Abstract
Upon heat shock, transcription of many stress-inducible genes is rapidly and dramatically stimulated by heat shock factor (HSF). A central region of the yeast HSF (designated HSFrr for “repression region”) was previously identified and proposed to be involved in repressing the activation domain under non-heat-shock conditions. Here, we used the phage display system to isolate proteins that interact with HSFrr. This should identify factors that modulate HSF activity or directly participate in HSF-mediated transcriptional activation. We constructed a randomly sheared yeast genomic library to express yeast proteins on the surface of λ phage. HSFrr binding phages were selected by cycles of affinity chromatography. DNA sequencing identified an HSFrr-interacting phage that contains the GAC1 gene. The GAC1 gene encodes the regulatory subunit for a type 1 serine/threonine phosphoprotein phosphatase, Glc7. Both gac1 and glc7 mutations had little effect on HSF activation of gene transcription of two heat shock genes, SSA4 and HSP82. In contrast, heat shock induction of CUP1 gene expression was completely abolished in a glc7 mutant and reduced in a gac1 mutant. The results demonstrate that the Glc7 phosphatase and its Gac1 regulatory subunit play positive roles in HSF activation of CUP1 transcription.
Organisms respond to elevated temperature by activating the transcription of stress-inducible genes whose products improve survival. The key transcriptional activator of the stress-inducible genes is heat shock factor (HSF). Upon heat shock, HSF undergoes two major changes in activity: an increase in DNA binding to its heat shock element sequences (HSEs) and the acquisition of its transcription-stimulatory activity. In higher organisms, the HSE binding activity of HSF is dramatically stimulated by heat shock as HSF monomers are converted to trimers (62). HSF trimers then bind tightly to HSEs, located upstream of heat shock gene promoters (reviewed in reference 33). The mechanism by which HSF triggers transcriptional activation is still unknown; however, DNA binding by HSF is apparently insufficient to cause heat shock activation, and another change in the conformation or modification of HSF is required (29). In budding yeasts (Saccharomyces cerevisiae and Kluyveromyces lactis), HSF binds to DNA in non-heat-shocked cells in vivo (20, 51) but the level of binding to the HSP82 promoter increases upon heat shock and decreases upon recovery (17). However, as in higher eukaryotes, a step beyond DNA binding is also required for yeast HSF to acquire its full activation potential (18).
Many proteins interact with HSF and thereby directly or indirectly regulate HSF transcriptional activity. First, HSF displays a number of critical inter- and intramolecular interactions. In heat-shocked cells, three HSF molecules interact to form homotrimers with strong DNA binding activity (42, 52). In uninduced cells, intramolecular interactions between the amino- and carboxyl-terminal coiled-coil domains are thought to prevent HSF from assuming the active trimer form (7, 43, 65). Second, the targeted modification of HSF appears to play a critical role in modulating activities of HSF. HSF is moderately phosphorylated under non-heat-shock conditions and becomes hyperphosphorylated upon heat shock (12, 53). The hyperphosphorylation of K. lactis HSF is involved in returning HSF to the inactive state after heat shock (25), whereas the constitutive moderate level of phosphorylation represses human HSF activity (30, 31). Also, correlative data suggest that hyperphosphorylation is involved in enhancing human HSF1 transcription activity (64). Therefore, protein kinases and phosphatases must interact with HSF, and they appear to regulate its activity. Third, DNA-bound HSF must communicate with the transcription machinery through protein contacts. Indeed, components of the general transcription machinery such as TATA binding protein also interact with HSF in vitro, and this interaction may be important for transcription in vivo (35). Fourth, chaperones such as Hsp90, Hsp70, and their partners can interact with HSF to inhibit transcription activation during the normal recovery from the heat shock response (1, 38, 44, 49, 66). Since a number of protein-protein interactions play important roles in regulating HSF activities, we sought to identify some of these proteins that can directly interact with HSF by using segments of yeast HSF to screen a phage display library.
A segment of the central region of yeast HSF, including part of the DNA binding domain and most of the trimerization domain, is designated HSFrr (for “repression region”). The repression region was found to be a negative regulator during non-heat-shock conditions, since a deletion of this region resulted in constitutive transcription activation by HSF (39). The mechanism for negative control by HSFrr might be mediated by intramolecular interaction with the HSF activation domain or intermolecular interaction with an as yet unidentified repressor protein that could modify HSF activity. We therefore decided to search first for HSF-interacting proteins that interact with HSFrr.
The phage display system allows the rapid selection and cloning of specific proteins that interact directly with a target protein in vitro (reviewed in reference 4). A library of phages where each phage encodes a different polypeptide fused to the coat or capsid protein of the phage is generated (54). The displayed polypeptides are available for interaction with a target protein that is bound to a solid support. Phages binding the target are selected, amplified, and reselected through multiple rounds of affinity purification (40). Although most applications of this method involve inserting small peptide-coding sequences into coat gene III of M13, a 50-kDa coding region has been successfully cloned without impairing Fd phage functions (36). A 50-kDa coding region has also been fused to the λ D gene and expressed successfully (23). When the peptide libraries are constructed in the coat gene III of phage M13, the expressed molecule(s) must be compatible with the bacterial export system. In contrast, when fused to the capsid gene D of bacteriophage λ, the displayed protein(s) or peptide(s) does not require secretion across the bacterial membrane and thereby provides more comprehensive screening for binding proteins (54).
In this paper, we describe the use of the λ phage display system to identify an HSFrr-interacting protein, Gac1. The Gac1 protein is a regulatory subunit for a type 1 serine/threonine phosphoprotein phosphatase, Glc7 (14, 16, 55). We were particularly intrigued by this interaction of HSF with a protein phosphatase, since the phosphorylation state of HSF appears to be critical in dictating its activity, as discussed above. We examined this interaction both in vitro and in vivo, and investigated its importance by examining HSF-activated gene expression in vivo in strains containing mutations in the GAC1 or GLC7 gene. These results provide strong support for a role of this phosphatase in modulating HSF transcriptional activity.
MATERIALS AND METHODS
Reagents, strains and plasmids.
Restriction enzymes, phage T4 polynucleotide kinase, and DNA polymerase I large fragment (Klenow) were from New England Biolabs, Inc. (Beverly, Mass.). Phage T4 DNA ligase, phage T4 DNA polymerase, and modified phage T7 DNA polymerase (Sequenase 2.0) were from Amersham Corp. (Arlington Heights, Ill.). S1 nuclease and Taq DNA polymerase were from Life Technologies (Gaithersburg, Md.). [γ-32P]ATP was from DuPont (Boston, Mass.). Oligonucleotides were synthesized at the Synthesis Facility of the Cornell University Biotechnology Program or at Life Technologies.
