Abstract
The observation of biological structures in live cells beyond the diffraction limit with super-resolution fluorescence microscopy is limited by the ability of fluorescence probes to permeate live cells and the effect of these probes, which are often toxic, on cellular behavior. Here we present a coherent confocal light scattering and absorption spectroscopic microscopy that for the first time enables the use of large numerical aperture optics to characterize structures in live cells down to 10 nm spatial scales, well beyond the diffraction limit. Not only does this new capability allow high resolution microscopy with light scattering contrast, but it can also be used with almost any light scattering spectroscopic application which employs lenses. We demonstrate that the coherent light scattering contrast based technique allows continuous temporal tracking of the transition from non-cancerous to an early cancerous state in live cells, without exogenous markers. We also use the technique to sense differences in the aggressiveness of cancer in live cells and for label free identification of different grades of cancer in resected tumor tissues.
Keywords: light scattering, coherence, confocal microscopy, spectroscopy, cancer
Graphical Abstract

Introduction
Recent developments in super-resolution fluorescence microscopy1–4 allow the observation of biological structures with spatial resolution beyond the diffraction limit5. However, it requires the introduction of fluorescence probes, which provide contrast to subcellular and subnuclear structures. In the case of live cells, these fluorescence probes are often limited by their ability to permeate the cell, low brightness, and their effect on cellular behavior, all of which are significant drawbacks6. Recently developed genetically fused fluorescent protein tags7–9 are substantially smaller than fluorescence probes, can be coexpressed with any desired protein, and provide straightforward access to specific and stoichiometric labeling10. However, they could sterically interfere with the protein function or influence protein mobility within the cell11 and their brightness and photostability are inferior to that of the traditional fluorescence probes. Another limitation of fluorescence based microscopic imaging is the difficulty in translating it to clinical use. Chemical fluorescence stains are usually toxic and live cell labeling approaches often require genetic modifications to the cell. To overcome these limitations, a source of subcellular contrast that is based on a different physical principle is needed.
Spectroscopic analysis of scattered light plays an increasingly important role in biology and medicine, as it can be used to inform on subcellular and extracellular biological structures12. Previously it was demonstrated that by monitoring microscopic subcellular structures with macroscopic spectral measurements, approaches such as light scattering spectroscopy (LSS) or elastic scattering spectroscopy (ESS) can detect pre-cancer in the esophagus13,14, pancreas15,16, breast17,18, cervix19–21, colon22, lung23 and other organs, independent of any visual cues. These methods can also monitor the response to chemotherapeutic treatment24,25, or serve as a source of highly specific subcellular native contrast in confocal microscopy in both point-scanning26–28 and line-scanning29 arrangements, as well as in widefield microscopy30,31.
Original confocal light absorption and scattering spectroscopic (CLASS) microscopy26 was capable of characterizing structures down to 10 nm. However, the size of its confocal pixel was still limited by the objective’s angle of acceptance. In the incoherent version of CLASS, when the numerical aperture (NA) of the objective’s angle exceeds NA=0.5, the spectra become virtually structureless. As it is the multispectral nature of CLASS that enables it to characterize structures beyond the diffraction limit, this capability is lost for high NA optics, which are needed for high resolution microscopy. In this paper we demonstrate that if the source coherence is preserved, the spectra detected by the microscope exhibit clear oscillatory structures, even for very large acceptance angles, such as NA=1.3. This finding will not only dramatically improve CLASS microscopy resolution, but could also be used with almost any light scattering spectroscopic application that employs lenses.
Several examples presented here serve as a demonstration of the new capabilities of coherent CLASS microscopy. We employed coherent CLASS microscopy on live cells to monitor the earliest subcellular morphological changes in organelles and structures as the cells become cancerous through the application of carcinogenic drugs. Such temporal tracking of live cell transition from non-cancerous to early cancer can be performed continuously on the time scale ranging from minutes to days. We also used the coherent CLASS microscope to sense differences in cancer aggressiveness in live cells and in resected esophageal adenocarcinoma tumor tissue samples from human subjects.
Methods
Optical Design of the Coherent CLASS Microscope
The coherent CLASS microscope optical design is presented in Figure 1a. It utilizes an Olympus IX81 inverted microscope frame and FluoView 1000 scanning unit. It also incorporates a broadband coherent supercontinuum source (Fianium WL-SC-400–4) that consists of a Ti:sapphire pump laser coupled to a photonic crystal fiber (PCF). The collimated light beam from the supercontinuum source is focused into a single mode fiber attached to the microscope entrance port. This broadband coherent light beam is scanned rapidly using a galvanometer scanning mirror. Following expansion and collimation with scan and tube lenses, the beam fills the back aperture of an objective. We used a high NA=1.35 achromatic microscope objective (Olympus UPLSAPO 60XW) that has little chromatic aberration (from 400 nm to 900 nm, the focus position differs by only 0.25 μm). The objective focuses light on the sample and the backscattered light is collected by the same objective. The scattered light is then de-scanned by the galvanometer scanning mirror and passes a 20/80 beam splitter on the way to the collection pinhole, blocking the majority of the light originating above and below the focal plane. This allows only the light scattered within a focal volume to be detected. An optical fiber collects and delivers this light to an imaging spectrograph (Princeton Instruments Acton SP-2300i) coupled with a scientific CMOS detector (Andor sCMOS Neo 5.5). The wavelength range of the supercontinuum laser, sCMOS quantum efficiency and spectrograph bandwidth give the coherent CLASS microscope a spectral range of 400 nm to 800 nm. The spatial resolution at a wavelength of 600 nm in the lateral and axial directions is 0.2 µm and 1.0 µm, respectively. Detection of the coherent CLASS spectrum for a single pixel location takes approximately 1.4 ms. Thus, a 128 x 128 image can be obtained in less than 25 seconds. We should note here that the imaging speed of the microscope is mainly limited by the interplay of the speed and sensitivity of the spectroscopy detector. Instead of the imaging spectrograph coupled with a scientific CMOS detector, we can also use a faster but slightly less sensitive high speed CMOS spectrometer (Avantes AvaSpec-ULS2048CL-EVO) capable of collecting spectra from a single pixel in approximately 0.1 ms and the entire frame in less than 2 seconds. The system is controlled with custom LabView (National Instrument, LabView 8.5) code. The synchronization between the scanning head and spectrometer is achieved by connecting the scanning head controller to the sCMOS camera with a TTL trigger cable.
Figure 1.
