with a high intensity level reached 240 min after transfection, which indicating that the mRNA molecules exited endosomes and enter the cytosol successfully.
3.6. In vivo delivery effectiveness by bioluminescence imaging
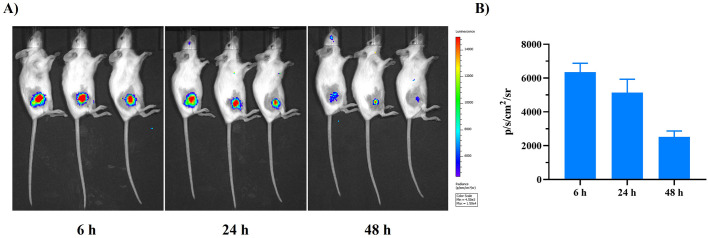
PVES/mRNA complexes (N/P = 32) encoding firefly luciferase were inoculated into mice through intramuscular administration to evaluate the efficiency of PVES as a mRNA in vivo delivery vector. The images (Fig. 5 ) showed obvious fluorescence at the administration site 6 h after injection. Real-time monitoring showed that photo flux faded to undetectable levels 48 h after injection.
Fig. 5.
In vivo delivery effectiveness by bioluminescence imaging. (A) Bioluminescent image to detect luciferase expression in mice 6 h, 24 h and 48 h after i.m. injection of PVES/luciferase mRNA. (B) Fluorescence signals in regions of interest (ROIs) were quantified using Living Image 3.0.
3.7. Verification of mRNA vaccine in vitro
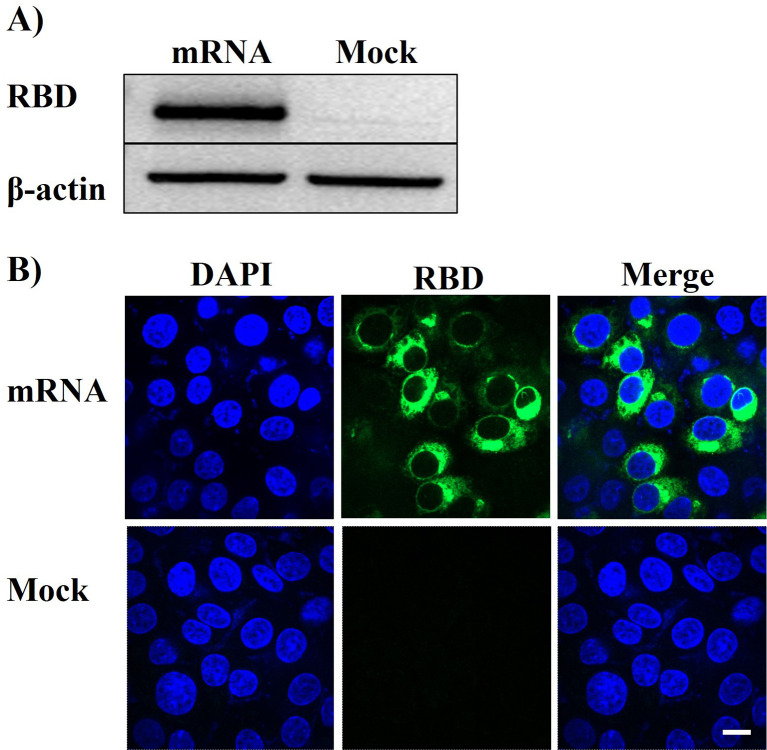
HEK-293T cells were transfected with PVES/mRNA vaccine, and the expression of RBD was confirmed by Western blot analysis and indirect immunofluorescence. The results (Fig. 6A ) showed that RBD was successfully expressed. In indirect immunofluorescence assay (Fig. 6B), the green fluorescence was observed in the cytoplasm. These results revealed that PVES can effectively encapsulate mRNA vaccine and transfect it into cells to express protein, which suggested it could be used for vaccine delivery.
Fig. 6.
In vitro expression of RBD protein after transfection with PVES/mRNA vaccine in HEK-293T cells. (A) Western Blot assay of RBD expression. (B) Indirect immunofluorescence analyses. Mock represents the negative control. Scale bar, 10 μm.
