Abstract
The product of the retinoblastoma susceptibility gene, pRB, is a nuclear phosphoprotein that controls cell growth by binding to and suppressing the activities of transcription factors such as the E2F family. Transactivation activity is inhibited when E2F is bound to hypophosphorylated pRB and released when pRB is phosphorylated by cyclin-dependent kinases (CDKs). To determine which of 16 potential CDK phosphorylation sites regulated the pRB-E2F interaction, mutant pRB proteins produced by site-directed mutagenesis were tested for the ability to suppress E2F-mediated transcription in a reporter chloramphenicol acetyltransferase assay. Surprisingly, no one CDK site regulated the interaction of pRB with E2F when E2F was bound to DNA. Instead, disruption of transcriptional repression resulted from accumulation of phosphate groups on the RB molecule.
The ability of the retinoblastoma protein (pRB) to control the cell cycle has been attributed to repression of transcription factors such as E2F, which regulates transcription of genes necessary for S-phase entrance (reviewed in reference 77). pRB may also be involved in transcriptional repression in a cell-type-specific manner by interacting with and repressing the activities of transcription factors such as the lymphoid cell-specific Elf-1 and PU.1 proteins (29, 88). Positive regulation of transcription by pRB is mediated through transcription factors such as MyoD, CCAAT enhancer binding protein (C/EBP) and NF-IL6, involved in the terminal differentiation of several cell types (12, 13, 28), as well as transcription factors such as Sp-1, ATF-2, and hBrm (11, 49, 50, 81, 85). However, the ability of pRB to regulate cell cycle events is not restricted to its action on polymerase II-regulated genes. Repression of polymerase I transcription by pRB is accomplished by the physical interaction of pRB with an upstream binding factor, resulting in the inability of this factor to bind to the ribosomal DNA promoter (87). pRB represses all polymerase III genes, including tRNA, 5S RNA, and TATA-box-containing genes such as U6, and does so by targeting the general polymerase III transcription factor TFIIIB (55). pRB is also thought to repress transcription through another mechanism involving recruitment of histone deacetylase to the promoter, resulting in deacetylation of histone proteins, thereby encouraging the formation of nucleosome structures (6, 65, 66).
E2F was the first cellular protein found to bind pRB and remains the best characterized of the pRB binding proteins (3, 10, 37, 71). There are five members of the E2F family of transcription factors, E2F-1 through E2F-5 (reviewed in reference 56). As heterodimeric complexes with the DP family of proteins, the E2F proteins regulate the activity of promoters containing E2F binding sites including genes encoding S-phase-regulatory proteins such as DNA polymerase α, thymidylate synthase, proliferating cell nuclear antigen, and ribonucleotide reductase, as well as genes regulating cell cycle progression such as those encoding cyclin A, cyclin E, cdc2, and B-myb (4, 21, 38, 54).
pRB belongs to a family of proteins including p107 and p130, which show considerable sequence homology in a region known as the pocket (24, 33, 60, 67). Each of these proteins is able to bind to E2F family members in a cell cycle-dependent manner and negatively regulate E2F-mediated transcription (9, 17, 26, 36, 40, 79, 86, 90, 93). pRB, p107, and p130 bind to different E2F molecules at various times during the cell cycle. pRB binds to E2F-1 through E2F-4 (59, 69), while p107 and p130 preferentially bind E2F-4 and E2F-5 (5, 27, 41, 75). pRB-E2F complexes are found mostly in the G1 phase, while p107–E2F-4 complexes persist throughout the cell cycle and contain cyclin E or cyclin A in different cell cycle phases. Complexes of E2F-4 and E2F-5 with p130 predominate in quiescent cells (reviewed in references 39 and 80).
The pRB family proteins bind to and alter the functions of E2F, including repression of transactivation (31), repression of apoptosis (45, 73), protection from degradation (35, 43), and determination of E2F-DNA binding site specificity (47, 83). pRB binds the transactivation domain of E2F to directly inhibit transactivation by E2F (26, 36, 40). When tethered to a promoter through E2F, pRB can also interact with surrounding transcription factors, preventing their interaction with the basal transcriptional machinery (89), or pRB itself may interact with the basal transcriptional machinery to inhibit transcription (7). The regions of pRB necessary for general repressor activity are the A and B domains, which form a transcriptional repressor motif regulated by cyclin-dependent kinases (CDKs) (15, 16). Direct interaction of pRB with E2F also prevents E2F-induced apoptosis. However, since the transactivation and apoptotic functions of E2F-1 are separable, the ability of pRB to inhibit E2F-induced apoptosis is not due to suppression of the E2F-1 transactivation function (45). E2F proteins are also cell cycle regulated by degradation via the ubiquitin proteasome pathway. An epitope in the C terminus is responsible for the instability of E2F, and direct binding of pRB to the C terminus of E2F protects it from degradation (35, 43). Finally, recent experiments using a technique known as CASTing have shown that different E2F-DP heterodimers bind distinct DNA E2F binding sites and that preference for a particular E2F-1–DP-1 site is changed upon binding of pRB to the complex (83).
pRB is a nuclear phosphoprotein whose activity is regulated throughout the cell cycle by phosphorylation by CDKs (1, 8, 14, 19, 20, 22, 25, 42, 44, 46, 48, 58, 61). Of the 16 potential p34cdc phosphorylation sites in the pRB molecule, at least 7 fully match the consensus sequence (70). Phosphorylation of pRB in mid-G1 is thought to be due to cyclin D-cdk4 and cyclin E-cdk2. Additional phosphorylation occurs by cyclin A-cdk2 during S phase and cyclin B-cdc2 during G2/M (reviewed in reference 39). Dephosphorylation of pRB is probably due to the activity of phosphoprotein phosphatase type 1 (63), which has been shown to associate with pRB in vivo, predominantly during mitosis (23).
The active form of pRB is thought to be hypophosphorylated, as both the oncoproteins of DNA tumor viruses and cellular proteins preferentially bind hypophosphorylated pRB (10, 62, 84, 88). Since pRB function is regulated by phosphorylation, the mechanism of pRB phosphorylation is a central question in cell growth control. Phosphorylation at particular sites of pRB may regulate its function, since the nuclearly-bound hypophosphorylated form of pRB lacks phosphorylation at specific sites compared to hyperphosphorylated pRB (68). Knudsen and Wang showed that phosphorylation of Ser807 and Ser811 of human pRB was required to release the interaction of c-Abl from pRB (52), and recent data suggest that the binding of pRB to free E2F is regulated by dual mechanisms involving phosphorylation either at a number of C-terminal sites or at two serines in the insert domain of pRB (53).
However, the structural requirements for pRB to bind stably to E2F on DNA are probably different from those for binding free E2F (78). pRB mutants containing the sequence NAAIRS replacing sequences highly conserved in p107 and p130 were made. The tertiary structures of these mutants were maintained, and the ability to bind free E2F or E2F bound to DNA was determined. pRB mutants which could form stable pRB-E2F-DNA complexes were also able to bind E2F free in solution. However, some pRB mutants which bound free E2F in solution were unable to form pRB-E2F-DNA complexes, suggesting that additional structural requirements are necessary for pRB to bind E2F on DNA (78).