The S. cerevisiae and Escherichia coli strains, plasmids, and phages used are listed in Table 1. Standard methods were used for restriction endonuclease digestion, ligation, and transformation of DNA (13). Plasmid pJTL042, containing a maltose binding protein (MBP)-HSFrr fusion, was constructed as described below. An EcoRI-BamHI fragment encoding HSF amino acids 207 to 395 was generated by PCR (10) with pURA3-yeast HSF (53) as a DNA template. The cycling program used was 30 cycles of 94°C for 90 s, 55°C for 2 min, and 72°C for 3 min. The PCR product was digested by EcoRI and BamHI and cloned into plasmid pMAL-c2 to form the MBP-HSFrr fusion. Plasmid pKCL002 containing a Gal4 DNA binding domain-HSFrr fusion was constructed by cloning the PCR product containing amino acids 207 to 395 of HSF (as described for the construction of pJTL042) into the EcoRI and BamHI sites of pGBDU-C2 (28).
TABLE 1.
Strains, plasmids, and phage used in this study
| Strain, plasmid, or phage | Genotype | Reference or source |
|---|---|---|
| S. cerevisiae strains | ||
| KT1098 | MATα leu2 ura3-52 trp1 glc7-1 | K. Tatchell |
| KT1099 | MATα leu2 ura3-52 trp1 | K. Tatchell |
| KT1100 | MATα leu2 ura3-52 trp1 gac1::LEU2 | K. Tatchell |
| E. coli strains | ||
| BL21 | F−ompT hsdS (rB− mB−) gal dcm | 56 |
| DH5α | φ80d Δ(lacZ)M15 Δ(argF-lac)U169 endA1 recA1 | Gibco BRL |
| NS2973 | Same as JM109 but λ imm434 nin5 | 54 |
| NS2974 | Same as JM109 but λ imm434 nin5 Cre+ | 54 |
| YMC | supF | 54 |
| Plasmids | ||
| pGBDU-C2 | Apr; URA3; 2μm; PADH1-GAL4 BD vector | 28 |
| pGEX-3X | Apr; lacIq Ptac GST | Pharmacia |
| pHH98 | Apr; TRP1; 2 μm; myc-GAC1(130–502) | 63 |
| pJTL029 | Apr; same as pRH825, except the NotI site has been changed to a SmaI site through SmaI linker ligation; loxP+ | This study |
| pJTL042 | Apr; MBP-HSFrr; 0.5-kb EcoRI-BamHI fragment containing amino acids 207 to 395 of HSF in the EcoRI and BamHI sites of pMAL-c2 | This study |
| pJTL047 | Apr; D-GAC1(162–406) construct selected by the phage display system; 0.7-kb fragment containing amino acids 162 to 406 of the Gac1 protein in the SmaI site of pJTL029 | This study |
| pJTL048 | Apr; GST-GAC1(162–406); 0.7-kb BamHI-EcoRI PCR fragment containing Gac1 amino acids 162 to 406 in the BamHI and EcoRI sites of pGEX-3X | This study |
| pKCL002 | Apr; URA3; 2μm; GAL4-HSFrr; 0.5-kb EcoRI-BamHI fragment containing amino acids 207 to 395 of HSF in the EcoRI and BamHI sites of pGBDU-C2 | This study |
| pMAL-c2 | Apr; lacIq PtacmalEΔ2-26-fx-lacZα | New England Biolabs |
| pRH825 | Apr; λ D gene cloned into pTrcHisA with a NotI site in the carboxyl terminus of D coding region; loxP+ | R. Hoess |
| pTrcHisA | Apr; trc promoter and lac operator | Invitrogen |
| pURA3-yHSF | Apr; URA3; 3.7-kb EcoRI HSFL3 fragment containing yHSF gene in the EcoRI site of YCp50 | 53 |
| YCp50 | Apr; URA3; CEN-ARS | 45 |
| Phage | ||
| λ D− loxP | Dam15 imm21 nin5 loxP+ | 54 |
Plasmid pJTL048 containing a glutathione S-transferase (GST)-Gac1 fusion was constructed as described below. A BamHI-EcoRI fragment encoding Gac1 amino acids 162 to 406 was generated by PCR (10) with pJTL047, a positive clone from phage display selection, as a DNA template. The PCR product was digested by BamHI and EcoRI and cloned into plasmid pGEX-3X to form the GST-Gac1 fusion.
Culture media.
Defined media for routine genetic manipulations were as described previously (13, 37). Synthetic complete (SC) medium consists of yeast nitrogen base (6.7 g/liter), appropriate amino acids, and 2% (wt/vol) glucose as described previously (2). Ampicillin was used at 100 μg/ml for liquid medium and 200 μg/ml for solid medium. Agar and dehydrated media were from Difco Laboratories (Detroit, Mich.). Other components were from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.).
Bacterial crude-extract preparation.
The crude extract containing overexpressed MBP-HSFrr was prepared from E. coli BL21 containing pJTL042. A 500-ml volume of this strain was grown to a density of 2 × 108 cells/ml (optical density at 600 mm [OD600] = 0.4 to 0.6) before the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The induced culture was aerated for another 3 h and harvested by centrifugation. The cell pellet was resuspended in 20 ml of Tris-buffered saline (TBS) and subjected to sonication for eight 20-s bursts followed by centrifugation at 15,000 × g for 10 min. The resulting supernatants were stored as 1.5-ml aliquots at −20°C.
Construction of the λ phage display library.
Yeast genomic DNA was randomly sheared (as described by Fleischmann et al. [15]) to an average size of 500 to 1,500 nucleotides, which is sufficient to encode an 18- to 55-kDa polypeptide. The blunt-ended DNA was cloned into the SmaI site, located in the carboxyl terminus of the D gene, of plasmid pJTL029. A total of 1.25 × 106 independent clones were generated. Plasmid pJTL029 contains a loxP sequence downstream of the genomic DNA insertion site SmaI. A Cre-loxP site-specific recombination system (54) was then used to incorporate plasmids containing the D fusion genes into the λ D− loxP (Table 1) phage genome. A more complete analysis and detailed description of the library can be found at our website (32a).
Phage display selection of HSFrr binding proteins. (i) Cycles of selection.
A chromatography selection, modified from methods described previously (22, 32), was performed to select for HSFrr binding phages. A 2-ml volume of the bacterial crude extract containing overexpressed MBP-HSFrr was added to 50 μl of amylose resin (New England Biolabs, Inc.) and incubated in a roller drum at room temperature for 30 min. The resin-bound MBP-HSFrr was washed three times with TBS. A 200-μl volume of the blocking reagent Blotto (TBS containing 5% nonfat dry milk and 0.05% Tween 20) was added, and the resin-protein complexes were incubated at room temperature for an additional 2 h. The Blotto was removed, and 200 μl of the phage library was added to the bead-protein mixture along with 200 μl of fresh Blotto. The mixture was continuously agitated overnight at 4°C. Unbound phages were removed, and the resin-protein-phage complexes were subjected to six consecutive 30-s washes with TBST (TBS containing 0.05% Tween 20) containing 10 mM MgCl2 followed by two washes with TBS containing 10 mM MgCl2. Protein-bound phages were then eluted twice with 50 μl of 10 mM maltose. The number of phages eluted from the resin was obtained by titer determination, and the eluted phages were amplified by growth in E. coli YMC. The percent recovery of bound phages was calculated as (output titer/input titer) × 100 for each round of selection. This selection process was repeated for five rounds to provide strong enrichment for phages that bind HSFrr. More than 108 phage particles were added for each round of selection.