3.8. Immune responses induced by PVES/mRNA vaccine complexes
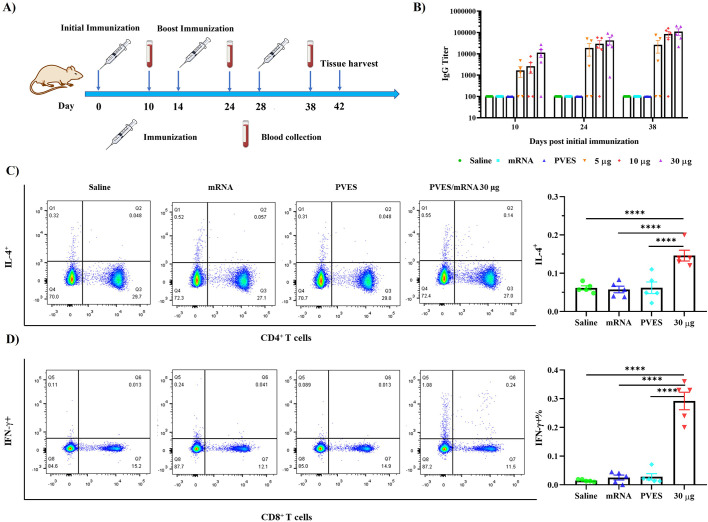
To further verify whether PVES vector can be used for mRNA vaccine delivery and induce immune response in vivo, groups of mice (n = 5) were immunized with different doses of PVES/mRNA vaccine complexes (5 μg, 10 μg and 30 μg/mouse). (Fig. 7A). Titers of RBD specific antibody were detected by ELISA assay to evaluate humoral immune response (Fig. 7B). As a result, three doses of PVES/mRNA complexes elicited significant antibody titers at 10 days after the first immunization. After boosting immunization, the antibody levels were rapid increased. The titers of RBD specific antibody immunized with the high dose were higher than those observed in mice immunized with lower doses. The mean endpoint titers after the third immunization of 30 μg group rose to >105 and were 1.3- and 4.2-fold higher than 10 μg and 5 μg groups. Therefore, we selected the 30 μg dose to immunize mice in the following studies.
Fig. 7.
Humoral and cellular immune responses of PVES/mRNA vaccine. (A) Schematic diagram of immunization, sample collection. (B) The SARS-CoV-2 Spike/RBD specific IgG antibody titer was determined by ELISA. The dashed line indicated the detection limit of the assay. mRNA indicated 30 μg mRNA control group, 5 μg, 10 μg and 30 μg represent PVES/mRNA complexes. Data were shown as mean ± SM. (C, D) Cellular immune responses were evaluated by intracellular cytokine staining assays, the proportions of IL-4-secreting CD4+ and IFN-γ-secreting CD8+ T cells were quantified. Data were shown as mean ± SM. Significance was calculated using one-way ANOVA (****, P < 0.0001).
To characterize the cellular immune responses induced by PVES/mRNA vaccine, intracellular cytokine staining (ICS) assays were performed (Fig. 7C, D). The result showed that after re-stimulation with RBD peptide pools in vitro, the average percentage of IFN-γ expressing CD8+ T cells and IL-4 expressing CD4+ T cells in PVES/mRNA group were considerably higher than control groups, which implied that PVES/mRNA vaccine was able to induce the RBD-specific CD8+ T cell and CD4+ T cell response.
3.9. Evaluation of in vivo toxicity
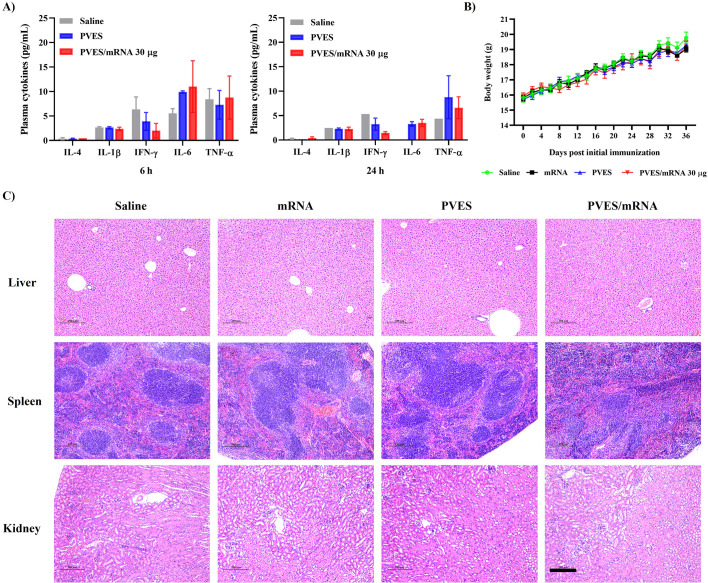
Since mRNA delivery vector often produce undesirable side effects and our study suggested that PVES would be a potential vector. Thus, we assessed the toxicity of PVES and PVES/mRNA complexes in vivo. During the entire vaccination period, the body weights of each group were monitored daily. After the immunization was completed, livers, spleens, and kidneys of three mice of each group (saline, mRNA, PVES and PVES/mRNA) were extracted to examine for histopathology. The results showed that there were no obvious local inflammation of injection site and significant decrease of body weights (Fig. 8B). mRNA, PVES and PVES/mRNA induced no obvious pathologic changes (Fig. 8C), which was similar to the results with saline control group.