In an effort to determine which of the 16 phosphorylation sites regulated the interaction of pRB with E2F on DNA, we tested the effects of RB proteins containing mutations in a number of phosphorylation sites on an important aspect of pRB-E2F function: repression of E2F transactivation by pRB. Disruption of the interaction of pRB with E2F bound to a promoter depended on accumulation of phosphate residues at multiple sites on pRB.
MATERIALS AND METHODS
Cell culture.
C33A cervical carcinoma cell line (from the American Type Culture Collection) and COS-7 cells were grown in alpha modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were grown in 5% CO2 at 37°C. Metabolic labeling was carried out with [32P]phosphoric acid at 800 μCi/ml in phosphate-free medium supplemented with 10% dialyzed fetal bovine serum for 4 to 5 h.
Plasmids.
Phosphorylation site mutant pRB constructs in the simian virus 40 (SV40) promoter-containing pECE vector were produced by site-directed PCR mutagenesis. ΔBXHA (wild-type murine pRB), Δp34HA, ΔP1,2HA, ΔP3,4HA, ΔAAASSHA, and ΔP1,2,3,4HA were described previously (32, 30). pECEHA-ΔK11 was a gift of Eldad Zacksenhaus (Toronto General Hospital, Toronto, Ontario, Canada). Ser243, Thr364/367, Ser800/804, and Thr814/819 were made by using ΔBXHA as a template and pairs of PCR primers containing single-base-pair mismatches in the codons of interest. An additional set of primers spanning 5′ and 3′ of the mutation and overlapping unique restriction sites within ΔBXHA was used. The PCR fragment was then cloned into the appropriate sites in the RB cDNA. ΔK6 was made by ligating a 0.79-kb DraIII-SauI fragment of ΔAAASSHA into a 4.91-kb DraIII-SauI fragment of ΔP1,2,3,4HA. ΔK7 was made by ligating a 0.79-kb DraIII-SauI fragment of pECEHA-ΔK11 into a 4.91-kb DraIII-SauI fragment of ΔBXHA. pECE constructs expressing cyclin E and cdk2 and the reporter construct E2(−80/−70)CAT were described previously (7).
Transfection and CAT assay.
C33A cells were transfected by the calcium phosphate or modified calcium phosphate-mediated transfection procedure (74). COS-7 cells were transfected by using either Lipofectamine (GIBCO) or Superfect (Qiagen). Chloramphenicol acetyltransferase (CAT) assays were performed 48 to 72 h posttransfection by the method of Sleigh (82). Three-microgram aliquots of the reporter construct E2(−80/−70)CAT were transfected along with indicated concentrations of the pRB construct and 5 μg each of cyclin E and cdk2 constructs. To prevent competition effects from the promoter carrying the effector genes on the reporter construct, an SV40 promoter-driven luciferase gene (pSVLuc) was used as filler DNA. One microgram of plasmid pRSVβgal or pCMVβgal was included in the transfection to normalize CAT activity for transfection efficiency. The CAT activity from the reporter construct in the presence of cyclin E and cdk2 is arbitrarily set to 100%. The ability of each pRB mutant to suppress transcription was determined by reduction in CAT activity. In experiments using roscovitine (Calbiochem), the drug was added at a final concentration of 700 nM 18 h following transfection and remained on the cells 40 to 48 h before harvesting.
Immunoprecipitation and Western blotting.
Transfected cells were lysed in 50 mM sodium fluoride–50 mM Tris-HCl (pH 8)–120 mM NaCl–0.5% Nonidet P-40, with the proteinase inhibitors leupeptin, aprotinin, phenylmethylsulfonyl fluoride (PMSF) added fresh. Immunoprecipitation of pRB was performed with 10 μl of antihemagglutinin (anti-HA) antibody 12CA5 (obtained from Paul Hamel, University of Toronto) followed by Western blotting with the anti-HA antibody. Coimmunoprecipitation of pRB mutants with E2F1 was performed with 5 μl of anti-pRB (Pharmingen 14001A) followed by Western blotting with anti-E2F1 antibody (Santa Cruz C-20).
Gel mobility shift assays.
Transfected cells were lysed on ice for 15 min in 10 mM HEPES (pH 7.9)–10 mM KCl–0.1 mM EDTA–0.1 mM EGTA–1 mM dithiothreitol (DTT), with 1 mM PMSF plus leupeptin and aprotinin (1 μg/ml, final concentration) added fresh. Cold Nonidet P-40 was added to a final concentration of 0.625%. Following centrifugation, the supernatant was removed and used to perform a β-galactosidase assay to correct for transfection efficiency. The pellet was dissolved in 20 mM HEPES (pH 7.9)–400 mM NaCl–1 mM EDTA–1 mM EGTA–1 mM DTT, with 1 mM PMSF plus leupeptin and aprotinin (1 μg/ml, final concentration) added fresh, and the nuclei were lysed by rocking for 15 min at 4°C. The binding of nuclear extracts of transfected cells to a 32P-labeled E2F oligonucleotide was performed in 10 mM HEPES (pH 7.2)–100 mM KCl–5 mM MgCl2–5% glycerol–0.5 mM DTT–20 μg of bovine serum albumin per ml–50 μg of salmon sperm DNA per ml for 15 min at room temperature, and complexes were resolved on a 4% native polyacrylamide gel in 1× Tris-borate-EDTA. The double-stranded E2F oligonucleotide of the sequence AGGTAAGTTTCGCGCCCTTTCCCA was end labeled with Klenow enzyme (GIBCO), and 0.25 to 0.5 ng was used for protein binding.
RESULTS
Characterization of pRB mutants.
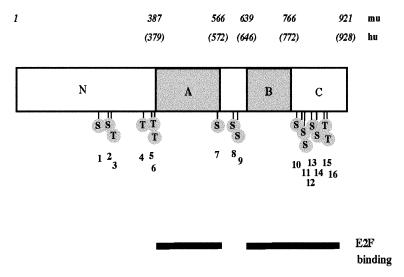
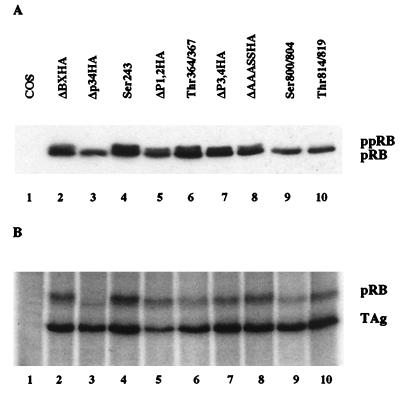
Murine RB DNA constructs containing mutations in one or two potential phosphorylation sites were produced by site-directed PCR and subcloned into the SV40 promoter-containing pECE vector. Each mutant construct and its corresponding mutation are shown in Table 1. The 16 potential murine phosphorylation sites were numbered in order to clarify comparison of murine and human pRB and to facilitate discussion of the literature. Human pRB is missing murine site 5 but contains an additional site at Thr5. Figure 1 shows a schematic of pRB and the location of each of its phosphorylation sites. Mutations were made in phosphorylation sites that were known to be phosphorylated in vivo in full-length pRB or a peptide fragment (51, 58, 92). To confirm the expression of each plasmid, proteins were expressed in COS-7 cells and immunoprecipitated with anti-HA antibody 12CA5. Immunoprecipitated proteins were visualized by Western blot analysis with antibody 12CA5. All RB plasmids were expressed at similar levels (Fig. 2A). However, three mutant constructs (Δp34HA, Ser800/804, and Thr814/819) did not show the mobility shift characteristic of the phosphorylated form of pRB in SDS-polyacrylamide gel electrophoresis. Despite the lack of mobility shift, all mutant proteins are at least partially phosphorylated, demonstrated by immunoprecipitation of orthophosphate-labeled proteins with the anti-HA antibody (Fig. 2B).