(ii) PCR assay of selected phages.
Two oligonucleotides, flanking the genomic DNA insertion site in plasmid pJTL029, were used to amplify the D-fusion fragment in the eluted phage lysates after each round of selection. The upstream primer, 5′-GGAATAAACCATGGTTGACCGTG-3′, is complementary to bases 18 through 40 upstream of the A in the D gene translation start codon, and the downstream primer, 5′-CAGCTTCGAATTCCTTAGCGGCCC-3′, is complementary to bases 27 through 4 downstream of the genomic DNA insertion site. A 10-μl volume of the eluted-amplified λ phage lysates (described above) was used as the DNA template. The cycling program used was 30 cycles of 94°C for 90 s, 55°C for 2 min, and 72°C for 3 min. The PCR products were visualized on 0.7% agarose gels.
(iii) Preparation of pure phage lysates.
After the homogeneity of the inserted DNA fragment population was determined by PCR, pure phage lysates were generated from the each enriched lysate. Phage lysates were diluted and plated onto tryptone agar to give 50 to 100 plaques per plate. Independent and well-isolated plaques were picked and added to 100 μl of phage dilution buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgSO4, 5 mM CaCl2, 50 mM NaCl). After a 1-h incubation at room temperature, phage lysates were prepared on strain YMC.
(iv) Sequencing.
Either the PCR products amplified from the phage lysates or the converted plasmids (see below) were sequenced. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.). A λ lysogen containing a D-fusion gene was excised and converted into a plasmid by Cre-loxP site-specific recombination (24). Phages were transduced into the Cre+ strain NS2974 at 30°C, and ampicillin-resistant colonies were selected. These colonies contained plasmids that were excised and converted from the λ phages. The structures of the ampicillin-resistant plasmids were confirmed by restriction mapping prior to transformation into the Cre− strain DH5α. The PCR products or plasmids were subjected to sequence analysis by the dideoxynucleotide chain termination method (47) with modified T7 DNA polymerase and [γ-32P]ATP-labeled primer.
Protein binding (pull-down) assays.
The binding assay and buffer conditions were performed as described previously (35). Briefly, 250 ng of MBP-HSFrr was added to 20 μl of GST- or GST-Gac1(162–406)-bound resins in the presence of 0.5 mg of bovine serum albumin per ml. After being mixed at 4°C for more than 2 h, the resin-bound protein complexes were washed twice with 1 ml of washing buffer (35) for 15 s. Bound proteins were eluted with 2× sodium dodecyl sulfate (SDS) loading dye and were electrophoresed on SDS–10% polyacrylamide gels. Western blot analysis was done by standard approaches (46). Briefly, proteins resolved in SDS–10% polyacrylamide gels were transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.). The blocking solution was from the BM chemiluminescenece system (Boehringer Mannheim, Indianapolis, Ind.). After brief washes with TBST, the membrane was incubated with a 1/10,000 dilution of anti-MBP antiserum (New England Biolabs, Inc.) overnight. After being washed three times with TBST, the membrane was incubated with a 1/20,000 dilution of goat anti-rabbit antibody coupled with peroxidase (Jackson ImmunoResearch Laboratories, West Grove, Pa.). The antigen-antibody complex was then detected with the enhanced chemiluminescence protein detection system from Amersham.
Immunoprecipitation.
The immunoprecipitation experiment and buffer conditions were as previously described (55) with a few modifications. Yeast cells were inoculated into 50 ml of SC medium to a density of 1.1 × 106 cells per ml. Growth continued with shaking at 30°C until the cultures had reached a density of 2 × 107 cells per ml. A total of 6.6 × 108 cells were used to produce protein extracts as described previously (55). Aliquots (90 μl) of the protein extract were mixed with 5 μl of anti-myc monoclonal antibody 9E10 and 5 μl of breaking buffer containing 1 mM phenylmethylsulfonyl fluoride and a 1:299 dilution of protease inhibitors (55). The reactions were mixed on a rotating wheel at 4°C for 2 h. The samples were centrifuged at 11,000 × g in a microcentrifuge at 4°C, and the supernatants were removed and added to 30 μl of protein G-agarose beads (Sigma P-4691). After being mixed at 4°C on a rotating wheel for 2 h, the resin-bound protein complexes were washed five times with 300 μl of washing buffer (55) for 5 s each at maximum speed and once with 300 μl of 0.5 M Tris (pH 7)–0.5 M NaCl. The beads were resuspended with 2× SDS loading dye and electrophoresed on SDS–10% polyacrylamide gels. Subsequent Western blot analysis was conducted as described above. To detect Gal4-HSFrr, the membrane was incubated with a 1/500 dilution of an anti-Gal4 DNA binding region antibody (Upstate Biotechnology, Lake Placid, N.Y.) and subsequently with a 1/20,000 dilution of a peroxidase-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories). To detect Myc-Gac1, the membrane was incubated with a 1/2,000 dilution of the anti-myc antibody (55) and subsequently with a 1/1,000 dilution of a peroxidase-conjugated sheep anti-mouse antibody (Amersham).
Culture growth conditions.
S. cerevisiae cultures were grown under different conditions and treated as described previously (5, 34, 57). Inocula for liquid cultures were aerated until they reached saturation in 2 ml of SC medium. Growth was initiated by inoculating saturated cultures into fresh SC medium to an OD650 of 0.5. Cultures of 7 ml were used for each condition tested. For heat shock induction, cultures were grown at room temperature (21 to 25°C) to the mid-exponential phase (OD650 = 1) and transferred to prewarmed flasks in a 39°C water bath, while the control cultures were maintained at room temperature. For copper induction, CuSO4 was added to mid-exponential-phase cultures to a final concentration of 500 μM and the cultures were incubated at 30°C for a further 45 min. For glucose starvation induction, cultures were grown at 30°C to the mid-exponential phase (OD650 = 1 to 1.5) and the cells were harvested, washed once with sterile distilled water, and resuspended to the same density in SC containing 0.05% glucose. The cultures were then aerated at 30°C for 3 h. For menadione induction, when the cultures at room temperature reached the mid-exponential phase, the control cultures were taken before induction treatment. Freshly prepared 50 mM menadione dissolved in ethanol was added to 7-ml cultures to a final concentration of 500 μM. Menadione induction was carried out for 70 min at room temperature. The cultures were chilled on ice for 5 min after induction for the time indicated. Cell pellets were obtained by centrifugation at 1,440 × g for 3 min, washed with 1 ml of cold water, and stored at −70°C.
RNA preparation.
Total RNA was prepared by a hot-acid-phenol extraction method (11). Cultures of 7 ml were harvested as described above. The cell pellets were resuspended with TES (10 mM Tris-Cl [pH 7.5], 10 mM EDTA, 0.5% SDS), acid-phenol was added, and the cultures were incubated at 65°C for 1 h. The RNA samples were then further extracted and precipitated as described (11). The concentration and purity of the RNA samples were determined from their absorbance at 260 and 280 nm, respectively.