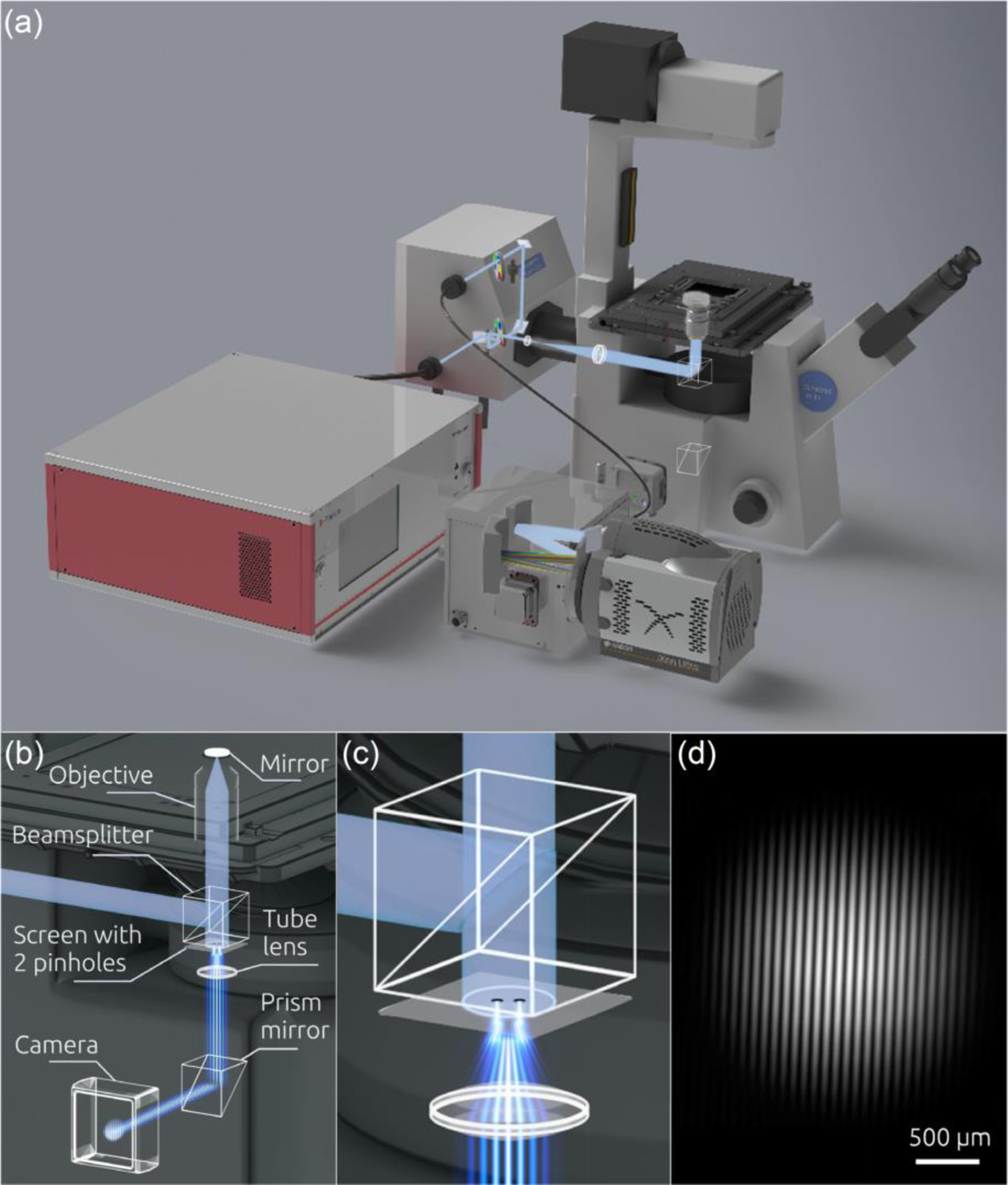
Coherent CLASS microscope. (a) Schematic of the light paths and optical elements of the coherent CLASS microscopy system. The system incorporates an inverted microscope frame, confocal scanning unit, broadband coherent supercontinuum source, imaging spectrograph, and thermo-electrically-cooled sCMOS detector. (b) Measurements of the degree of coherence in the CLASS microscope with a Young interferometer. (c) Close-up view of the Young interferometer. (d) Experimentally measured spatial distribution of light intensity in the coherent CLASS microscope based Young interferometer.
It is important to note that in reality, light is only partially coherent. There are various factors that affect the degree of coherence, including the light source coherence, properties of the optical components, and the pinhole size. In our case, the degree of coherence has been increased by employing a supercontinuum broadband laser source coupled to the system with a single mode optical fiber. To measure the spectral degree of coherence μ12 of the light field in the coherent CLASS microscope, we incorporated a Young interferometer setup into the light path of the microscope (see Figure 1b). To do this, we employed a screen with two 80 μm pinholes separated by 1.15 mm, placing it directly under a microscope beamsplitter cube. The light from the supercontinuum source was initially passed through a 10 nm bandpass filter, mimicking the effect of the spectrometer detection in the coherent CLASS microscopy setup. The beam was reflected from a mirror positioned in the focal plane of the microscope, and the reflected beam was then collimated by the objective, before illuminating the screen containing the pinholes (Figure 1c). The interferogram produced in the far field of the Young mask was recorded with the CCD detector (Figure 1d). From the interferogram, using the standard formula for the spectral degree of coherence5, we found that in the coherent CLASS microscope μ12 = 0.92.
Light Scattering Spectra in Coherent Confocal Microscopy
In case of an objective with a large acceptance angle, the informative part of the spectrum in the conventional CLASS microscopy is often destroyed due to angular averaging of the scattering signal (Figure 2a). However, in the coherent case, this informative part is preserved due to in-phase constructive interference (Figure 2b).
Figure 2.
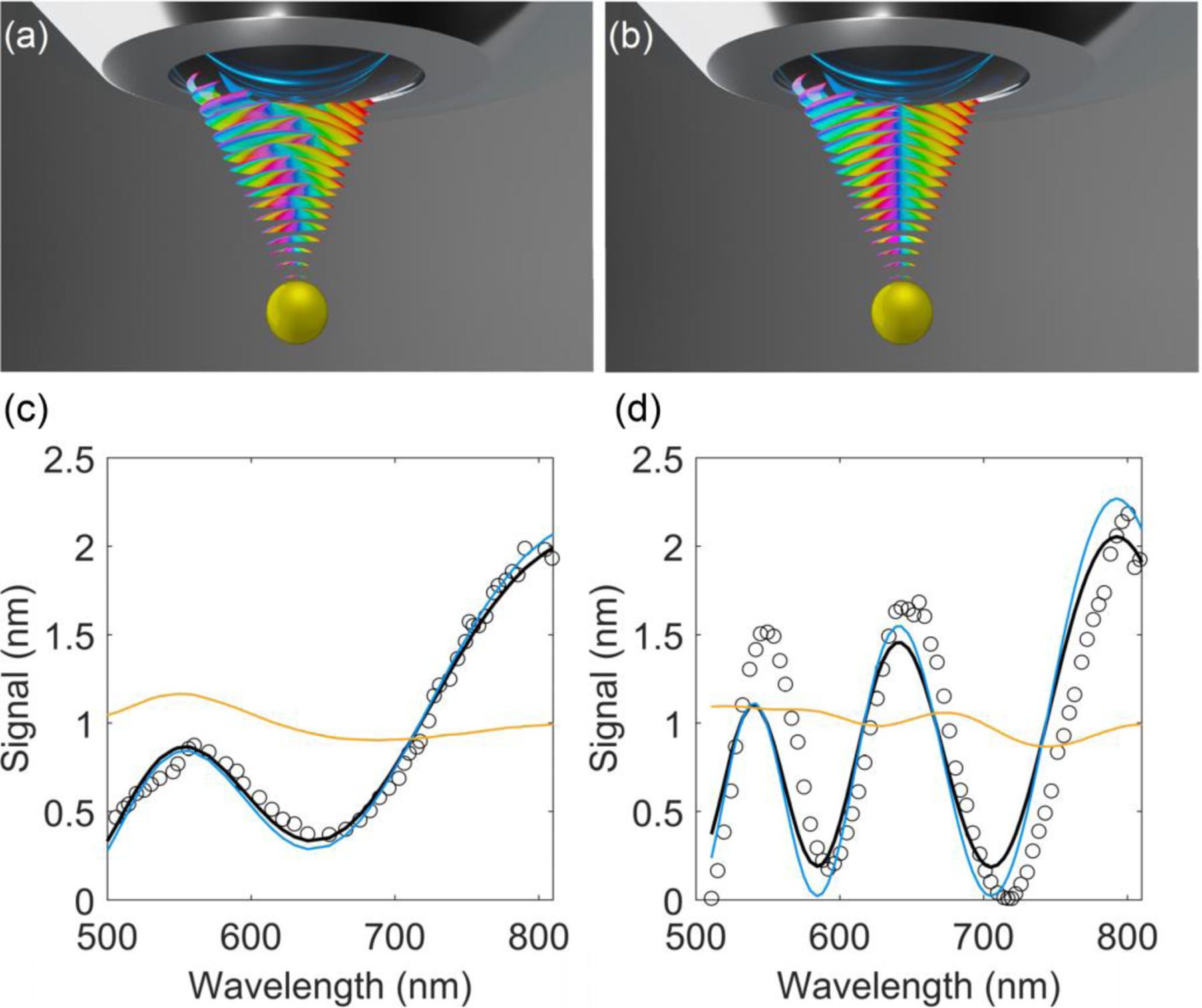
Principles of coherent CLASS microscopy. The schematic shows the interplay between phase interference and angular averaging in the incoherent (a) and coherent (b) cases. Spectra of light scattered by polystyrene microspheres with nominal diameters of (c) 540 ± 20 nm and (d) 1050 ± 20 nm in aqueous solution of glycerol (circles) experimentally measured using the coherent CLASS microscope with a large numerical aperture objective, NA=1.3. Also shown are simulations of the incoherent (solid orange line) and coherent (solid blue line) CLASS microscopy spectra for the same microspheres under the same measurement conditions. The black solid lines are the best fits of the spectra represented by Eq. (4) for both microsphere sizes. The resulting degree of coherence of the CLASS spectra is 0.93.