TABLE 1.
Summary of pRB phosphorylation site mutantsa
| Mutant | Phosphorylation siteb
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N terminus
|
A
|
Spacer
|
C terminus
|
|||||||||||||
| 1 Ser230 Ser224 | 2 Ser249 Ser243 | 3 Thr252 Thr246 | 4 Thr356 Thr350 | 5 Thr364 | 6 Thr373 Thr367 | 7 Ser567 Ser561 | 8 Ser608 Ser601 | 9 Ser612 Ser605 | 10 Ser780 Ser773 | 11 Ser788 Ser781 | 12 Ser795 Ser788 | 13 Ser807 Ser800 | 14 Ser811 Ser804 | 15 Thr821 Thr814 | 16 Thr826 Thr819 | |
| ΔBX-HA(wt) | S | S | T | T | T | T | S | S | S | S | S | S | S | S | T | T |
| Δp34HA | A | R | A | A | A | A | A | E | ||||||||
| Ser243 | A | |||||||||||||||
| ΔP1,2HA | A | R | ||||||||||||||
| Thr364/367 | R | A | ||||||||||||||
| ΔP3,4HA | A | A | ||||||||||||||
| ΔAAASSHA | A | A | ||||||||||||||
| Ser800/804 | A | R | ||||||||||||||
| Thr814/819 | R | A | ||||||||||||||
| ΔP1,2,3,4HA | A | R | A | A | ||||||||||||
| ΔK6 | A | R | A | A | A | A | ||||||||||
| ΔK7 | A | A | A | A | E | A | A | |||||||||
| ΔK11 | A | R | A | A | A | A | A | A | E | A | A | |||||
Human pRB contains a 16th phosphorylation site at Thr5.
Sites are aligned with sequences of human (top row) and murine (bottom row) pRB at the indicated positions.
FIG. 1.
Diagram of murine pRB. Numbers above the schematic designate the amino acids. The A and B regions required for binding the SV40 large T antigen are shown. Positions of the potential phosphorylation sites (S [serine] and T [threonine]), numbered sequentially, in the protein are shown. The bars at the bottom represent the regions required for pRB to bind E2F on DNA.
FIG. 2.
(A) Expression of pRB mutant constructs. Ten micrograms of RB plasmid was transfected into COS-7 cells. Quantities of cell lysates corrected for transfection efficiency by using cotransfected pCMVβgal were immunoprecipitated with 10 μl of anti-HA antibody 12CA5 followed by Western blotting with anti-HA antibody. The underphosphorylated (pRB) and hyperphosphorylated (ppRB) forms of pRB are indicated. (B) Orthophosphate labeling of pRB mutants. At 36 h after transfection of pRB mutants, COS-7 cells were incubated in 1.2 ml of phosphate-deficient medium, containing 800 μCi of 32PO4 per ml at 37°C for 4.5 h. Phosphorylated pRB was immunoprecipitated with anti-HA antibody 12CA5, loaded on an SDS–7.5% polyacrylamide gel, and visualized by autoradiography. Coprecipitated T antigen (TAg) is indicated.
SV40 large T antigen coimmunoprecipitates with all of the pRB mutants (Fig. 2B). Mutation of the phosphorylation sites does not affect binding of hypophosphorylated pRB to large T antigen. When the amounts of lysates loaded are corrected for transfection efficiency, there is no apparent difference among the mutants in the ability to bind large T antigen (data not shown).
The Δp34HA construct contains mutations in 8 of the 16 potential phosphorylation sites (sites 3, 4, 8, 9, 11, 12, 13, and 14) and is relatively refractory to phosphorylation as previously reported (30). The electrophoretic mobility shift of pRB with hyperphosphorylation requires an intact Ser804 (site 14) and Pro805, and phosphorylation of Ser804 (site 14) is necessary for the shift (30). This explains the lack of shift in the Δp34HA and Ser800/804 (sites 13 and 14) mutants. However, Thr814/819 (sites 15 and 16) also produces a protein that does not shift but contains both the hypophosphorylated and hyperphosphorylated species in a single band, demonstrated by coimmunoprecipitation of the hypophosphorylated form with large T antigen (data not shown) and immunoprecipitation of the orthophosphate-labeled form with the anti-HA antibody. This result suggests that phosphorylation at the three most C-terminal phosphorylation sites (sites 14, 15, and 16) of pRB may be responsible for the electrophoretic mobility shift of the phosphorylated species.
Coimmunoprecipitation of pRB mutants with E2F1.
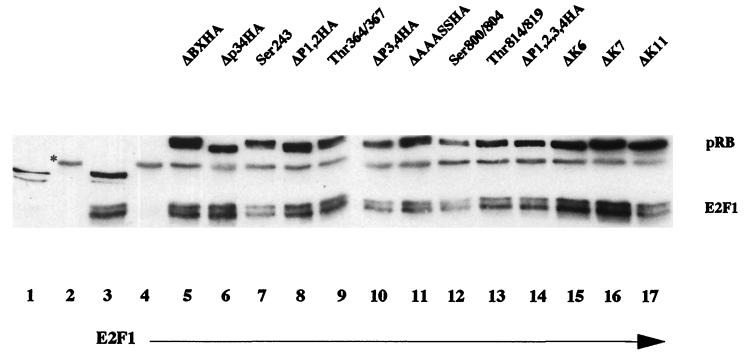
Each of the pRB mutants were tested for the ability to bind E2F1 by immunoprecipitation using anti-pRB followed by Western blot analysis with anti-E2F1 antibody (Fig. 3). Equal amounts of RB protein were immunoprecipitated from C33A lysates corrected for transfection efficiency with β-galactosidase. Similar amounts of E2F1 bound to mutant and wild-type hypophosphorylated pRB, suggesting that mutation of the phosphorylation sites does not significantly alter binding of hypophosphorylated pRB to free E2F1. The binding of the hypophosphorylated form of each pRB mutant to both large T antigen and free E2F1 suggests that the structural integrity and the major functions of pRB are maintained regardless of the charge of the amino acid substitution.
FIG. 3.
Coimmunoprecipitation of E2F1 with pRB mutants. Ten micrograms of RB plasmid was transfected with 0.5 μg of pCMV-E2F1 in C33A cells. Quantities of cell lysates corrected for transfection efficiency by using cotransfected pCMVβgal were immunoprecipitated with 5 μl of anti-pRB antibody (Pharmingen 14001A). pRB and coprecipitating proteins were separated on an SDS–7.5% polyacrylamide gel. The top half of the nitrocellulose was Western blotted with anti-HA antibody 12CA5, and the bottom half was Western blotted with anti-E2F1 (Santa Cruz C-20). The asterisk represents immunoglobulin G. Lane 1, untransfected C33A lysate (10% of the quantity used in immunoprecipitation); lane 2, immunoprecipitation of untransfected C33A lysate; lane 3, C33A lysate transfected with pCMV-E2F1 alone (10% of the quantity used in immunoprecipitation); lanes 4 to 17, immunoprecipitation of lysates transfected as indicated.