Fig. 8.
In vivo safety evaluation of PVES and PVES/mRNA vaccine. (A) Saline, PVES and PVES/mRNA (30 μg) were intramuscularly injected into mice at the dose of 180 μg for PVES. Cytokines in plasma were recorded at 6 h and 24 h post injection. (B) Body weight changes during the immune period. (C) H&E staining of liver, spleen, kidney pathology on 42 days post initial immunization. Scale bar, 200 μm.
Moreover, the levels of several cytokines (IL-1β, IFN-γ, IL-6, TNF-α and IL-4) in plasma at 6 h and 24 h after immunization with saline, PVES and PVES/mRNA were determined. As a result (Fig. 8A), the levels of all the cytokines at 6 h were higher than 24 h. At 6 h, the levels of IL-4, IL-1β, IFN-γ and TNF-α in PVES and PVES/mRNA groups were equal to or lower than saline group. The level of IL-6 in PVES and PVES/mRNA groups was slightly higher than saline group. At 24 h, the levels of IL-4, IL-1β and IFN-γ in PVES and PVES/mRNA groups were equal to or lower than saline group. The level of IL-6 and TNF-α in PVES and PVES/mRNA groups were slightly higher than saline group.
4. Discussion
Vitamin E (VE, α-tocopherol) is not only a daily nutrient, but also an important pharmaceutical agent. VE has been included in FDA inactive ingredients list for intravenous, oral and topical use. In addition, VE has been used as an immune supplement in human, as an emulsion adjuvant component in several veterinary vaccines as well as an adjuvant used in an H1N1 pandemic vaccine (Pandemrix). [19,20]. VE is also utilized as a drug delivery vehicle via the form of tocopherol polyethylene glycol succinate (TPGS) micelles [21,22]. So far, most studies of VE in delivery field have focused on cancer therapy, there are few reports on nucleic acid. Only in 2016, Liu et al. developed a series of vitamin E modified PEI 1.8 k for DNA delivery and the results showed that VE labeling greatly enhance the cellular uptake of GFP DNA plasmid and could successfully deliver pDNA to the liver and lung of living mice [23].
PEI is a water-soluble cationic polymer and VE is a hydrophobic molecule, the covalent conjunction of VE to PEI forms amphiphilic copolymers, which could self-assemble to produce stable micelles [24,25]. It was proved by the homogeneous spheroidal nanoparticles observed by TEM, and the lower polydispersity index (PDI < 0.2) in hydrodynamic diameter. The particle size of PVES was about 180 nm, while it was about 144 nm after condensing negatively charged mRNA. The lower size indicated that PVES/mRNA complexes could form more stable and compact nanoparticles by electrostatic interaction, which is important to allow cellular uptake by endocytosis and avoid rapid clearance by the reticuloendothelial system (RES) in system delivery applications [26].
The micelle-based delivery systems have unique versatility to deliver a variety of payloads including drugs, proteins, peptides, DNA, siRNA etc. [[27], [28], [29]]. As expected, PVES micelle showed high transfection efficiency in four cell lines without significant cytotoxicity. The transfection efficiency (N/P = 32) was close to positive control PEI 25 k and Lipofectamine 3000 on HEK-293T cells. At low N/P ratios (N/P < 32), PVES exhibited less efficient transfection than PEI 25 k. This may result from reduction in the number of primary amines for derivatives modified with VE. Furthermore, in both HeLa and Vero cells, the transfection efficiencies of PVES were much higher than PEI 25 k, which further confirmed the good delivery efficiency of PVES. Dendritic cells (DCs) are specialized antigen-presenting cells that play a central role in initiating and regulating immunity and DCs were usually difficult to transfect [30]. We evaluated the transfection efficiency of PVES/mRNA to DC 2.4 cells, and the results showed that transfection was less efficient but still significantly higher than PEI 1.8 k/mRNA control.