To compare incoherent and coherent CLASS microscopy spectra for a single spherical scatterer of diameter δ, we follow the previously developed approach27 where we expand both incident and scattering electromagnetic waves into an infinite series of directional plane waves. The directions of these plane waves are constrained by the objective’s numerical aperture. The incoherent CLASS spectrum is then
| (1) |
where λ is the wavelength, and are unit vectors giving the propagation’s direction of the incident and the scattered light, respectively, is the normalized objective pupil function of both light delivery and collection, is the objective’s solid angle associated with its NA. Finally, is the Mie scattering amplitude of the wave with wavelength λ scattered by a scatterer with size δ in a direction , with the incident wave coming from direction , and it can be expressed as
| (2) |
where Pn are ordinary Legendre polynomials, θk is the angle between and , and coefficient An depends on the particle’s size and optical properties32.
On the other hand, the coherent CLASS spectrum is
| (3) |
In the case of backscattering where the solid angle of the objective is very small, both coherent and incoherent spectra are reduced to the same expression . However, for large solid angles, angular averaging provides very different results for the coherent and incoherent cases. To test that this effect preserves the informative oscillatory part of the spectra, we used coherent CLASS microscopy with a large acceptance angle (NA=1.3) objective to measure spectra of light scattered by polystyrene microspheres in the aqueous solution of glycerol. This provided optical parameters close to that of the organelles surrounded by cytoplasm. The spectra of microspheres with nominal diameters of 540 ± 20 nm (Figure 2c) and 1050 ± 20 nm (Figure 2d) were measured. We also performed numerical simulations of the same spectra for the conventional and coherent CLASS microscopes with the same objective. The majority of the oscillatory structure remained only in the coherent CLASS microscope spectra, confirming that this type of detection would produce a significantly higher spectral contrast. The best fits were obtained for the microspheres, with the diameters of 550 nm and 1060 nm suggesting that the accuracy of the coherent CLASS microscopy measurements was up to 10 nm. Furthermore, sensitivity of coherent CLASS microscopy to small changes in scatterer size is also significantly better than that of incoherent CLASS microscopy (Supporting Information, Figure S1). It is important to emphasize that while the dimensions of the coherent CLASS microscopy confocal volume are diffraction limited (0.2 μm by 1.0 μm) and define the spatial resolution of the technique, its multispectral nature enables CLASS microscopy to characterize structures much smaller than a wavelength within that diffraction limited volume.
The comparison of the experimental and simulated spectra gives another opportunity to evaluate the CLASS spectra coherence. Though it is rather obvious that the coherent CLASS spectra describe the experiment much better than the incoherent ones, we can also quantify this by introducing the spectrum
| (4) |
with μs representing the degree of coherence of the CLASS spectra. By finding the best fits of the spectra represented by Eq. (4) to the experimental data for both sizes of microspheres simultaneously, we found the degree of coherence in the CLASS spectra to be μs = 0.93. This value is in excellent agreement with the spectral degree of coherence μ12 = 0.92 measured using the Young interferometer setup, indicating a high degree of coherence achieved in the coherent CLASS microscope.
Human Esophageal Cancer Cell Lines and Resected Esophageal Samples
Human esophageal cancer cell lines (OE33 and OE19), were acquired from Sigma-Aldrich (St. Louis, MO). These cells were grown in RPMI1640 medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum and 2 mM Glutamine. The non-dysplastic Barrett’s esophagus cell line CP-A was purchased from ATCC (Manassas, VA) and cultured with MCDB-153 medium supplemented with 20 mg/L adenine, 0.4 µg/ml hydrocortisone, 1x ITS Supplement (Sigma-Aldrich, St. Louis, MO), 8.4 µg/L cholera toxin, 20 ng/ml recombinant human epidermal growth factor, 140 µg/ml bovine pituitary extract, 4 mM glutamine and 5% fetal bovine serum. Incubation of all cells was done in an atmosphere of 5% CO2 at 37 °C, where the cells were then sub-cultured to around 70–80% confluency. Glass bottom dishes (MatTek Corporation, Ashland, MA) were used to culture cells in order to facilitate coherent CLASS microscopy measurements. When the monolayer confluency was reached, fresh RPMI1640 or MCDB media without phenol red were employed just before microscopy measurements. This replacement of the culture medium eliminated light absorption from the media itself.
Total RNA was extracted from OE19, OE33, CP-A, and NNK treated CP-A cells with the RNeasy Mini Kit (Qiagen, Valencia, CA) and was used in the real-time PCR experiments with a previously reported primer set33. All PCR reactions were done with a QuantStudio 6 Real-Time PCR System (Thermofisher, MA) and a SYBR® Green PCR Master Mix (Thermofisher, MA) according to the manufacture’s protocol. The data was analyzed for triplicate samples and the mean ± standard deviation (SD) was used.
Paraffin-embedded tissue blocks for the selected subjects were obtained from the BIDMC Department of Pathology archives. The BIDMC Institutional Review Board reviewed the protocol, and the necessary approvals were obtained. The database for patients who underwent esophagectomy at BIDMC was reviewed and subjects with no prior chemotherapy or radiation therapy were selected. All tissue blocks were reviewed by an expert GI pathologist for determination of the degree of esophageal tumor differentiation and to describe the cellularity of the sample at various sites of interest. Prior to spectroscopic measurements, the tissue blocks were initially deparaffinized in xylene, followed by rehydration in graded alcohols34. The tissue areas targeted, corresponding to the paraffin blocks, were marked on H&E-stained sections.
Results
Cancer Aggressiveness Evaluation with Coherent CLASS Microscopy
Several organelles, such as the mitochondria, Golgi bodies, or vesicles produced by cells, such as exosomes, are known to be altered in cancer. Mitochondria decrease in size during cancer progression35 and are also known to change the type of energy metabolism to accommodate a high metabolic rate of cancer cells36. The number and size of the Golgi bodies37 and PML nuclear bodies38 also change with cancer progression. Our own recent studies have shown that exosomes are markedly altered in cancer cells and may serve as a mechanism of tumorigenesis39.
To evaluate the ability of the coherent CLASS microscopy system to detect differences in cancer aggressiveness in live cells through subcellular structure sizing, we employed two types of human esophageal adenocarcinoma cell lines: a less aggressive moderately differentiated OE19 cell line known to have on average 56.8 ± 1.8 chromosomes and a more aggressive poorly differentiated OE33 cell line which has 100.2 ± 4.4 chromosomes40. To confirm that OE33 cells indeed represent more aggressive cancer when compared to OE19 cells, we quantified expression levels of several genes that are associated with cancer aggressiveness using qRT-PCR.
The expression level of the auxiliary protein of DNA synthase, known as proliferating cell nuclear antigen (PCNA) is used as a marker of proliferation41 and is known to increase at progressive cancer stages42,43. The expression level of PCNA was approximately two times higher in OE33 cells than in OE19 cells (Figure 3). We also observed significantly higher expression levels of the B-cell lymphoma 2 oncogene (Bcl-2), which inhibits apoptotic cell death44, and of the nuclear factor κB (NF-κB), which is important to the development and progression of cancer, such as proliferation, migration and apoptosis45. OE33 cells also exhibited a significantly higher expression level of the signal transducers and activators of transcription (STAT1), a cytoplasmic protein that acts as both a signal messenger and transcription factor, participating in the cellular response to cytokines and growth factors46. Increased activation of STAT1 correlates with an increase in tumor progression for several types of cancer47.