Repression of E2(−80/−70)-CAT by pRB mutants.
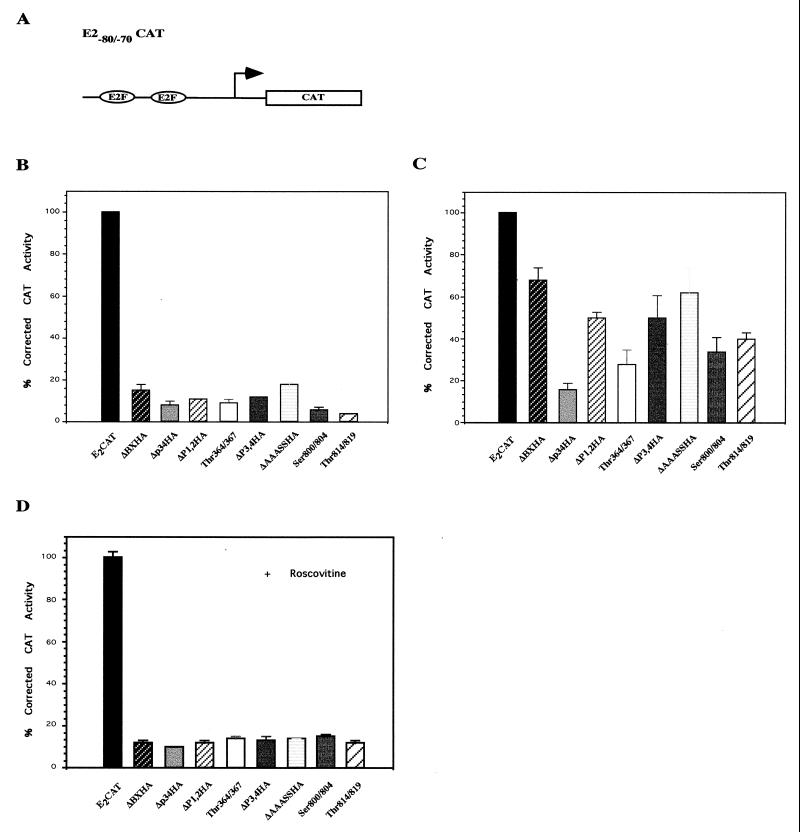
The ability of each of the phosphorylation site pRB mutants to repress E2F-mediated transcription was determined by using a CAT reporter construct [E2(−80/−70)-CAT] containing the adenovirus early EIIaE promoter, which contains two E2F binding sites (Fig. 4A). Three micrograms of RB plasmid was cotransfected with E2(−80/−70)-CAT into C33A cells either in the absence or in the presence of cyclin E-cdk2. This cell line was chosen because it contains a nonfunctional mutant RB protein (76), and therefore any effect seen on promoter activity is solely due to exogenously added pRB. One hundred percent activity represents reporter activity in the absence of RB plasmid. At high concentrations, there is no significant difference among the double-site mutants in the ability to repress E2F-mediated transcription compared to the wild type (Fig. 4B). In the absence of phosphorylation, both the wild type and the Δp34HA mutant repress E2F-mediated transcription to 12 to 15%. However, when cyclin E and cdk2 plasmids are added to the transfection in order to increase the amount of kinase activity present to phosphorylate pRB to high stoichiometry in vivo, significant differences between the mutants and wild-type pRB are seen (Fig. 4C). In the presence of kinase activity, wild-type pRB represses reporter activity 70 to 75% whereas Δp34HA fully represses activity to 15%, similar to repression in the absence of kinase. This is indicative of the ability of wild-type pRB to be inactivated by phosphorylation in the presence of kinase activity and the inability of Δp34HA to be inactivated due to mutation of eight potential p34cdc phosphorylation sites. Mutations Thr364/367 (sites 5 and 6), in the N terminus proximal to the region required to bind to E2F (72), and mutations Ser800/804 (sites 13 and 14) and Thr814/819 (sites 15 and 16), in the C terminus of pRB within the E2F binding region, produced proteins which repressed transcription 50% better than the wild type in the presence of kinase activity. Mutants Ser243 (data not shown), ΔP1,2HA, ΔP3,4HA, and ΔAAASSHA were similar to the wild type in the presence of kinase.
FIG. 4.
Repression of the E2 (−80/−70)-CAT reporter construct. (A) Schematic of the E2(−80/−70)CAT reporter construct. Three micrograms of E2(−80/−70)-CAT was transfected into C33A cells with 3 μg of each pRB construct (B) as well as 5 μg each of cyclin E and cdk2 constructs (C). One microgram of CMVβgal was included in the transfection to normalize CAT activity for transfection efficiency. SVLuc was used as filler DNA to prevent promoter competition. The activity obtained with the reporter construct in the absence of RB plasmid (B) and in the presence of cyclin E and cdk2 (C) is arbitrarily set to 100%. The ability of each pRB mutant to suppress transcription was determined by reduction in CAT activity. Error bars represent standard errors of two to four experiments. (D) C33A cells were transfected as for panel B, with roscovitine added at a final concentration of 700 nM 18 h following transfection. The activity obtained with the reporter construct in the presence of roscovitine is arbitrarily set to 100%.
To confirm that the differences in the ability of the pRB mutants to be released from repression in the presence of cyclin E-cdk2 is a consequence of variations in the capability of each mutant to become phosphorylated as a result of the mutation, each mutant’s ability to repress E2F-mediated transcription in the absence of endogenous kinase activity was determined. To inhibit endogenous cdk2-cdc2 activity, we used roscovitine, which competes for the ATP binding domain of kinases, at a concentration of 700 nM. In the presence of roscovitine, all pRB phosphorylation mutants repressed E2F activity to similar levels (Fig. 4D). This finding suggests that the small difference in repression of mutants Ser800/804 and Thr814/819 compared to wild-type pRB in the absence of cyclin E-cdk2 seen in Fig. 4B is due to endogenous kinase activity and confirms that the variation seen among the mutants in the presence of kinase is a direct result of phosphorylation of pRB.
Studies defining the mechanism of phosphorylation of pRB in the G1 phase of the cell cycle suggests that both cyclin D and cyclin E are required (34, 64). Therefore, we tested the ability of cyclin D-cdk4 kinase activity to release pRB from its interaction with E2F on the E2(−80/−70)-CAT promoter. Exogenously added cyclin D1 or D1-cdk4 expression plasmids did not relieve E2F-mediated repression by wild-type pRB in C33A cells (data not shown), whereas cyclin E-cdk2 complexes did relieve E2F-mediated repression by pRB (Fig. 4C). This is consistent with our previously published data which showed the inability of cyclin D1 to relieve repression by GALmRB in C33A cells (7). Furthermore, C33A cells contain a significant amount of p16 protein, as seen by Western blot analysis (data not shown), which could inhibit the activity of exogenously added cyclin D-cdk4.