S1 nuclease protection assay.
We used the S1 nuclease protection assay to measure the amount of specific RNAs obtained. The sequences of the oligonucleotide probe for the CUP1 gene is 5′GCAGCTACCACATTGGCATTGGCACTCATGACCTTCcggggt-3′, which is complementary to nucleotides +66 to +31 relative to the CUP1 gene translation start. The sequence of the oligonucleotide probe for the SSA4 gene is 5′GGCCGTTGTCTGGTGCTCCAGTGGGGCCTGCTCCAGCACCCGGAACgtttaa-3′, which is complementary to nucleotides +1906 to +1861. The sequence of the oligonucleotide probe for the HSP82 gene is 5′-CCTATTCAAGGCCATGATGTTCTACCTAATCTACCTCTTCCcgggat-3′, which is complementary to nucleotides +2155 to +2115. The lowercase letters are additional oligonucleotides that are not complementary to the RNA, so that bands resulting from RNA-DNA duplexes are easily distinguished from the band representing the probe. The S1 nuclease protection assay was carried out as previously described (19) except for a few modifications. A 15-μg portion of total RNA was used for each S1 nuclease assay, with 150 U of S1 nuclease, at 37°C for 60 min. The final DNA pellet was resuspended in 3 μl of Tris-EDTA and 4 μl of formamide loading dye, and 3-μl volumes of the samples were loaded on to the 10% denaturing polyacrylamide–urea gel. The gels were quantitatively analyzed with a PhosphorImager and the Image Quant program (Molecular Dynamics, Sunnyvale, Calif.).
We electrophoresed each RNA sample on an ethidium bromide-stained agarose gel to demonstrate that equal amounts of RNA were used in each S1 nuclease protection assay. We also used an oligonucleotide probe for the ADH1 or ACT1 gene to serve as an internal control for each S1 nuclease protection assay. However, we found this method to be less reliable than using quantification derived from reading the absorbance at 260 and 280 nm.
RESULTS
Selection of HSFrr-interacting proteins by phage display.
Amino acid residues 208 to 394 of yeast HSF were previously shown to repress the activation function of HSF (Fig. 1) (39). To gain insight to the mechanism of HSF transcription regulation, we selected a protein(s) that interacts with this region of HSF from a λ phage display library that contains randomly sheared yeast genomic DNA fused to the D capsid gene, whose product resides on the surface of phage λ (see Materials and Methods). This selection process was repeated for five rounds to provide strong selection for phages that bind HSFrr.
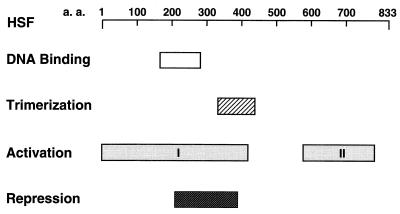
FIG. 1.
Functional domains of HSF. The regions involved in DNA binding, trimerization, activation, or repression are depicted. Activation domain I is involved in transient activity, while domain II is involved in sustained activity. The repression region (HSFrr) was used for the phage display analysis. a.a., amino acids.
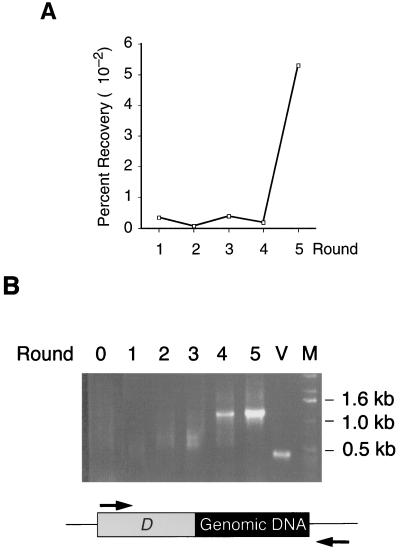
The number of phages bound to the resin from each round of selection was monitored by titer determination and PCR analysis. The titers of bound phages after each elution are shown in Fig. 2A. The percentages of recovered phage after each round of selection remained low in the first four rounds, but increased dramatically (30-fold) in the fifth round, indicating that HSF binding phages were being selected. To confirm that the selection was indeed enriching for specific DNAs, we conducted PCR analysis to detect the inserted DNA fragment(s) downstream of the D gene in the eluted phage lysate. The results are shown in Fig. 2B. The PCR product of the vector control (lane V) shows a 0.4-kb D gene fragment. The starting phage library (lane 0) shows a smear due to different sizes of DNA inserts. We found no obvious enrichment of DNA fragments during the first three selections. However, the fourth round of selection shows enrichment of a particular 1.2-kb band. By the fifth round, this band is the predominant species in the PCR assay. These results indicate a striking enrichment of a single phage that binds the HSF repression region.
FIG. 2.
Enrichment of HSFrr binding phages. (A) Percentages of recovered phage [(output titer/input titer) × 100] after each round of selection. (B) PCR analysis of the eluted phage lysate from each selection. The numbers indicate the different rounds of selection. Lane 0 shows the PCR analysis on the starting material, the phage library lysate. Lane V shows the PCR analysis of vector pJTL029, which has no DNA inserted downstream of the λ D gene. Lane M contains the 1-kb DNA ladder (Life Technologies). The PCR products were analyzed by electrophoresis on a 0.7% agarose gel. The schematic represents the D-yeast genomic DNA fusion construct, and the PCR primers are shown as arrows.
Nineteen pure phage lysates were generated from individual plaques of the fifth-round selection and examined by PCR. A 1.2-kb DNA band was amplified in all except one of the lysates, indicating that only one prominent species of D-fusion phage was enriched in the selection (data not shown). The one phage lysate that does not contain the 1.2-kb D-fusion fragment was later discarded because DNA sequencing analysis showed that it does not encode an in-frame D-fusion protein.
The HSFrr-interacting phage encodes the Gac1 protein.
DNA-sequencing analysis showed that the selected phage contains amino acids 162 to 406 of the GAC1 gene fused in frame to the carboxyl terminus of the λ D gene. The GAC1 gene encodes a 794-amino-acid regulatory subunit for a type 1 serine/threonine phosphoprotein phosphatase (16). This phosphatase, glycogen synthase phosphatase (Glc7), contributes to dephosphorylation and activation of glycogen synthase (14). HSF is also regulated at the level of phosphorylation, so that the interaction of this regulatory or targeting subunit of a phosphatase with HSF could be critical in modulating HSF function.
HSFrr and Gac1 proteins can physically interact.