Figure 3.
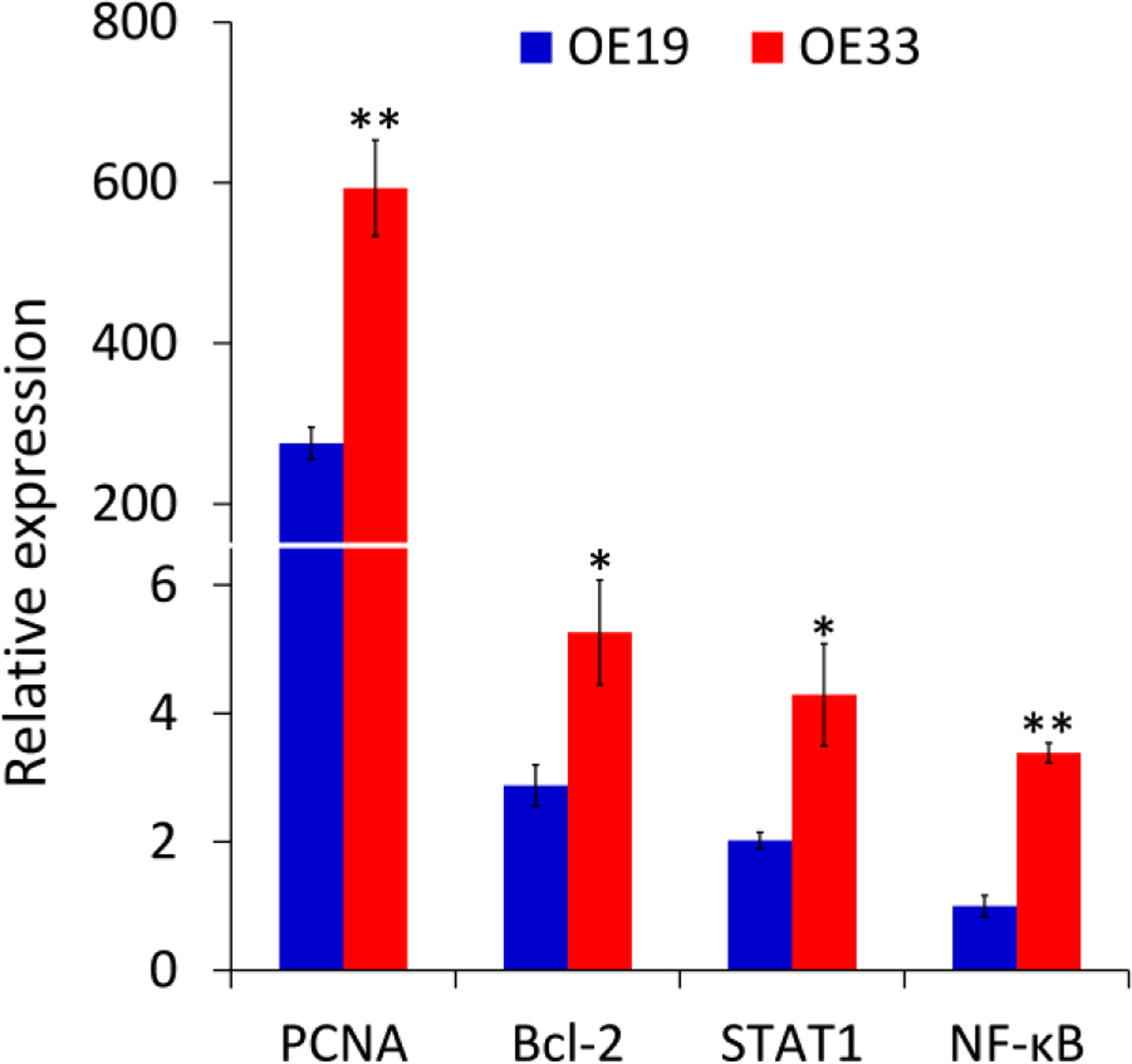
Expression of oncogenes in OE19 and OE33 cells measured by qRT-PCR. The vertical axis is relative expression level normalized against the expression level of NF-κB in OE19. All samples were run in triplicate with GAPDH used as a reference gene (*P < 0.05, **P < 0.01 by two-tailed t-test).
The measurements of live cultured esophageal adenocarcinoma cell lines with the coherent CLASS microscope were obtained in a miniature incubator (Bioscience Tools) at 37°C. An Olympus 60x 1.35 NA UPLSAPO oil immersion objective was used to obtain the best achievable resolution in a thin optical section (0.2 µm optical resolution in Z) inside the cells. Figure 4a shows the confocal reflectance images of four typical moderately differentiated cells (OE19, first row) and four typical poorly differentiated cells (OE33, second row). The two cell types are not readily distinguishable based on the reflectance image alone. We also measured the particle sizes from the confocal reflectance images by thresholding the image and performing connected-component labeling48. The confocal particle size was calculated based on the area of each of the identified particles or particle clusters (vertical axis in Figure 4b). This confocal particle/cluster size determination does not show any difference between the two cell types.
Figure 4.
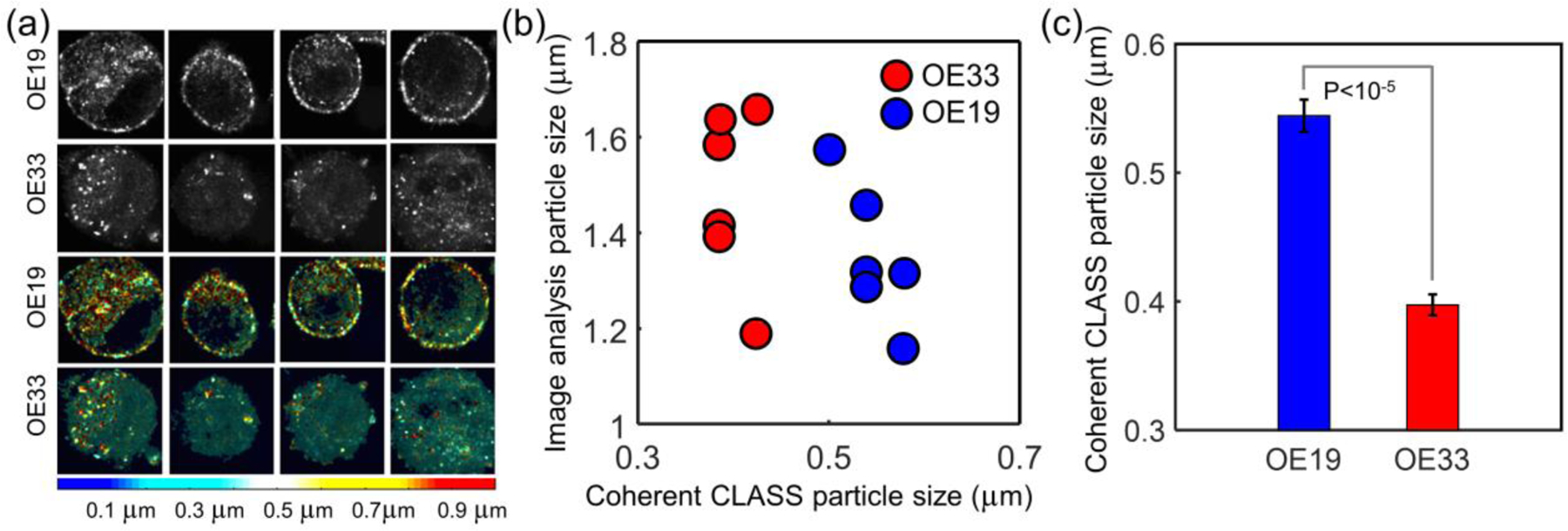
Coherent CLASS microscopy of live esophageal adenocarcinoma cancer cells. (a) Confocal reflectance images of four moderately differentiated cultured cells (OE19, first row) and four poorly differentiated cells (OE33, second row) and coherent CLASS microscopy images of the same cells (third and fourth rows, respectively) with color representing particle size (color bar). (b) Comparison of coherent CLASS microscope-derived particle size and average particle/cluster size obtained from confocal image analysis. (c) Average coherent CLASS microscope-derived particle sizes and standard errors (P < 0.00001, by two-tailed t-test). Scale bar equals 5 µm.