Cyclin E-cdk2 regulation of the pRB mutants on E2F-mediated transcription.
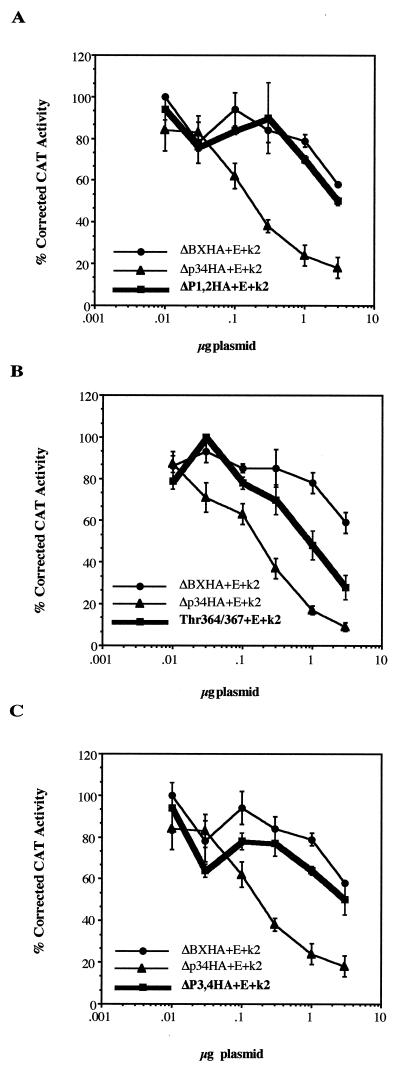
We have previously shown, by titration of the transfected amount of RB plasmid, that the Δp34HA mutant is a 50-fold better repressor of the E2(−80/−70)-CAT promoter in P19 cells than wild-type pRB (31). To confirm that the differences in repression seen by the pRB mutants were a consequence of their ability to be phosphorylated, various concentrations of the RB plasmids were cotransfected with E2(−80/−70)-CAT in the presence of constant amounts of cyclin E-cdk2 kinase. If phosphorylation at one particular site was required to relieve the interaction of pRB with E2F on DNA, then mutation of that site would result in a protein that could not be released from E2F by kinase activity, which would be more active in repression of E2F-mediated transcription than the wild type. The Δp34HA mutant is approximately 25-fold better at suppressing E2F activity in C33A cells than wild-type pRB in the presence of cyclin E-cdk2 kinase (only 0.125 μg of plasmid is required to repress to 60% of transcriptional activity, versus 3 μg for wild-type pRB) (Fig. 5A).
FIG. 5.
Cyclin E-cdk2 regulation of pRB mutants on E2F-mediated transcription. Various amounts of RB plasmid were cotransfected with 3 μg of E2(−80/−70)-CAT construct into C33A cells in the presence of 5 μg each of cyclin E (E) and cdk2 (k2) constructs. Promoter activity was determined as for Fig. 3. (A) ΔP1,2HA; (B) Thr364/367; (C) ΔP3,4HA. Error bars represent the standard errors of two to four experiments.
(i) N-terminal mutants.
Ser243 and ΔP1,2HA contain one-site (site 2) and two-site (sites 3 and 4) mutations, respectively, in the N terminus of pRB upstream of the E2F binding site (Table 1). Both Ser243 (data not shown) and ΔP1,2HA repress E2F-mediated transcription like wild-type pRB in the presence of cyclin E-cdk2 kinase activity, suggesting that these sites do not regulate the interaction of pRB with E2F on DNA (Fig. 5A). Thr364/367 contains mutations in two phosphorylation sites (sites 5 and 6) immediately N terminal of the A-domain portion of the E2F binding region and repressed E2F activity approximately 4.5-fold better than wild-type pRB (Fig. 5B). However, this repression did not meet the level of repression by Δp34HA (25-fold), which has eight phosphorylation sites (sites 3, 4, 8, 9, 11, 12, 13, and 14) mutated, while Thr364 and Thr367 are wild type. This finding suggested that accumulation of a number of phosphate groups may regulate transcriptional repression by pRB rather than phosphorylation of one particular residue, or one or more of the mutated sites in Δp34HA may be responsible for the greater suppressive ability. To test these possibilities, double-site mutants were made in the spacer region and C terminus of pRB, including the sites mutated in Δp34HA.
(ii) Spacer region mutant.
ΔP3,4HA contains mutations of Ser601 (site 8) and Ser605 (site 9) in the spacer between the A and B regions of pRB required for binding large T antigen. Recent data suggest that phosphorylation at both of these sites in the context of full-length human pRB can inhibit binding to free E2F (53). If phosphorylation at only these two sites completely removed pRB from E2F, then mutation of these sites would result in a protein that was unable to be released from E2F upon phosphorylation and that would repress transcription better than wild-type pRB. In the present assay, ΔP3,4HA repressed E2F-mediated transcription similarly to wild-type pRB, suggesting that phosphorylation at other sites on ΔP3,4HA can disrupt its interaction with E2F and that these two sites are not sufficient to regulate E2F binding on DNA (Fig. 5C). This result is consistent with another report which showed that mutation of the human sites 8 and 9 (human Ser608 and Ser612) repressed E2F-mediated transcription to the same level as wild-type pRB (2). However, Knudsen and Wang (53) demonstrated that human sites 8 and 9 did regulate the interaction of pRB with free E2F. Therefore, the sites that regulate the interaction of pRB with free E2F may be different from those that regulate the binding of pRB to E2F bound to DNA or those that regulate the binding of pRB-E2F to DNA.
(iii) C-terminal mutants.
Double-site mutations were made in six (sites 11, 12, 13, 14, 15, and 16) of the seven exon 23 phosphorylation sites, and the ability of each mutant to repress E2F activity in the presence of cyclin E-cdk2 activity was determined. Mutation of Ser781/788 (sites 11 and 12) in the ΔAAASSHA mutant resulted in a protein which functioned like wild-type pRB (data not shown), suggesting that neither of these sites is involved in regulating the pRB-E2F interaction when bound to DNA. However, mutation of Ser800/804 (sites 13 and 14) and Thr814/819 (sites 15 and 16) produced proteins that repressed transcription four- and threefold better than wild-type pRB in the presence of kinase activity (Table 2). Of the eight sites mutated in Δp34HA, only two (sites 13 and 14 in the Ser800/804 mutant) had an effect on E2F-mediated transcription when mutated together without other mutations, suggesting a role in regulating the pRB-E2F interaction on DNA. The other six sites had no effect when mutated in pairs (ΔP1,2HA, ΔP3,4HA, and ΔAAASSHA). However, the level of repression obtained with the Ser800/804 mutant does not explain the greater suppressive ability of the Δp34HA mutant, suggesting that the other six mutated sites are also involved in some way in regulating the interaction, if not on their own then in combination with other sites. To test this possibility, mutant proteins combining those mutations which showed no effect on their own were produced and tested for the ability to repress E2F-mediated transcription.
TABLE 2.