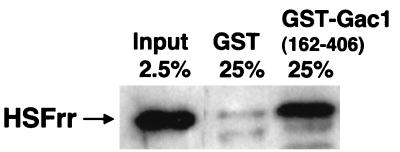
A simple pull-down binding assay confirmed the direct physical interaction between HSFrr and Gac1 proteins in vitro. Purified MBP-HSFrr was mixed with either GST or GST-Gac1(162–406), both of which were bound to glutathione-agarose resin. The resins were washed twice before the protein was eluted with SDS loading dye. The eluted MBP-HSFrr was visualized by Western blotting with antibody against MBP. The results are shown in Fig. 3. Approximately 2.5% of the MBP-HSFrr input was retained on GST-Gac1(162–406), while only a barely detectable portion of MBP-HSFrr was retained by GST. The results indicate that HSFrr can bind directly to Gac1.
FIG. 3.
Physical interaction between HSFrr and Gac1(162–406) in a pull-down assay. Resin-bound GST or GST-Gac1(162–406) was equilibrated with purified MBP-HSFrr as indicated. After being washed, bound proteins were eluted with SDS loading dye, and a fraction of each sample (shown as a percentage) was electrophoresed on SDS–10% polyacrylamide gels. The bound MBP-HSFrr was visualized by Western blotting with an antibody against MBP.
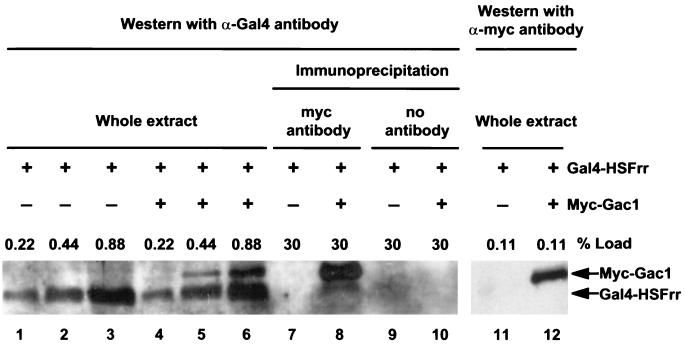
We also used an immunoprecipitation analysis to demonstrate that Gac1 and HSFrr interact in a yeast extract. Protein extracts from gac1 mutant strain KT1100 containing either pGal4-HSFrr (pKCL002) alone or both pGal4-HSFrr (pKCL002) and pmyc-GAC1(130–502) (pHH98) were prepared. The Gac1(130–502) region spans the Gac1 region (162 to 406) that was selected by interacting with HSFrr in the phage display system. The Myc-Gac1(130–502) was precipitated by anti-myc antibody 9E10, and the precipitates were analyzed by Western blotting with antibody against the Gal4 DNA binding region. The results are shown in Fig. 4. Approximately 1% of the Gal4-HSFrr input was coimmunoprecipitated with Myc-Gac1(130–502) by antibody against myc (Fig. 4, lane 8). A critical control shows that Gal4-HSFrr was not precipitated by anti-myc antibody in the absence of Myc-Gac1 (lane 7). Surprisingly, the Myc-Gac1 protein could cross-react with anti-Gal4 antibody (lanes 5, 6, and 8), but it did so to a lesser degree than it cross-reacted with anti-myc antibody (lane 12). Several experiments were performed to confirm that the band we describe as Myc-Gac1 was indeed Myc-Gac1 and that the anti-Gal4 antibody could cross-react with the Myc-Gac1 protein in yeast extract (data not shown). The reason for the cross-reactivity is not known. Furthermore, when the Myc-Gac1 protein present in immunoprecipitates or whole extract was detected by anti-myc antibody, no degradation product was detected (Fig. 4, lane 12, and data not shown), indicating that the Gal4-HSFrr signal in lane 8 was not a degradation product of Myc-Gac1. In summary, these results demonstrate that Gac1 protein interacts with HSFrr both as purified recombinant proteins and as proteins in crude yeast extracts.
FIG. 4.
Physical interaction between HSFrr and Gac1(130–502) in an immunoprecipitation assay. Whole protein extracts prepared from KT1100 containing pGal4-HSFrr alone (lanes 1 to 3, 7, 9, and 11) or both pGal4-HSFrr and pmyc-GAC1(130–502) (lanes 4 to 6, 8, 10, and 12) were used for the immunoprecipitation analysis. After precipitation by anti-myc antibody, a fraction of input extract and precipitates (shown as percentages) was electrophoresed on an SDS–10% polyacrylamide gel. The precipitates were visualized by Western blotting with anti-Gal4 DNA binding region antibody (lanes 1 to 10) or with anti-myc antibody (lanes 11 and 12).
Role of the Gac1 and Glc7 proteins in HSF activation in vivo.
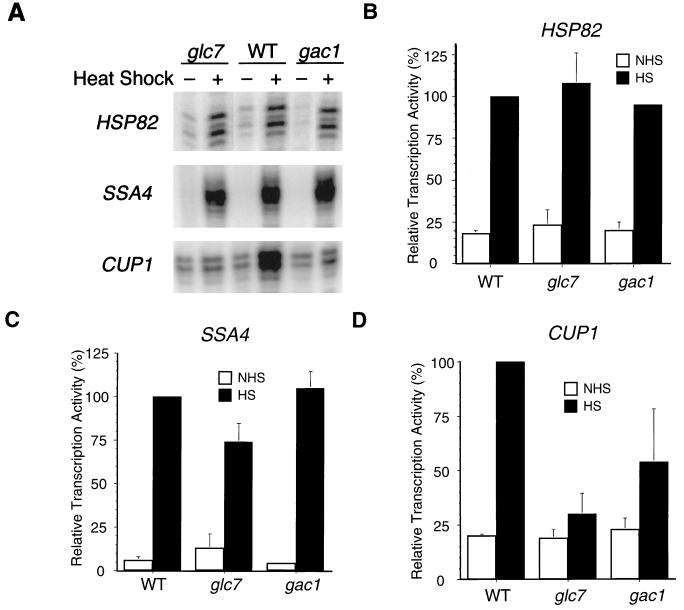
Since the phosphorylation state of HSF appears critical to its activity in stress-gene regulation, we determined whether Gac1 and Glc7 are important in regulating HSF transcriptional activation in vivo. Transcription of a variety of stress-inducible genes is activated by HSF. We measured the mRNA levels of a representative set of stress-inducible genes including SSA4 (an Hsp70 family gene), HSP82 (an Hsp90 family gene), and CUP1 (a stress-inducible gene) from control cultures or cultures that had been heat shocked for 10 min in wild-type, glc7, and gac1 mutant strains. The GLC7 gene is essential for cell viability: deletion of this gene results in death (14). The glc7-1 allele is a nonlethal mutation that encodes an R73C point mutation of Glc7 protein, which results in its diminished phosphatase activity (6, 14, 41). The gac1 mutation is a LEU2 insertion that causes a defect in glycogen synthase activity (16). Figure 5 shows that both gac1 and glc7 mutations have little effect on SSA4 or HSP82 transcription induction in response to heat shock. Interestingly, heat shock induction of CUP1 gene expression is abolished in the glc7 mutant and reduced to 50% of the wild-type expression in the gac1 mutant (Fig. 5A and D). Similar effects of the glc7 and gac1 mutations were also observed after a 30-min heat shock treatment (data not shown). The residual CUP1 transcription induction by heat shock in the gac1 mutant could result from redundant activities of Gac1 homologs (see Discussion). The results indicate that the Glc7 and Gac1 proteins are critical in the HSF transcriptional activation of CUP1 gene expression in response to heat shock. The results also indicate that the Gac1 and Glc7 proteins do not substantially affect HSF activation on heat shock genes such as SSA4 and HSP82.