However, an overlay of the coherent CLASS microscope particle size reconstruction on the reflectance image (third and fourth rows of Figure 4a) readily shows that the poorly differentiated cells have a larger number of small size organelles (blue and cyan colors). The color bar at the bottom of Figure 4a represents the size in micrometers extracted from the coherent CLASS spectra. The coherent CLASS microscope reconstructed particle size (horizontal axis in Figure 4b) shows perfect separation between cells with a different degree of differentiation. The average organelle size for OE19 cells was 545 nm ± 12 nm while the average organelle size for OE33 cells was 398 nm ± 8 nm, respectively. This agrees well with the LSS measurements performed on the same cell lines (Supporting Information: LSS Measurements in Cancer Cell Lines and Figure S2). This difference in the average organelle sizes of the two cell types extracted from the coherent CLASS microscope images can be seen in Figure 4c and is highly significant (P < 0.00001, two-tailed t-test).
To summarize, the relative fraction of the smaller subcellular organelles in the more aggressive OE33 esophageal adenocarcinoma tumor cells is consistently larger in all six samples compared to less aggressive OE19 esophageal adenocarcinoma tumor cells (also six samples), while the respective average organelle size is smaller. Average organelle size, which is readily reconstructable from coherent CLASS microscopy data, can serve as a reliable and highly significant native marker of local tumor heterogeneity and metastatic potential.
In addition to the measurements in live cell cultures, we also investigated the potential of coherent CLASS microscopy to identify different grades of cancer in tissue by performing measurements in resected esophageal tumor tissue samples. Measurements were obtained on several well and moderately differentiated esophageal adenocarcinoma and squamous tumors, along with well and poorly differentiated gastric tumors (see Methods).
To correlate the coherent CLASS microscopy findings with pathology, confocal reflectance images of unstained tissue were obtained from the same area as the H&E histology images (Figure 5a,b). In turn, coherent CLASS data was obtained from the regions indicated with the red dashed squares in the confocal reflectance images (Figure 5c,d). Coherent CLASS data was processed in two ways: in Figure 5e,f the most prominent particle size contributing to the spectrum is mapped in color onto the reflectance image. These images indicate that the reflectance signal is predominantly associated with organelle sizes that are < 1 µm, an observation that is consistent with our fiber probe measurements. Figure 5g,h compare the relative concentration of small to large particles (less than 0.84 µm and between 1.5 µm and 4 µm). Well differentiated adenocarcinoma tissue (Figure 5g) exhibits a significantly higher proportion of small particles, as compared to moderately differentiated adenocarcinoma, again indicating an increase in size accompanied with cancer progression. These images clearly identify microscopic differences that correlate with the pathology of the tissue. This imaging method can be implemented in vivo endoscopically, thereby providing an opportunity to non-invasively monitor cancer tumors in terms of their cellular composition. Importantly, this confocal tissue imaging is achieved without the use of any contrast agents or dyes and can be rapidly performed over large tissue areas, as information about subcellular organelle sizes and nuclear sizes is spectrally encoded in the image (see for example Figure 5e–h).
Figure 5.
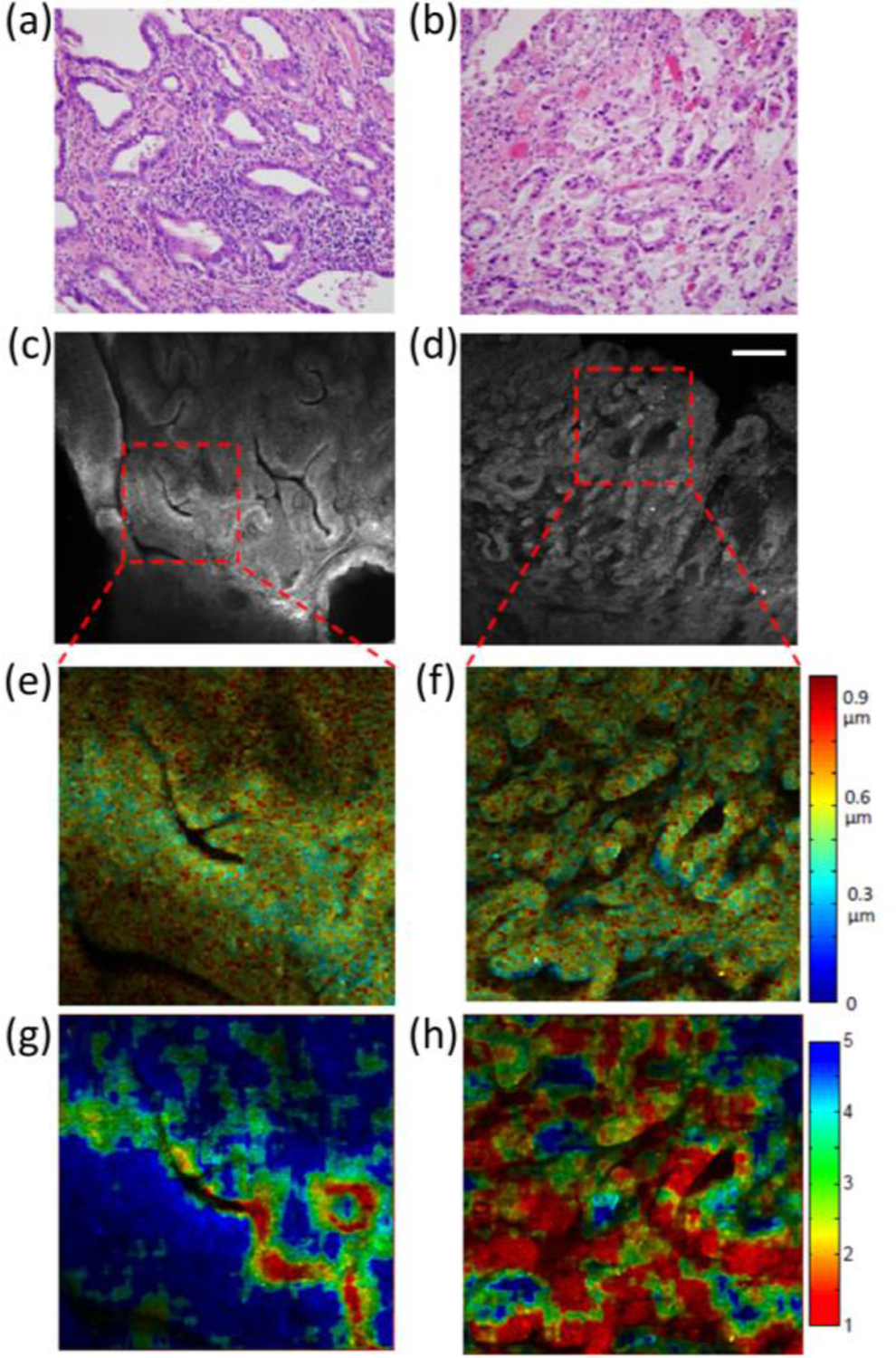
Coherent CLASS microscopy of esophageal adenocarcinoma tumor samples. The left column shows images from a well-differentiated tumor, while the right column shows images from a moderately differentiated tumor. (a, b) show H&E stained, 200x magnification histopathology images. (c, d) show confocal reflectance images of unstained tissue with the region of CLASS data collection indicated with the red dashed square. (e, f) show contributions of the main size, color-mapped over the grayscale reflectance images. (g, h) show the ratio of small (<0.84 µm) to intermediate (between 1.5 and 4 µm) sizes. Scale bar equals 200 µm.