In addition, we found that the transfection efficiency of PEI 25 k in HeLa, Vero and DC2.4 cells was much lower than in HEK-293T cells. In order to further analyze if the decline in transfection efficiency related to cell viability, we assessed the cytotoxicity of PVES/mRNA and PEI 25 k/mRNA complexes at N/P of 32 and 40 in HEK-293T, HeLa, Vero and DC2.4 cells. The result showed that the cell viabilities of PVES/mRNA and PEI 25 k/mRNA complexes were high in HEK-293T cells. This explain why the transfection efficiency was similar for PVES and PEI 25 k in HEK-293T cells. The lower cell viabilities of PEI 25 k/mRNA on other three cell lines explains why the transfection efficiency of PEI 25 k/mRNA is much lower than in HEK-293 T cells. It mainly related to cell viability and not to transfection efficacy. We further compared the cytotoxicity of PVES and PEI 25 k, As expected, in dose-dependent cytotoxicity test, HEK-293T cells maintained significantly high viability after transfection with PVES relative to PEI 25 k at relatively higher concentrations, which shows the safety advantage of PVES.
Then, we evaluated in vivo delivery capability of PVES/mRNA in mice. To visualize mRNA expression, a firefly luciferase reporter encoding mRNA was used in PVES/mRNA complexes. Following intramuscular injection, robust expression of firefly luciferase was detected in the injection site in BALB/c mice 6 h after injection. This indicated that PVES may be a good delivery vector in vivo. Whether it could deliver mRNA vaccine was until unknown because the size of mRNA vaccine is different from eGFP mRNA and firefly luciferase mRNA. Next, we assessed the protein expression in vitro after transfection with PVES/mRNA vaccine and the results showed that mRNA vaccine could successfully translate into antigen in cytoplasm. Subsequently, we determined the antigen-specific antibody IgG after immunized with different doses of PVES/mRNA vaccine complexes. The result showed that three doses could induce SARS-Cov-2 RBD IgG in sera after being firstly vaccinated. Remarkably, two booster doses resulted in rapid increase of IgG. Thus, we speculate that PVES delivering mRNA vaccine has dose dependent effect on RBD specific antibody titers, indicating that it has the potential to be an efficient vaccine vehicle. Additionally, PVES/mRNA vaccine is administrated with the most common used intramuscular injection for human use.
Cell-mediated immune responses play a critical role in combating viral infections. They are comprised of T-cell responses, which fundamentally differ from antibody (humoral) responses in the way they bring about infection control [31]. SARS-CoV-2 mRNA vaccines have been reported that they could induce strong cell immune responses, especially type 1 T cellular responses [5,32]. Here, RBD specific CD4+ and CD8+ T cell responses were evaluated by intracellular cytokine staining assay. PVES/mRNA vaccine elicited antigen-specific CD8+ T cells expressing type 1 (Th1) immune response cytokine (IFN-γ) and CD4+ T cells expressing type II cytokine (IL-4). Therefore, PVES/mRNA vaccine could induce a substantial T cell response against SARS-CoV-2 RBD antigen aside from humoral immune responses. In addition, the level of IFN-γ/CD8+ T cells in immunized group was approximately 10-fold higher than control group whereas IL-4/CD4+ T cells was only 2-fold, which revealed it induced a Th1-biased cellular immune response.
The safety of vaccine has been a public concern. Recently, there are two reports about the thrombosis and thrombocytopenia after ChAdOx1 COVID-19 vaccination [33,34]. In the case of mRNA vaccine, some researchers suspect the immune system response to the delivery vehicle is causing the side effects [12]. Thus, we evaluated the safety of PVES vector and PVES/mRNA vaccine and the results showed that following immunization, no local inflammation response at the injection site or other adverse effects were observed during the observation periods. After all the experiments were completed, the histopathology of liver, spleen and kidney showed no obvious pathological alteration of groups of PVES, mRNA and PVES/mRNA compared to the control group.
However, after immunization with PVES or PVES/mRNA vaccine, some cytokines levels such as IL-6 and TNF-α were slightly higher than control group at 24 h. They might return to normal levels because the levels at 24 h were much lower than 6 h. Moreover, we determined the cytokines levels at 48 h post immunization and the levels were lower than 24 h. Because of almost all the cytokine levels were less than the minimum detection limit and undetectable, we did not present here. Thus, we believe PVES is a potential mRNA vaccine delivery system without serious toxicity.