FIG. 5.
S1 nuclease protection assay of HSP82, SSA4, and CUP1 transcription induction by heat shock. (A) Total RNAs were isolated from wild-type (WT) (KT1099), glc7 (KT1098), and gac1 (KT1100) strains. Cultures were grown to mid-exponential phase at room temperature and allowed to continue to grow at room temperature (−) or subjected to heat shock at 39°C for 10 min (+). The RNAs were annealed to an oligonucleotide probe complementary to the HSP82, SSA4, or CUP1 gene as indicated. (B to D) Quantification of each of the three types of experiments is shown for HSP82 (B), SSA4 (C), CUP1 (D); the labels NHS and HS refer, respectively, to non-heat-shock and heat-shock conditions. The S1 nuclease protection assays were quantitatively analyzed with a PhosphorImager and the Image Quant program. The pixel counts of each reaction are normalized to heat-shock-treated wild-type samples (100%). The numbers represent the mean of three independent experiments; standard deviations are indicated by error bars.
Glc7- and Gac1-mediated activation on CUP1 transcription is specific for HSF regulation.
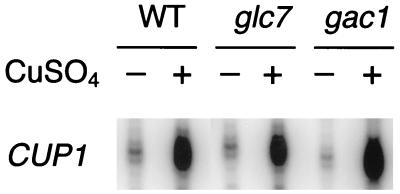
The CUP1 gene encodes metallothionein and can also be transcriptionally induced by copper via a transcription activator, Ace1 (3, 26, 59). To demonstrate that Glc7 and Gac1 affect CUP1 transcription specifically via their modulation of HSF activity, we measured CUP1 transcription in response to copper induction, which is an HSF-independent pathway (50, 57). Neither the glc7 nor the gac1 mutation has an effect on copper induction of CUP1 gene transcription (Fig. 6). This result demonstrates that the reduction of CUP1 transcription in response to heat shock caused by the glc7 and gac1 mutations is not a consequence of a general impairment of CUP1 promoter function.
FIG. 6.
S1 nuclease protection assay of CUP1 transcription induction by copper. Total RNAs were isolated from wild-type (WT) (KT1099), glc7 (KT1098), and gac1 (KT1100) strains. Cultures were grown to mid-exponential phase at 30°C and either harvested (−) or induced by 500 μM CuSO4 for 45 min (+). The RNAs were annealed to an oligonucleotide probe complementary to the CUP1 gene.
Roles of Glc7 and Gac1 on other HSF-dependent pathways of CUP1 transcription regulation.
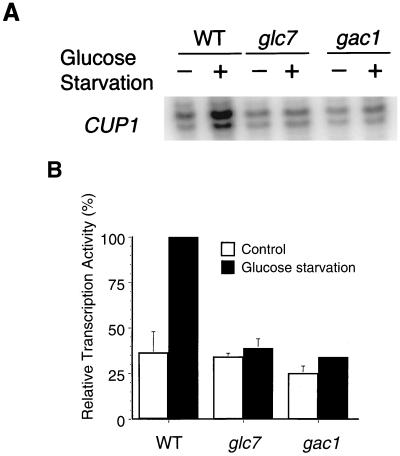
CUP1 gene transcription is activated by HSF in response to heat shock, glucose starvation, or oxidative stress induction (34, 57). Since Glc7 and Gac1 play positive roles in CUP1 transcription in response to heat shock, we determined whether they are also involved in other HSF-dependent pathways of CUP1 transcription regulation. CUP1 gene transcription in response to glucose starvation was measured in glc7 and gac1 mutants. Cultures were starved for glucose in 0.05% glucose medium for 3 h, and RNA was assayed by the S1 nuclease protection method. As shown in Fig. 7, induction of CUP1 transcription was eliminated in the glc7 mutant and greatly reduced in the gac1 mutant. The results indicated that Glc7 and Gac1 proteins also positively regulate HSF-dependent glucose starvation induction of CUP1 gene transcription.
FIG. 7.
S1 nuclease protection assay of CUP1 transcription induction by glucose starvation. (A) Total RNAs were isolated from wild-type (WT) (KT1099), glc7 (KT1098), and gac1 (KT1100) strains. Cultures were grown to mid-exponential phase at 30°C and either harvested (−) or washed and switched to 0.05% glucose medium for 3 h (+). The RNAs were annealed to an oligonucleotide probe complementary to the CUP1 gene as indicated. (B) S1 nuclease assays were quantified as in the experiment in Fig. 5. The numbers represent the mean of two independent experiments; standard deviations are indicated by error bars.
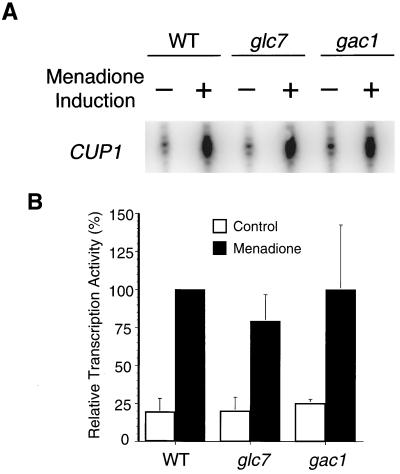
CUP1 transcription induction by oxidative stress in glc7 or gac1 mutants was also measured. Menadione, a derivative of vitamin K3 that generates superoxide anion through redox cycling, was used to generate oxidative stress as described previously (34). Cultures were treated with 500 μM menadione for 70 min before RNA extraction for S1 nuclease protection assay. As shown in Fig. 8, CUP1 transcription induction by menadione was not affected by either the glc7 or the gac1 mutation. These results indicate that Glc7 and Gac1 are important for HSF-activated transcription of CUP1 that is stimulated by heat shock or glucose starvation. In contrast, the activation of CUP1 transcription by HSF-dependent oxidative stress is dependent on neither Glc7 nor Gac1.
FIG. 8.
S1 nuclease protection assay of CUP1 transcription induction by menadione. (A) Total RNAs were isolated from wild-type (WT) (KT1099), glc7 (KT1098), and gac1 (KT1100) strains. Cultures were grown to mid-exponential phase at room temperature and either harvested (−) or induced with 500 μM menadione for 70 min (+). The RNA was annealed to an oligonucleotide probe complementary to the CUP1 gene as indicated. (B) S1 nuclease assays were quantified as in the experiment in Fig. 5. The mean and standard deviation (error bars) are for four independent experiments.