Temporal Monitoring of Cancer Progression in Live Cells
To evaluate the ability of the coherent CLASS microscope imaging technique to characterize changes in live cells in transition from non-cancerous to early cancer via subcellular structure sizing, we studied the progression of non-dysplastic Barrett’s CP-A cells to cancer using the Nicotine-derived nitrosamine ketone (NNK) carcinogen exposure model33. NNK is a potent carcinogen found in tobacco, which is known to induce cancer in epithelial cell lines of most organs.
Cell culture was done at 37°C in a 5% CO2 atmosphere, as described in the Methods section, and the measurements on live cultured non-dysplastic Barrett’s CP-A cell lines were obtained in a miniature incubator, as described above. Five live cells were monitored through the NNK treatment by recording each cell’s position using a motorized microscope stage. After an initial (0 hour) measurement on the five cells, NNK was added to the media solution. The high resolution confocal reflectance images overlaid on the transmission images (Figure 6a) show that CP-A cells have a similar appearance before and right after the 24-hour carcinogen treatment.
Figure 6.
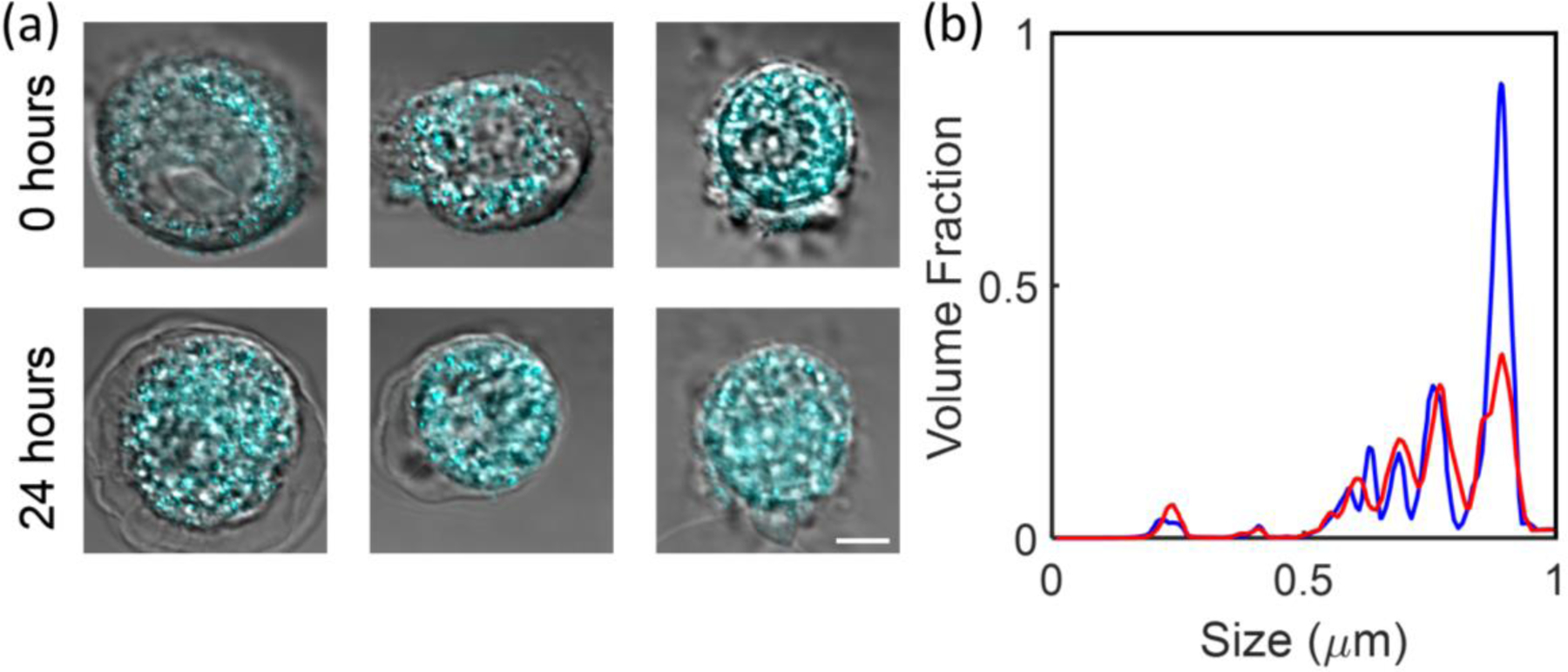
Coherent CLASS microscopy quantifies cancer progression in live cells. (a) High resolution confocal reflectance microscopy images (cyan) overlaid on the transmission images of three esophageal CP-A cells before treatment and the same three cells after 24-hour NNK carcinogen treatment. (b) Size distributions reconstructed from coherent CLASS microscope data, averaged over five cells before treatment (blue) and after 24-hour NNK carcinogen treatment (red). Scale bar equals 5 µm.
The coherent CLASS microscope allows reconstruction of the scatterer size map, which can be presented as a distribution of scatterer sizes before and after the treatment (Figure 6b). This distribution clearly shows a decrease of the 800 nm size particles and an increase in the 250 nm size particles. This shift in size distributions is concomitant with an approximately 30% increase in STAT1 expression level, which is indicative of cancer progression41. The observed increase in the relative fraction of the smaller subcellular organelles after 24-hour NNK carcinogen treatment is consistent with Figure 4c, where an increase in the relative fraction of smaller subcellular organelles was observed in the more aggressive adenocarcinoma tumor cells.
Discussion
Coherent CLASS microscopy reveals significant and highly repeatable differences in the scatterer size distributions in live cells, which clearly correlate with cancer progression and aggressiveness. These measurements, along with the measurements in tissue, suggest that light scattering spectroscopic imaging could serve as a reliable native marker of local tumor heterogeneity and metastatic potential. An important example of such tumors is esophageal cancers, where more than 50% are detected at metastatic stages, at which point they are no longer operable, and the five-year survival rate is a dismal 5%49. Historically, surgically removing the tumor containing section of the esophagus was the only curative treatment. Recently, several studies have demonstrated that combination therapies using chemotherapy drugs prior to surgical removal significantly improve survival rates50,51. These treatments rely on accurate assessments of the cancer type and likelihood of progression. Esophageal cancer, like most forms of cancer, is very diverse and can be decomposed into many types of diseases. A single esophageal cancer tumor is likely to be composed of a large variety of cancer cells, which continually evolve in response to their environment. Due to this highly diverse nature of the disease, there is a broad spectrum of responsiveness to treatment as well as patient prognosis. Presently, the prognosis is determined based on the overall condition of the patient and the stage of the disease. However, nearly all aspects of assessing the stage of the disease are prone to large errors. Here, we propose to develop a technology that can improve the accuracy of grading a cancer tumor. The grade of the cancer tumor is an important consideration in nearly all aspects of cancer patient care. At the same time, the technology could potentially be extended toward monitoring cancer treatment and determining the cancer phenotype non-invasively.
Presently, the grade of a tumor is determined through biopsies and histology. This assessment is prone to errors due to the large heterogeneity of the cancer tumor and the small size of the biopsy sample. Ideally, the measurement should be performed in vivo, preferably in real time, with the capacity to assess the entire tumor. This would allow the monitoring of patient treatments, and the ability to develop a personalized treatment approach. Additionally, in vivo tumor assessment offers the opportunity to measure the microenvironment of the tumor, which is known to influence tumor growth rate and could be taken into account when determining patient prognosis and treatment.
The capability to accurately evaluate tumor heterogeneity would yield far-reaching advances in planning and monitoring cancer therapies. The coherent CLASS microscopy presented here provides such a tool. The absence of stains in coherent CLASS microscopy makes time-course cancer progression studies simple to implement and would be compatible with in vivo human observations. In addition, the study of cellular structures in early pre-cancer can help clarify how and which cellular structures are altered at the earliest stages of carcinogenesis and lead to early prevention and detection targets. These changes should also be linked to known genetic markers to improve our understanding of cell function. For example, genetic instability is one of the initial characteristics of early pre-cancer and has been shown to be associated with gross cellular morphology52,53. Eventually, these markers can be used to predict cancer development54, while at the same time improving our understanding of how the disease arises.