Activities of mutants
| Mutant | 60% Activity (μg)a | Fold differenceb |
|---|---|---|
| ΔBXHA(wt) | 3 | 1 |
| Δp34HA | 0.12 | 25 |
| Ser243 | 2 | 1.5 |
| ΔP1,2HA | 1.8 | 1.7 |
| Thr364/367 | 0.65 | 4.6 |
| ΔP3,4HA | 1.56 | 1.9 |
| ΔAAASSHA | 3 | 1 |
| Ser800/804 | 0.7 | 4.3 |
| Thr814/819 | 1 | 3 |
| ΔP1,2,3,4HA | 1.3 | 2.3 |
| ΔK6 | 0.12 | 25 |
| ΔK7 | 0.13 | 23 |
| ΔK11 | 0.14 | 22 |
The amount of RB plasmid required to repress promoter activity to 60% in the presence of cyclin E and cdk2 kinase, determined from the graphs of E2-CAT activity.
Difference in the amount of plasmid required to repress promoter activity for each mutant compared to wild-type pRB.
Cumulative effect of phosphorylation of pRB on E2F-mediated transcription.
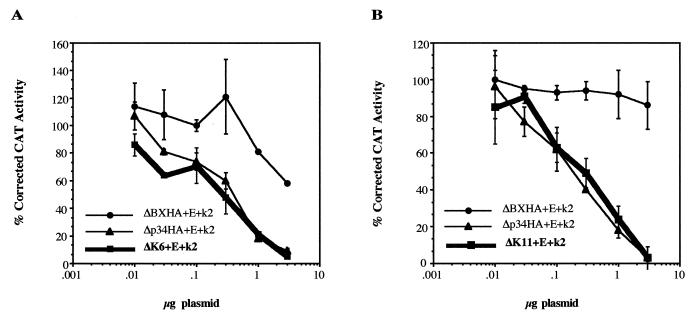
ΔP1,2HA and ΔP3,4HA were combined to form ΔP1,2,3,4HA, containing mutations in four phosphorylation sites (3, 4, 8, and 9). Each of these sites had no effect on E2F-mediated transcription when mutated in pairs. However, when four sites were mutated, the mutant RB protein repressed E2F-mediated transcription 2.3-fold in the presence of kinase (Table 2). When these four sites were mutated in combination with Ser781/788 (sites 11 and 12), the resulting protein, ΔK6, repressed E2F-mediated transcription like the eight-site mutant Δp34HA (Fig. 6A). This effect is not limited to these particular sites, as mutation of seven sites, 10 to 16, in the C terminus of pRB, ΔK7, also repressed transcription to the same extent as Δp34HA (Table 2). This result is consistent with Knudsen and Wang’s data which also showed that mutation of the seven C-terminal sites in the human RB-LP protein remained bound to E2F1 and repressed transcription in the presence of cyclins D1, E, and A (53). These data suggest that accumulation of phosphate groups on pRB may contribute to disruption of its interaction with E2F bound to a promoter. However, there is a limit to the number of sites that need to be phosphorylated, as mutation of additional sites in Δp34HA did not produce a protein with additional suppressive ability. ΔK11 contains mutations in 11 of the 16 potential phosphorylation sites (3, 4, and 8 to 16) and repressed E2F-mediated transcription to the same extent as Δp34HA (Fig. 6B).
FIG. 6.
Regulation of E2F-mediated transcription by ΔK6 (A) and ΔK11 (B). Various amounts of RB plasmid were cotransfected with 3 μg of E2(−80/−70)-CAT construct into C33A cells in the presence of 5 μg each of cyclin E (E) and cdk2 (k2) constructs. Promoter activity was determined as for Fig. 4.
Analysis of binding of pRB mutants to DNA-bound E2F.
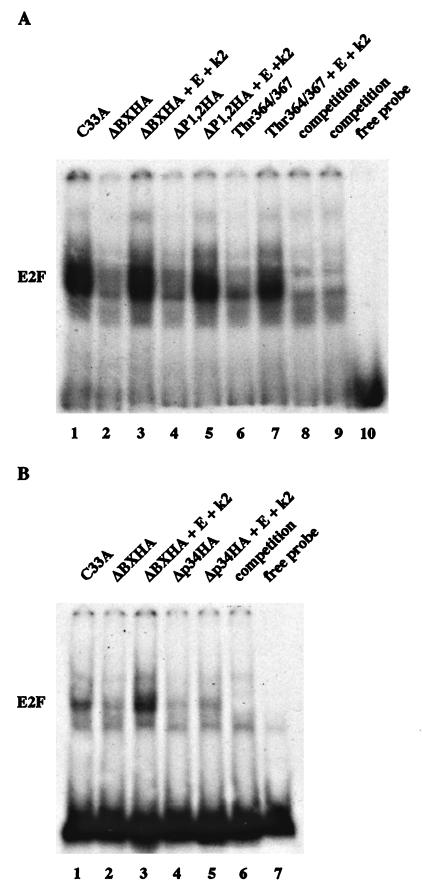
To confirm that phosphorylation of pRB was regulating E2F binding to the promoter, we used gel mobility shift assays to analyze the binding of pRB mutants to E2F protein bound to a 32P-labeled oligonucleotide containing an E2F site. Figure 7A, lane 1, shows the E2F activity of C33A cells that binds to the oligonucleotide in the absence of transfected proteins. This E2F activity disappears when wild-type pRB is transfected, suggesting that hypophosphorylated pRB is binding to E2F and removing it from the DNA (lane 2). We find that mouse pRB removes E2F from the DNA, unlike human pRB, which binds to DNA-bound E2F and supershifts to a slower-mobility complex. When cyclin E and cdk2 are cotransfected with wild-type pRB, the E2F binding activity reappears, suggesting that phosphorylated pRB is no longer interacting with E2F and E2F is now free to bind to the E2F site (lane 3). pRB mutants which acted like the wild type in the reporter assay, such as ΔP1,2HA, behaved similarly to wild-type pRB (lanes 4 and 5). Mutant Thr364/367 was 4.6-fold more potent than wild-type pRB at suppressing E2F activity in the presence of kinase in the reporter assay, and the amount of free E2F bound to the oligonucleotide in the gel shift was reduced three- to fourfold when mutant Thr364/367 was coexpressed with cyclin E-cdk2 (compare lanes 3 and 7). Finally, mutant Δp34HA was 25-fold more potent than wild-type pRB at suppressing E2F activity in the presence of kinase, as evidenced by the absence of free E2F activity bound to the oligonucleotide whether or not Δp34HA was cotransfected with cyclin E-cdk2 (Fig. 7B; compare lanes 3 and 5). The effect exerted by the various phosphorylation site mutants on binding DNA-bound E2F is consistent with our data from the reporter assays and suggests that phosphorylation of pRB regulates its binding to DNA-bound E2F and consequently the E2F-mediated promoter repression.
FIG. 7.
Gel mobility shift of pRB mutants. Three micrograms of RB plasmid was transfected into C33A cells in the absence or presence of 5 μg of cyclin E (E) and cdk2 (k2) plasmids as indicated above the lanes. Nuclear extracts were prepared as described in Materials and Methods. Quantities of extracts corrected for transfection efficiency by using cotransfected pCMVβgal were added to a 32P-labeled oligonucleotide containing an E2F site; complexes were resolved on a 4% native polyacrylamide gel. Competition of the E2F binding activity was performed with a 50-fold excess of unlabeled oligonucleotide.