DISCUSSION
HSF is a transcriptional activator of a variety of stress-inducible genes. To gain insight into the mechanisms of HSF activation, we used the phage display system to select for HSF-interacting proteins. Using a repression region of HSF, we selected a phage that displayed the Gac1 protein, which is the regulatory subunit of the phosphatase Glc7. The Gac1 protein interacts with the catalytic subunit of glycogen synthase phosphatase, Glc7, both physically and genetically (55, 61). Pull-down and immunprecipitation assays confirmed that Gac1 and HSF also physically interact. Analysis of yeast mutants with mutations in GLC7 and GAC1 showed that these genes play positive roles in HSF activation of CUP1 transcription in response to heat shock. This activation shows an interesting specificity in that these mutations do not affect the transcription of two other HSF-activated heat shock genes tested.
The phosphorylation state of HSF changes upon heat shock. It is phosphorylated under non-heat-shock conditions and becomes hyperphosphorylated upon heat shock (12, 53). Therefore, the hyperphosphorylation has long been postulated to be critical to HSF transcriptional activation. There is evidence suggesting that hyperphosphorylation is involved in transcription activation of human HSF1 (64). However, to date, no evidence links hyperphosphorylation of yeast HSF to its enhanced transcriptional activity upon heat shock (25). Our results indicate that Gac1 and Glc7 positively regulate HSF transcription activation on CUP1 gene transcription in response to heat shock. The simplest model is that Gac1, through its interaction with the Glc7 phosphatase and HSF, targets the dephosphorylation of HSF during heat shock. This specific dephosphorylation would activate HSF and allow it to stimulate transcription from the CUP1 promoter.
Interestingly, the Gac1 and Glc7 proteins are also known to act positively to stimulate the enzymatic activity of glycogen synthase (6, 14, 21, 41). This glycogen synthase activity resides in the cytoplasm. In contrast, yeast HSF, which we propose here to also be a target of Gac1 and Glc7, is thought to reside predominantly in the nucleus (as inferred from its constitutive occupancy on the HSEs of heat shock promoters [20, 25, 27, 51]). While cellular localization studies of the Glc7 protein show that significant levels of Glc7 are present in the cytoplasm, it is found predominantly in the nucleus (58). Therefore, the cellular distribution of Glc7 is compatible with proposal that it activates HSF.
The concept that a phosphatase activates HSF invites speculation that a kinase(s) might repress HSF activity. The Calderwood group has determined that phosphorylation of mammalian HSF1 by glycogen synthase kinase 3 and by mitogen-activated protein kinases result in repression of HSF transcription activity (9). The transcriptional activity of HSF could be subject to the competing activities of specific phosphatases and kinases that are in turn differentially modulated by cellular signals.
Glc7 does not act alone but appears to be targeted to its substrates through interactions with Gac1 and perhaps other Gac1-like proteins. Three yeast proteins, Gip2, Pig1, and Pig2, have protein sequence similarity to Gac1 (8, 61). The amino acid region from 162 to 406 of the Gac1 protein, which we found to interact with HSFrr, contains two conserved sequence motifs that are shared among these Gac1 homologs. These other homologs may also function with Glc7 to activate HSF, and this could explain the residual heat shock activation of CUP1 transcription in the gac1 mutant but not in the glc7 mutant. Pig1, Pig2, and Gac1 all interact with glycogen synthase (8, 58). The conserved regions in these Gac1 homologs might serve a common function of directing the Glc7 phosphatase to its protein targets (8). We hypothesize that Gac1 serves as a bridge to bring the catalytic subunit of the phosphatase, Glc7, to its HSF substrate.
Why does the Glc7-activated HSF affect only CUP1 gene transcription but no other heat shock genes? It was previously known that CUP1 gene regulation by HSF is distinct from regulation of other heat shock genes. CUP1 gene transcription is highly induced at 39°C but not at 37°C, a temperature at which other heat shock genes are induced (57). It was also found that CUP1 activation by HSF requires its carboxyl-terminal activation domain, while the carboxyl-terminal activation domain of HSF is dispensable for transcription induction of other heat shock genes (such as SSA1) (57). The HSE of CUP1 has a different sequence arrangement from other heat shock genes such as SSA3 or SSA4, and it contributes to some of the distinct regulation of CUP1 by HSF (48). Two hypotheses could explain the specific requirement of Glc7 activity on HSF activation of CUP1 gene transcription. First, the HSF may require Glc7-Gac1 modification to acquire the ability for DNA binding to the CUP1 promoter. Second, HSF may acquire an active conformation after binding to the CUP1 promoter that is different from the conformation used when it binds to the heat shock gene promoter, and this conformation change might require Glc7-Gac1 modulation. However, the difference in HSEs between CUP1 and other heat shock genes may not account for the entire effect of Glc7, because HSP82 contains an HSE that is similar to that of CUP1 and HSP82 activation is independent of Glc7 regulation. There may be other factors, such as the core promoter structure, involved in Glc7 regulation of HSF activity for CUP1 gene transcription.
Tu and Carlson showed that the Glc7 protein is required for glucose repression (60). A specific allele, glc7-T152K, of the GLC7 gene results in relief of glucose repression of SUC2 gene expression. Interestingly, the glc7-1 allele, which we used here, did not relieve the SUC2 glucose repression (60). Furthermore, we demonstrated in this study that CUP1 gene activation in response to glucose starvation is abolished in glc7-1 and gac1 mutants. Not only does glc7-1 fail to cause relief of glucose repression, but also this mutation leads to the failure of glucose starvation to activate CUP1 transcription. Therefore, the Glc7-modulated HSF activation of CUP1 by glucose starvation is clearly distinct from the glucose repression of other genes such as SUC2. This conclusion is in agreement with studies by Tamai et al. showing that the genes known to regulate glucose repression of SUC2 play no role in the activation of CUP1 transcription by glucose starvation (57).
Finally, we found that Glc7 has an effect on CUP1 gene transcription in response to heat shock and glucose starvation but not oxidative stress induction. Liu and Thiele determined that the phosphorylation patterns of HSF subjected to heat shock and oxidative stress are distinct (34). This may explain why CUP1 induction in response to oxidative stress is independent of Glc7 and Gac1 functions. It is possible that specific residues are subjected to dephosphorylation of Glc7 and that this occurs only when HSF receives stimuli from heat shock or glucose starvation signaling pathways. Our study indicates that the mechanism of HSF transcription on different genes is sophisticated and capable of distinguishing among different types of stress signals.
ACKNOWLEDGMENTS
We are especially grateful to R. Hoess for generating the starting vector to generate the λ phage library and for advice about conducting the phage display selection. We are grateful to K. Tatchell for the gift of strains, antibody, and plasmid and for communication of results prior to publication. We thank D. Thiele for freely exchanging information. We thank K. Leptos for constructing plasmid pKCL002. J. T. Lin thanks P. Mason for advice on the pull-down assay and E. Guzmán for assisting with the production of the photographic figures. We thank the members of the Lis laboratory for general interest and helpful discussions. We thank P. Mason and D. Lee for their comments on the manuscript.
This work was supported by Public Health Service grant GM25232 (awarded to J. T. Lis) from the National Institutes of Health. J. T. Lin was supported by NIH postdoctoral fellowship award 1F32GM17989-01.