In this paper we reported on a microscopy technique that utilizes spectroscopic properties of light scattering as both the source of morphological information and the intrinsic contrast in cells and biological tissue. Another important source of the intrinsic contrast in the microscopy of biological structures is their refractive index. Several microscopic techniques, such as optical diffraction tomography microscopy55–58 and optical coherence tomography microscopy employ refractive index to achieve label free high resolution imaging. Optical diffraction tomography microscopy is capable of quantitative evaluation of the spatial distribution of the refractive index in live cells56 or even in thick transparent tissue samples58 in the transillumination geometry. This makes it a very promising tool for studying individual unlabeled cells or unlabeled pathology samples, provided back reflection geometry typical for in vivo imaging in tissue is not required. Compared to conventional microscopy, optical coherence tomography microscopy, also often called optical coherence microscopy (OCM)59–62, provides improved imaging depth at high imaging speeds. In comparison, coherent CLASS microscopy is capable of complementing microscopic imaging information with functional morphological information, such as reconstructing the size distribution of the organelles and other scatterers in the confocal volume. This can be done despite the fact that these organelles and scatterers are significantly smaller than the wavelength and cannot be resolved due to the diffraction limit.
In conclusion, we have demonstrated that coherent CLASS microscopy allows continuous temporal tracking of the transition from a non-cancerous state to an early cancerous state in live cells, without using exogenous markers. We have also shown that the technique can detect differences in the aggressiveness of cancer in live cells, and can be used for label free identification of different grades of cancer in resected tumor tissues.
Supplementary Material
Acknowledgments
This work was supported by US National Science Foundation grants EFRI-1830878, CBET-1605116, and CBET-1948722 and US National Institutes of Health grants R01 CA228029, R01 EB003472, R01 CA205431, R01 EB025173, and R01 CA218382.
Footnotes
Associated content
Supporting Information
The Supporting Information is available free of charge.
Simulations of the incoherent and coherent CLASS microscopy spectra for a large NA objective, coherent CLASS microscopy spectra of light scattered by polystyrene microspheres, and LSS measurements in cancer cell lines.
The authors declare no competing financial interest.
References
- 1.Betzig E; Patterson GH; Sougrat R; Lindwasser OW; Olenych S; Bonifacino JS; Davidson MW; Lippincott-Schwartz J; Hess HF Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson MG Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell SW Far-field optical nanoscopy. Science 2007, 316, 1153–1158. [DOI] [PubMed] [Google Scholar]
- 4.Rust MJ; Bates M; Zhuang X Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Born M; Wolf E Principles of Optics: Electromagnetic Theory of Propagation, Interference and Diffraction of Light; Elsevier, 2013. [Google Scholar]
- 6.Fernández-Suárez M; Ting AY Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol 2008, 9, 929–943. [DOI] [PubMed] [Google Scholar]
- 7.Lukinavicius G; Umezawa K; Olivier N; Honigmann A; Yang G; Plass T; Mueller V; Reymond L; Correa IR Jr.; Luo ZG; Schultz C; Lemke EA; Heppenstall P; Eggeling C; Manley S; Johnsson K A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem 2013, 5, 132–139. [DOI] [PubMed] [Google Scholar]
- 8.Lukinavicius G; Reymond L; D’Este E; Masharina A; Gottfert F; Ta H; Guther A; Fournier M; Rizzo S; Waldmann H; Blaukopf C; Sommer C; Gerlich DW; Arndt HD; Hell SW; Johnsson K Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 2014, 11, 731–733. [DOI] [PubMed] [Google Scholar]
- 9.Grimm JB; English BP; Choi H; Muthusamy AK; Mehl BP; Dong P; Brown TA; Lippincott-Schwartz J; Liu Z; Lionnet T; Lavis LD Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods 2016, 13, 985–988. [DOI] [PubMed] [Google Scholar]
- 10.van de Linde S; Heilemann M; Sauer M Live-cell super-resolution imaging with synthetic fluorophores. Annu. Rev. Phys. Chem 2012, 63, 519–540. [DOI] [PubMed] [Google Scholar]
- 11.Schermelleh L; Ferrand A; Huser T; Eggeling C; Sauer M; Biehlmaier O; Drummen GPC Super-resolution microscopy demystified. Nat. Cell Biol 2019, 21, 72–84. [DOI] [PubMed] [Google Scholar]
- 12.Boustany NN; Boppart SA; Backman V Microscopic imaging and spectroscopy with scattered light. Annu. Rev. Biomed. Eng 2010, 12, 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu L; Pleskow DK; Chuttani R; Vitkin E; Leyden J; Ozden N; Itani S; Guo L; Sacks A; Goldsmith JD; Modell MD; Hanlon EB; Itzkan I; Perelman LT Multispectral scanning during endoscopy guides biopsy of dysplasia in Barrett’s esophagus. Nat. Med 2010, 16, 603–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovat LB; Johnson K; Mackenzie GD; Clark BR; Novelli MR; Davies S; O’Donovan M; Selvasekar C; Thorpe SM; Pickard D; Fitzgerald R; Fearn T; Bigio I; Bown SG Elastic scattering spectroscopy accurately detects high grade dysplasia and cancer in Barrett’s oesophagus. Gut 2006, 55, 1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L; Pleskow DK; Turzhitsky V; Yee EU; Berzin TM; Sawhney M; Shinagare S; Vitkin E; Zakharov Y; Khan U; Wang F; Goldsmith JD; Goldberg S; Chuttani R; Itzkan I; Qiu L; Perelman LT Light scattering spectroscopy identifies the malignant potential of pancreatic cysts during endoscopy. Nat. Biomed. Eng 2017, 1, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian H; Pradhan P; Liu Y; Capoglu IR; Rogers JD; Roy HK; Brand RE; Backman V Partial-wave microscopic spectroscopy detects subwavelength refractive index fluctuations: an application to cancer diagnosis. Opt. Lett 2009, 34, 518–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter CM; Pogue BW; Jiang S; Dehghani H; Wang X; Paulsen KD; Wells WA; Forero J; Kogel C; Weaver JB; Poplack SP; Kaufman PA Image-guided optical spectroscopy provides molecular-specific information in vivo: MRI-guided spectroscopy of breast cancer hemoglobin, water, and scatterer size. Opt. Lett 2007, 32, 933–935. [DOI] [PubMed] [Google Scholar]
- 18.Austwick MR; Clark B; Mosse CA; Johnson K; Chicken DW; Somasundaram SK; Calabro KW; Zhu Y; Falzon M; Kocjan G; Fearn T; Bown SG; Bigio IJ; Keshtgar MRS Scanning elastic scattering spectroscopy detects metastatic breast cancer in sentinel lymph nodes. J. Biomed. Opt 2010, 15, 047001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drezek RA; Richards-Kortum R; Brewer MA; Feld MS; Pitris C; Ferenczy A; Faupel ML; Follen M Optical imaging of the cervix. Cancer 2003, 98, 2015–2027. [DOI] [PubMed] [Google Scholar]
- 20.Sokolov K; Drezek R; Gossage K; Richards-Kortum R Reflectance spectroscopy with polarized light: is it sensitive to cellular and nuclear morphology. Opt. Express 19995, 302–17. [DOI] [PubMed] [Google Scholar]
- 21.Drezek R; Guillaud M; Collier T; Boiko I; Malpica A; Macaulay C; Follen M; Richards-Kortum R Light scattering from cervical cells throughout neoplastic progression: influence of nuclear morphology, DNA content, and chromatin texture. J. Biomed. Opt 2003, 8, 7–16. [DOI] [PubMed] [Google Scholar]
- 22.Roy HK; Liu Y; Wali RK; Kim YL; Kromine AK; Goldberg MJ; Backman V Four-dimensional elastic light-scattering fingerprints as preneoplastic markers in the rat model of colon carcinogenesis. Gastroenterology 2004, 126, 1071–1081. [DOI] [PubMed] [Google Scholar]