Taken together, PVES may be a promising mRNA delivery vector. Further study will be carried out such as evaluation of the adjuvant properties and improving targeting.
5. Conclusion
In summary, we developed a mRNA vaccine delivery system based on modified PEI 1.8 k with vitamin E succinate. In vitro, PVES could transfect mRNA into multiple cell lines including HEK-293T, HeLa, Vero and DC2.4 cells and the cytotoxicity was much lower than positive control PEI 25 k. We also provided in vivo evidence that PVES/mRNA administered intramuscularly could efficiently deliver vaccine to induce potent humoral and cellular immune responses and show no obvious toxicity, which demonstrated the potential of PVES as a safe and effective delivery carrier for mRNA vaccine.
Credit author statement
JY and SW conceived the project. JR synthesized, prepared and characterized PVES and performed transfection in vitro, cytotoxicity, cellular uptake, western blot, bioluminescence imaging, indirect immunofluorescence and ELISA assays. YC performed vaccination and evaluation of toxicity in vivo. HL performed intracellular cytokine staining assays. JY, LL and XW designed and synthesized SARS-CoV-2 mRNA vaccine. JR and JY analyzed all the data. JR wrote the manuscript. JY and SW edited and revised the manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (Grant No. 81830101) and the National Science and Technology Major Project (Grant No. 2017ZX09303013-003).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2021.08.061.
Appendix A. Supplementary data
Supplementary material
References
- 1.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics – developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Coolen A.L., Lacroix C., Mercier-Gouy P., Delaune E., Monge C., Exposito J.Y., Verrier B. Poly (lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials. 2019;195:23–37. doi: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 4.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., Tan S., Hao J., Wang Q., Tan W., Gao G.F., Yan J. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021;12:776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang T., Li C., Wang X., Zhao D., Zhang M., Gao H., Liang Z., Xiao H., Liang X.J., Weng Y., Huang Y. Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle. Bioact. Mater. 2020;5:1053–1061. doi: 10.1016/j.bioactmat.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 8.Lostalé-Seijo I., Montenegro J. Synthetic materials at the forefront of gene delivery. Nat. Rev. Chem. 2018;2:258–277. doi: 10.1038/s41570-018-0039-1. [DOI] [Google Scholar]
- 9.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery system. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 10.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 11.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadman M. Public needs to prep for vaccine side effects. Science. 2020;370:1022. doi: 10.1126/science.370.6520.1022. [DOI] [PubMed] [Google Scholar]
- 13.Ndeupen S., Qin Z., Jacobsen S., Estanbouli H., Bouteau A., Igyártό B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. bioRxiv. 2021:430128. doi: 10.1101/2021.03.04.430128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiper M., Tounsi N., Kole R., Kichler A., Zuber G. Self-aggregating 1.8 kDa polyethylenimines with dissolution switch at endosomal acidic Ph are delivery carriers for plasmid DNA, mRNA, siRNA and exon-skipping oligonucleotides. J. Control. Release. 2017;246:60–70. doi: 10.1016/j.jconrel.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Pandey A.P., Sawant K.K. Polyethylenimine: a versatile, multifunctional non-viral for nucleic acid delivery. Mater. Sci. Eng. C. 2016;68:904–918. doi: 10.1016/j.msec.2016.07.066. [DOI] [PubMed] [Google Scholar]
- 16.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M., Li M., Zhang Z., Gong T., Sun X. Induction of HIV gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug. Deliv. 2016;23:2596–2607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 18.Li M., Zhao M., Fu Y., Li Y., Gong T., Zhang Z., Sun X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Release. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y., Luo J., Tan S., Otieno B.O., Zhang Z. The applications of vitamin E TPGS in drug delivery. Eur. J. Pharm. Sci. 2013;49:175–186. doi: 10.1016/j.ejps.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Lodaya R.N., Kanitkar A.P., Friedrich K., Henson D., Yamagata R., Nuti S., Mallett C.P., Bertholet S., Amiji M.M., O’Hagan D.T. Formulation design, optimization and in vivo evaluations of an α-tocopherol-containing self-emulsified adjuvant system using inactivated influenza vaccine. J. Control. Release. 2019;316:12–21. doi: 10.1016/j.jconrel.2019.10.042. [DOI] [PubMed] [Google Scholar]