DISCUSSION
The phosphorylation sites of pRB are found dispersed throughout the protein, with six sites (sites 1 to 6) in the N terminus, one (site 7) within the A domain, two (sites 8 and 9) in the spacer region, and seven (sites 10 to 16) in the C terminus. The reason there are so many phosphorylation sites is unknown. Do they each regulate different functions of pRB? There is an accumulating body of data which suggests that phosphorylation at specific sites of pRB regulates different protein-protein interactions. Knudsen and Wang showed that phosphorylation of sites 13 and 14 of human pRB was required to release the interaction of c-Abl from pRB, whereas phosphorylation of sites 15 and 16 was required to release the interaction of large T antigen from pRB (52). However, none of these sites regulated the interaction of pRB with E2F in a bandshift assay (52). Recent data also suggest that the binding of pRB to free E2F is regulated by dual mechanisms involving phosphorylation either at a number of C-terminal sites or at sites 8 and 9 in the insert domain of pRB (53).
Our data suggest that regulation of the pRB-E2F interaction on DNA by phosphorylation of pRB occurs by accumulation of phosphate groups on the pRB molecule. There are a number of possible explanations for how phosphorylation may interrupt the pRB-E2F interaction: (i) the conformation of the E2F binding domain of pRB may be changed by phosphorylation at these sites; (ii) the conformation of the domain is unchanged but addition of negative charges may eliminate hydrophobic contacts with E2F; and (iii) addition of the phosphate groups may cause a preferential interaction of the E2F binding domain with another part of the RB molecule, therefore releasing E2F. Recent data obtained from the structure of pRB bound to an LXCXE peptide (57) suggest that release of the LXCXE peptide from pRB upon phosphorylation of pRB probably occurs by competition of the phosphorylated pRB polypeptide segment for the LXCXE peptide binding site. There is a six-lysine basic patch on the rim of the LXCXE binding site that may help the binding of the phosphorylated peptide segment of pRB (57). Although the binding site for E2F is probably different from the LXCXE binding site (the A-B interface rather than the B box), it is possible that this competitive mechanism is also responsible for releasing pRB from E2F when bound to DNA. The greater suppressive activity in the presence of cyclin E kinase activity seen with RB molecules containing six or more phosphorylation sites mutated compared to wild-type pRB may be due to the ability of the mutated pRB polypeptide segment to remain bound to E2F even when the other unmutated sites are phosphorylated. Data obtained from mutants ΔK6 and ΔK7 also suggest that it does not matter whether the phosphorylated polypeptide of pRB is at the N terminus or C terminus of pRB.
Studies defining the mechanism of phosphorylation of pRB in the G1 phase of the cell cycle suggest that both cyclin D and cyclin E are required (34, 64). In yeast, phosphorylation of pRB is mediated through Cdc28. Three G1 cyclins, Cln1, Cln2, and Cln3, regulate the activity of Cdc28 throughout G1. Phosphorylation of pRB requires a combination of Cln3 and either Cln1 or Cln2. Mammalian cyclin E is able to substitute for a cln2 mutant whereas cyclin D1 is able to substitute for a cln3 mutant, suggesting collaboration between cyclin D1 and cyclin E in phosphorylating pRB (34). This phenomenon is also seen in mammalian cells. Use of inhibitors of cyclin D-cdk4/6 and cyclin E-cdk2 kinase activity, p16ink4A and cdk2DN, respectively, with U2-OS cells containing wild-type pRB showed that pRB is only partially phosphorylated by cyclin D kinase activity; complete phosphorylation requires cyclin E kinase action (64). In addition, phosphorylation of pRB only by cyclin D kinase did not release pRB from its interaction with E2F1 (64). This finding is consistent with our data showing that exogenously added cyclin D1 or D1-cdk4 expression plasmids did not relieve E2F-mediated repression in C33A cells (data not shown), whereas cyclin E-cdk2 complexes did relieve E2F-mediated repression by pRB (Fig. 4C). Lundberg and Weinberg (64) suggest that phosphorylation by cyclin D may cause a conformational change in pRB which allows phosphorylation by cyclin E-cdk2 or may release pRB from certain nuclear structures which hinder access of cyclin E to pRB. It is possible that in our experimental system there is enough endogenous cyclin D kinase activity in C33A cells to provide the necessary conformation change or to disrupt nuclear tethering so that the exogenous cyclin E-cdk2 can phosphorylate pRB and relieve its interaction with E2F.
pRB mutants ΔP1,2HA, ΔP3,4HA, and ΔAAASSHA resulted in proteins that repressed transcription like the wild type both in the absence and in the presence of cyclin E kinase. This suggests that cyclin E kinase does not act on the sites mutated in these mutant proteins. Several groups have worked to determine which of the phosphorylation sites of pRB are phosphorylated by cyclin A, E, or D kinase complexes. Zarkowska and Mittnacht determined that the human sites 3 and 4 (ΔP1,2HA) are phosphorylated by cyclin D but not by cyclin E or A (91). Others reported that sites 3 and 4 are phosphorylated by both cdk2 and cdk4 kinases (18). Site 9 (ΔP3,4HA) and site 12 (ΔAAASSHA) are also regulated by cyclin E (18). Based on the latter study, cyclin E does act on those sites mutated in the ΔP1,2HA, ΔP3,4HA and ΔAAASSHA mutants. Since mutations of these sites resulted in pRB that repressed transcription like the wild type in the presence of cyclin E-cdk2 kinase activity, one conclusion may be that none of these sites are involved in regulating the pRB-E2F interaction on DNA.
An alternative model suggests that when pRB is bound to E2F on a promoter, certain phosphorylation sites may be masked and therefore unavailable for phosphorylation. This masking could occur as a result of a change in conformation of the pRB molecule when bound to E2F on the promoter or as a result of the binding of another protein such as a general transcription factor to pRB when it is found bound to the DNA through E2F. This may explain why phosphorylation of sites 8 and 9 in the spacer region mutant ΔP3,4HA is able to release pRB from free E2F (53) but is not important in the release of pRB from E2F bound to a promoter (Fig. 5C). It is possible that sites 11 and 12 (ΔAAASSHA) are also masked and unavailable for phosphorylation but sites 5 and 6 (Thr364/367), sites 13 and 14 (Ser800/804), and sites 15 and 16 (Thr814/819) are accessible when bound to E2F at the promoter. However, mutation of these sites in pairs resulted in pRB proteins which repressed E2F-mediated transcription only 3- to 5-fold better than the wild type whereas mutation of these sites in combination (six or more sites) resulted in proteins which repressed E2F-mediated transcription 25-fold better than the wild type in the presence of kinase activity (Δp34HA, ΔK6, ΔK7, and ΔK11). Therefore, phosphorylation at particular sites may be important to induce slight conformational changes so that other phosphorylation sites are unmasked and become available for phosphorylation.
ACKNOWLEDGMENTS
This work was supported in part by the National Cancer Institute of Canada with funds from the Terry Fox Run, the Medical Research Council of Canada, and Apotex Inc. through the University-Industry Program.