REFERENCES
- 1.Abravaya K, Myers M P, Murphy S P, Morimoto R I. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 2.Adams A, Gottschling D E, Kaiser C, Stearns T. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 3.Buchman C, Skroch P, Welch J, Fogel S, Karin M. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol Cell Biol. 1989;9:4091–4095. doi: 10.1128/mcb.9.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton D R. Phage display. Immunotechnology. 1995;1:87–94. doi: 10.1016/1380-2933(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 5.Butler G, Thiele D J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon J F, Pringle J R, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Barlev N A, Westergaard O, Jakobsen B K. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 1993;12:5007–5018. doi: 10.1002/j.1460-2075.1993.tb06194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng C, Huang D, Roach P J. Yeast PIG genes: PIG1 encodes a putative type 1 phosphatase subunit that interacts with the yeast glycogen synthase Gsy2p. Yeast. 1997;13:1–8. doi: 10.1002/(SICI)1097-0061(199701)13:1<1::AID-YEA49>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Chu B, Soncin F, Price B D, Stevenson M A, Calderwood S K. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 10.Coen D M. Enzymatic amplification of DNA by PCR: standard procedures and optimization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. Boston, Mass: Wiley Interscience; 1994. pp. 15.0.1–15.7.5. [Google Scholar]
- 11.Collart M A, Oliviero S. Preparation of yeast RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. Boston, Mass: Wiley Interscience; 1993. pp. 13.12.1–13.12.2. [Google Scholar]
- 12.Cotto J J, Kline M, Morimoto R I. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J Biol Chem. 1996;271:3355–3358. doi: 10.1074/jbc.271.7.3355. [DOI] [PubMed] [Google Scholar]
- 13.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 14.Feng Z H, Wilson S E, Peng Z Y, Schlender K K, Reimann E M, Trumbly R J. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J Biol Chem. 1991;266:23796–23801. [PubMed] [Google Scholar]
- 15.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 16.Francois J M, Thompson-Jaeger S, Skroch J, Zellenka U, Spevak W, Tatchell K. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giardina C, Lis J T. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giardina C, Lis J T. Sodium salicylate and yeast heat shock gene transcription. J Biol Chem. 1995;270:10369–10372. doi: 10.1074/jbc.270.18.10369. [DOI] [PubMed] [Google Scholar]
- 19.Greene J M, Struhl K. S1 analysis of messenger RNA using single-stranded DNA probes. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. Boston, Mass: Wiley Interscience; 1988. pp. 4.6.7–4.6.9. [DOI] [PubMed] [Google Scholar]
- 20.Gross D S, English K E, Collins K W, Lee S. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990;216:611–632. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 21.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 22.Harrison J L, Williams S C, Winter G, Nissim A. Screening of phage antibody libraries. Methods Enzymol. 1996;267:83–108. doi: 10.1016/s0076-6879(96)67007-4. [DOI] [PubMed] [Google Scholar]
- 23.Hoess, R. H. Personal communication.
- 24.Hoess R H, Ziese M, Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoj A, Jakobsen B K. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 1994;13:2617–2624. doi: 10.1002/j.1460-2075.1994.tb06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huibregtse J M, Engelke D R, Thiele D J. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc Natl Acad Sci USA. 1989;86:65–69. doi: 10.1073/pnas.86.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakobsen B K, Pelham H R B. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 1991;10:369–376. doi: 10.1002/j.1460-2075.1991.tb07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurivich D A, Sistonen L, Kroes R A, Morimoto R I. Effect of sodium salicylate on the human heat shock response. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- 30.Kline M P, Morimoto R I. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knauf U, Newton E M, Kyriakis J, Kingston R E. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 32.Kretzschmar T, Zimmermann C, Geiser M. Selection procedures for nonmatured phage antibodies: a quantitative comparison and optimization strategies. Anal Biochem. 1995;224:413–419. doi: 10.1006/abio.1995.1059. [DOI] [PubMed] [Google Scholar]
- 32a.Lin, J. T., and J. T. Lis. March 1999, posting date. Phage display library. [On-line.] http://www.bio.cornell.edu/biochem/lis/lis.html.
- 33.Lis J T, Xiao H, Perisic O. Modular units of heat shock regulatory regions: structure and function. In: Morimoto R, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 411–428. [Google Scholar]
- 34.Liu X D, Thiele D J. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- 35.Mason P B J, Lis J T. Cooperative and competitive protein interactions at the hsp70 promoter. J Biol Chem. 1997;272:33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 36.McCafferty J, Johnson R H, Chiswell D J. Phage-enzymes: expression and affinity chromatography of functional alkaline phosphatase on the surface of bacteriophage. Protein Eng. 1991;4:955–961. doi: 10.1093/protein/4.8.955. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 38.Mosser D D, Duchaine J, Massie B. The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol Cell Biol. 1993;13:5427–5438. doi: 10.1128/mcb.13.9.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker C S. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990;62:807–818. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- 40.Parmley S F, Smith G P. Antibody-selectable filamentous Fd phage vectors affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- 41.Peng Z Y, Trumbly R J, Reimann E M. Purification and characterization of glycogen synthase from a glycogen-deficient strain of Saccharomyces cerevisiae. J Biol Chem. 1990;265:13871–13877. [PubMed] [Google Scholar]
- 42.Perisic O, Xiao H, Lis J T. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 base pair recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 43.Rabindran S K, Haroun R I, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 44.Rabindran S K, Wisniewski J, Li L, Li G C, Wu C. Interaction between heat shock factor and hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol Cell Biol. 1994;14:6552–6560. doi: 10.1128/mcb.14.10.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoro N, Johansson N, Thiele D J. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18:6340–6352. doi: 10.1128/mcb.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y, Mosser D D, Morimoto R I. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silar P, Butler G, Thiele D J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorger P K, Lewis M J, Pelham H R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- 52.Sorger P K, Nelson H C M. Trimerization of a yeast transcriptional activator via coiled-coil motif. Cell. 1989;59:807–814. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 53.Sorger P K, Pelham H R B. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 54.Sternberg N, Hoess R H. Display of peptides and proteins on the surface of bacteriophage λ. Proc Natl Acad Sci USA. 1995;92:1609–1613. doi: 10.1073/pnas.92.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart J S, Frederick D L, Varner C M, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 57.Tamai K T, Liu X, Silar P, Sosinowski T, Thiele D J. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatchell, K. Personal communication.
- 59.Thiele D J. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol. 1988;8:2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu J, Song W, Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westwood J T, Wu C. Activation of Drosophila heat shock factor: conformational change associated with monomer to trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, X., H. Hart, P. Roach, and K. Tatchell. Unpublished data.
- 64.Xia W, Voellmy R. Hyperphosphorylation of heat shock transcription factor 1 is correlated with transcriptional competence and slow dissociation of active factor trimers. J Biol Chem. 1997;272:4094–4102. doi: 10.1074/jbc.272.7.4094. [DOI] [PubMed] [Google Scholar]
- 65.Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zou J, Guo Y, Guettouche T, Smith D F, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]