- 21.Muddineti O.S., Ghosh B., Biswas S. Current trends in the use of vitamin E-based micellar nanocarriers for anticancer drug delivery. Expert. Opin. Drug. Deliv. 2017;14:715–726. doi: 10.1080/17425247.2016.1229300. [DOI] [PubMed] [Google Scholar]
- 22.Gao M., Deng J., Chu H., Tang Y., Wang Z., Zhao Y., Li G. Stereoselective stabilization of polymeric vitamin E conjugate micelles. Biomacromolecules. 2017;18:4349–4356. doi: 10.1021/acs.biomac.7b01409. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Feng M., Liang D., Yang J., Tang X. Vitamin E-labeled polyethylenimine for in vitro and in vivo gene delivery. Biomacromolecules. 2016;17:3153–3161. doi: 10.1021/acs.biomac.6b00776. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh B., Biswas S. Polymer micelles in cancer therapy: state of the art. J. Control. Release. 2021;332:127–147. doi: 10.1016/j.jconrel.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Ghezzi M., Pescina S., Padula C., Santi P., Favero E.D., Cantù L., Nicoli S. Polymeric micelles in drug delivery: an insight of the techniques for their characterization and assessment in biorelevant conditions. J. Control. Release. 2021;332:312–336. doi: 10.1016/j.jconrel.2021.02.031. [DOI] [PubMed] [Google Scholar]
- 26.Gwak S.J., Nice J., Zhang J., Green B., Macks C., Bae S., Webb K., Lee J.S. Cationic, amphiphilic copolymer micelles as nucleic acid carriers for enhanced transfection in rat spinal cord. Acta Biomater. 2016;35:98–108. doi: 10.1016/j.actbio.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshmukh A.S., Chauhan P.N., Noolvi M.N., Chaturvedi K., Ganguly K., Shukla S.S., Nadagouda M.N., Aminabhavi T.M. Polymeric micelles: basic research to clinical practice. Int. J. Pharm. 2017;532:249–268. doi: 10.1016/j.ijpharm.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kaur J., Mishra V., Singh S.K., Gulati M., Kapoor B., Chellappan D.K., Gupta G., Dureja H., Anand K., Dua K., Khatik G.L., Gowthamarajan K. Harnessing amphiphilic polymer micelles for diagnostic and therapeutic applications: breakthroughs and bottlenecks. J. Control. Release. 2021;334:64–95. doi: 10.1016/j.jconrel.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Elsabahy M., Song Y., Eissa N.G., Khan S., Hamad M.A., Wooley K.L. Morphologic design of sugar-based polymer nanoparticles for delivery of antidiabetic peptides. J. Control. Release. 2021;334:1–10. doi: 10.1016/j.jconrel.2021.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Sehgal K., Dhodapkar K.M., Dhodapkar M.V. Targeting human dendritic cells in situ to improve vaccines. Immunol. Lett. 2014;162:59–67. doi: 10.1016/j.imlet.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zajac A.J., Harrington L.E. Immune response to viruses: cell-mediated immunity. Refer. Mod. Biomed. Sci. 2014 doi: 10.1016/B978-0-12-801238-3.02604-0. [DOI] [Google Scholar]
- 32.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaňo D., Amanat F., Muramatsu H., Oguin T.H., III, Ojha A., Zhang L., Mu Z., Parks R., Manzoni T.B., Roper B., Strohmeier S., Tombácz I., Arwood L., Nachbagauer R., Karikó K., Greenhouse J., Pessaint L., Porto M., Taylor T.P., Strasbaugh A., Campbell T.A., Lin P.J.C., Tam Y.K., Sempowski G.D., Farzan M., Choe H., Saunders K.O., Haynes B.F., Andersen H., Eisenlohr L.C., Weissman D., Krammer F., Bates P., Allman D., Locci M., Pardi N. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz N.H., Sørvoll I.H., Michelsen A.E., Munthe L.A., Lund-Johansen F., Ahlen M.T., Wiedmann M., Aamodt A.H., Skattør T.H., Tjønnfjord G.E., Holme P.A. Thrombosis and thrombocytopenia after ChAdOx1 nCoV-19 vaccination. N. Engl. J. Med. 2021 doi: 10.1056/nejmoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCov-19 vaccination. N. Engl. J. Med. 2021 doi: 10.1056/nejmoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material