We thank Paul Hamel, Eldad Zachsenhaus, and Rod Bremner for helpful discussions. V. Brown especially thanks Sanja Pajovic for technical help with the gel mobility shift assays, P. Hamel for the gift of ΔBXHA, Δp34HA, ΔP1,2HA, ΔP3,4HA, and ΔP1,2,3,4HA, and E. Zachsenhaus for the gift of ΔK11.
REFERENCES
- 1.Akiyama T, Ohuchi T, Sumida S, Matsumoto K, Toyoshima K. Phosphorylation of the retinoblastoma protein by cdk2. Proc Natl Acad Sci USA. 1992;89:7900–7904. doi: 10.1073/pnas.89.17.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelman D, Perry S, Hollingsworth R, Gregory R J, Driscoll B, Fung Y K, Bookstein R. Engineered mutants of pRB with improved growth suppression potential. Oncogene. 1997;15:2855–2866. doi: 10.1038/sj.onc.1201465. [DOI] [PubMed] [Google Scholar]
- 3.Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-1, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1070. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 4.Bandara L R, Buck V M, Zamanian M, Johnston L H, Thangue L B L. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 1993;12:4317–4324. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchkovich K, Duffy L A, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 9.Cao L, Faha B, Dembski M, Tsai L-H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 10.Chellappan S, Hiebert S, Mudryi M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 11.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y-H F, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1 mediated transcription by liberating Sp1 from a negative regulator. Mol Biol Cell. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P-L, Riley D J, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 13.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P-L, Scully P, Shew J-Y, Wang J Y J, Lee W-H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 15.Chow K N B, Dean D C. Domains A and B in the RB pocket interact to form a transcriptional repressor motif. Mol Cell Biol. 1996;16:4862–4868. doi: 10.1128/mcb.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow K N B, Starostik P, Dean D C. The Rb family contains a conserved cyclin-dependent-kinase-regulated transcriptional repressor motif. Mol Cell Biol. 1996;16:7173–7181. doi: 10.1128/mcb.16.12.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 18.Connell-Crowley L, Harper J W, Goodrich D W. Cyclin D1/cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeCaprio J A, Fusuke Y, Ajchenbaum F, Griffin J D, Livingston D M. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc Natl Acad Sci USA. 1992;89:1795–1798. doi: 10.1073/pnas.89.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C-M, Livingston D M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 21.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 23.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 24.Ewen M, Xing Y, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping, and expression of p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 25.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J-Y, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 26.Flemington E K, Speck S H, Kaelin W G., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Gu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 29.Hagemeier C, Bannister A J, Cook A, Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamel P, Gill R M, Phillips R A, Gallie B L. Regions controlling hyperphosphorylation and conformation of the retinoblastoma gene product are independent of domains required for transcriptional repression. Oncogene. 1992;7:693–701. [PubMed] [Google Scholar]
- 31.Hamel P, Gill R M, Phillips R A, Gallie B L. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamel P A, Cohen B L, Sorce L M, Gallie B L, Phillips R A. Hyperphosphorylation of the retinoblastoma gene product is determined by domains outside the simian virus 40 large-T-antigen-binding regions. Mol Cell Biol. 1990;10:6586–6595. doi: 10.1128/mcb.10.12.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannon G J, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with cdk2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 34.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 35.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 36.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 38.Helin K, Wu C-L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 39.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 40.Hiebert S W, Chellappan S P, Horowitz J M, Nevins J R. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 41.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van’t Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann F, Martelli F, Livingston D M, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 44.Horton L E, Qian Y, Templeton D J. G1 cyclins control of the retinoblastoma gene product growth regulation activity via upstream mechanisms. Cell Growth Differ. 1995;6:395–407. [PubMed] [Google Scholar]
- 45.Hsieh J-K, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–1852. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 46.Hu Q, Lees J A, Buchkovich K J, Harlow E. The retinoblastoma protein physically associates with the human cdc2 kinase. Mol Cell Biol. 1992;12:971–980. doi: 10.1128/mcb.12.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 48.Kato J-Y, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 49.Kim S-J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S-J, Wagner S, Liu F, Reilly M A, Robbins P D, Green M R. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 51.Kitagawa M, Higashi H, Jung H-K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J-Y, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-cdk4 is different from that for phosphorylation by cyclin A/E-cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 52.Knudsen E S, Wang J Y J. Differential regulation of retinoblastoma protein function by specific cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 53.Knudsen E S, Wang J Y J. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 55.Larminie C G C, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LaThangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 57.Lee J-O, Russo A A, Pavletich N P. Structure of the retinoblastoma tumor-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 58.Lees J A, Buchkovich K J, Marshak D R, Anderson C W, Harlow E. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. The adenovirus E1a-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 61.Lin B T-Y, Gruenwald S, Morla A O, Lee W-H, Wang J Y J. Retinoblastoma cancer suppressor gene product is a substrate of the cell cycle regulator cdc2 kinase. EMBO J. 1991;10:857–864. doi: 10.1002/j.1460-2075.1991.tb08018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludlow J W, DeCaprio J A, Huang C, Lee W-H, Paucha E, Livingston D M. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell. 1989;56:57–65. doi: 10.1016/0092-8674(89)90983-5. [DOI] [PubMed] [Google Scholar]
- 63.Ludlow J W, Glendening C L, Livingston D M, DeCaprio J A. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 66.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Villain J P L, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 67.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 68.Mittnacht S, Lees J A, Desai D, Harlow E, Morgan D O, Weinberg R A. Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 1994;13:118–127. doi: 10.1002/j.1460-2075.1994.tb06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 71.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 72.Qian Y, Luckey C, Horton L, Esser M, Templeton D J. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol Cell Biol. 1992;12:5363–5372. doi: 10.1128/mcb.12.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qin X-Q, Livingston D M, Kaelin W G, Jr, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 75.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheffner M, Munger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sellers W R, Kaelin W G. RB as a modulator of transcription. Biochim Biophys Acta. 1996;1288:M1–M5. doi: 10.1016/0304-419x(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 78.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 80.Sidle A, Palaty C, Dirks P, Wiggan O, Kiess M, Gill R M, Wong A K, Hamel P A. Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation. Crit Rev Biochem Mol Biol. 1996;31:237–271. doi: 10.3109/10409239609106585. [DOI] [PubMed] [Google Scholar]
- 81.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 82.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eukaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 83.Tao Y, Kassatly R F, Cress W D, Horowitz J M. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Templeton D J, Park S H, Lanier L, Weinberg R A. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Udvadia A J, Rogers K T, Higgins P D R, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1 mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 87.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma gene. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C-Y, Petryniak B, Thompson C B, Kaelin W G, Leiden J M. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science. 1993;260:1330–1335. doi: 10.1126/science.8493578. [DOI] [PubMed] [Google Scholar]
- 89.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 90.Zamanian M, Thangue N B L. Transcriptional repression by the Rb-related protein p107. Mol Cell Biol. 1993;4:389–396. doi: 10.1091/mbc.4.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- 92.Zarkowska T, U. S, Harlow E, Mittnacht S. Monoclonal antibodies specific for underphosphorylated retinoblastoma protein identify a cell cycle regulated phosphorylation site targeted by CDKs. Oncogene. 1997;14:249–254. doi: 10.1038/sj.onc.1200824. [DOI] [PubMed] [Google Scholar]
- 93.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]