- 23.Bargo PR; Prahl SA; Goodell TT; Sleven RA; Koval G; Blair G; Jacques SL In vivo determination of optical properties of normal and tumor tissue with white light reflectance and an empirical light transport model during endoscopy. J. Biomed. Opt 2005, 10, 034018. [DOI] [PubMed] [Google Scholar]
- 24.Cerussi A; Hsiang D; Shah N; Mehta R; Durkin A; Butler J; Tromberg BJ Predicting response to breast cancer neoadjuvant chemotherapy using diffuse optical spectroscopy. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 4014–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalut KJ; Ostrander JH; Giacomelli MG; Wax A Light scattering measurements of subcellular structure provide noninvasive early detection of chemotherapy-induced apoptosis. Cancer Res 2009, 69, 1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itzkan I; Qiu L; Fang H; Zaman MM; Vitkin E; Ghiran IC; Salahuddin S; Modell M; Andersson C; Kimerer LM; Cipolloni PB; Lim K-H; Freedman SD; Bigio I; Sachs BP; Hanlon EB; Perelman LT Confocal light absorption and scattering spectroscopic microscopy monitors organelles in live cells with no exogenous labels. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 17255–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang H; Qiu L; Vitkin E; Zaman MM; Andersson C; Salahuddin S; Kimerer LM; Cipolloni PB; Modell MD; Turner BS; Keates SE; Bigio I; Itzkan I; Freedman SD; Bansil R; Hanlon EB; Perelman LT Confocal light absorption and scattering spectroscopic microscopy. Appl. Opt 2007, 46, 1760–1769. [DOI] [PubMed] [Google Scholar]
- 28.Huang P; Hunter M; Georgakoudi I Confocal light scattering spectroscopic imaging system for in situ tissue characterization. Appl. Opt 2009, 48, 2595–2599. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y; Li X; Kim YL; Backman V Elastic backscattering spectroscopic microscopy. Opt. Lett 2005, 30, 2445–2447. [DOI] [PubMed] [Google Scholar]
- 30.Pasternack RM; Qian Z; Zheng JY; Metaxas DN; White E; Boustany NN Measurement of subcellular texture by optical Gabor-like filtering with a digital micromirror device. Opt. Lett 2008, 33, 2209–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding H; Nguyen F; Boppart SA; Popescu G Optical properties of tissues quantified by Fourier-transform light scattering. Opt. Lett 2009, 34, 1372–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weise W; Zinin P; Wilson T; Briggs A; Boseck S Imaging of spheres with the confocal scanning optical microscope. Opt. Lett 1996, 21, 1800–1802. [DOI] [PubMed] [Google Scholar]
- 33.Arredondo J; Chernyavsky AI; Grando SA Nicotinic receptors mediate tumorigenic action of tobacco-derived nitrosamines on immortalized oral epithelial cells. Cancer Biol. Ther 2006, 5, 511–517. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W; van Weerden WM; de Ridder CMA; Erkens-Schulze S; Schönfeld E; Meijer TG; Kanaar R; van Gent DC; Nonnekens J Ex vivo treatment of prostate tumor tissue recapitulates in vivo therapy response. Prostate 2019, 79, 390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson GR; Wardell SE; Cakir M; Yip C; Ahn Y-R; Ali M; Yllanes AP; Chao CA; McDonnell DP; Wood KC Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat. Commun 2018, 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warburg O Origin of cancer cells. Science 1956, 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg-Lerner A; Benyair R; Hizkiahou N; Nudel N; Maor R; Kramer MP; Shmueli MD; Zigdon I; Lev MC; Ulman A; Sagiv JY; Dayan M; Dassa B; Rosenwald M; Shachar I; Li J; Wang Y; Dezorella N; Khan S; Porat Z; Shimoni E; Avinoam O; Merbl Y Golgi organization is regulated by proteasomal degradation. Nat. Commun 2020, 11, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W; Bao S PML-mediated signaling and its role in cancer stem cells. Oncogene 2014, 33, 1475–1484. [DOI] [PubMed] [Google Scholar]
- 39.Melo SA; Sugimoto H; O’Connell JT; Kato N; Villanueva A; Vidal A; Qiu L; Vitkin E; Perelman LT; Melo CA; Lucci A; Ivan C; Calin GA; Kalluri R Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fichter CD; Herz C; Munch C; Opitz OG; Werner M; Lassmann S Occurrence of multipolar mitoses and association with Aurora-A/-B kinases and p53 mutations in aneuploid esophageal carcinoma cells. BMC Cell Biol 2011, 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MA; Vanbibber MM; Safatle-Ribeiro AV; Ribeiro U Jr.; Reynolds JC Expression of p53, PCNA, and C-erbB-2 in Barrett’s metaplasia and adenocarcinoma. Dig. Dis. Sci 1997, 42, 2453–2462. [DOI] [PubMed] [Google Scholar]
- 42.Chen H; Wang LD; Guo M; Gao SG; Guo HQ; Fan ZM; Li JL Alterations of p53 and PCNA in cancer and adjacent tissues from concurrent carcinomas of the esophagus and gastric cardia in the same patient in Linzhou, a high incidence area for esophageal cancer in northern China. World J. Gastroenterol 2003, 9, 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCabe ML; Dlamini Z The molecular mechanisms of oesophageal cancer. Int. Immunopharmacol 2005, 5, 1113–1130. [DOI] [PubMed] [Google Scholar]
- 44.Cain JE; Di Giovanni V; Smeeton J; Rosenblum ND Genetics of renal hypoplasia: insights into the mechanisms controlling nephron endowment. Pediat. Res 2010, 68, 91–98. [DOI] [PubMed] [Google Scholar]
- 45.Dolcet X; Llobet D; Pallares J; Matias-Guiu X NF-kB in development and progression of human cancer. Virchows Arch 2005, 446, 475–482. [DOI] [PubMed] [Google Scholar]
- 46.Khodarev N; Ahmad R; Rajabi H; Pitroda S; Kufe T; McClary C; Joshi MD; MacDermed D; Weichselbaum R; Kufe D Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene 2010, 29, 920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hix LM; Karavitis J; Khan MW; Shi YH; Khazaie K; Zhang M Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. J. Biol. Chem 2013, 288, 11676–11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro LG; Stockman GC Computer vision; Pearson Education, Inc., 2001. [Google Scholar]
- 49.Enzinger PC; Mayer RJ Esophageal cancer. N. Engl. J. Med 2003, 349, 2241–2252. [DOI] [PubMed] [Google Scholar]
- 50.Kleinberg L; Gibson MK; Forastiere AA Chemoradiotherapy for localized esophageal cancer: regimen selection and molecular mechanisms of radiosensitization. Nat. Clin. Pract. Oncol 2007, 4, 282–294. [DOI] [PubMed] [Google Scholar]
- 51.Allum WH; Bonavina L; Cassivi SD; Cuesta MA; Dong ZM; Felix VN; Figueredo E; Gatenby PA; Haverkamp L; Ibraev MA; Krasna MJ; Lambert R; Langer R; Lewis MP; Nason KS; Parry K; Preston SR; Ruurda JP; Schaheen LW; Tatum RP; Turkin IN; van der Horst S; van der Peet DL; van der Sluis PC; van Hillegersberg R; Wormald JC; Wu PC; Zonderhuis BM Surgical treatments for esophageal cancers. Ann. N. Y. Acad. Sci 2014, 1325, 242–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung PP; Fritsch ES; Blugeon C; Souciet J-L; Potier S; Lemoine S; Schacherer J; de Montigny J Ploidy influences cellular responses to gross chromosomal rearrangements in Saccharomyces cerevisiae. BMC Genomics 2011, 12, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel JJ; Amon A New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol 2012, 28, 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maley CC; Galipeau PC; Finley JC; Wongsurawat VJ; Li X; Sanchez CA; Paulson TG; Blount PL; Risques R-A; Rabinovitch PS; Reid BJ Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet 2006, 38, 468–473. [DOI] [PubMed] [Google Scholar]
- 55.Lauer V New approach to optical diffraction tomography yielding a vector equation of diffraction tomography and a novel tomographic microscope. J. Microsc 2002, 205, 165–176. [DOI] [PubMed] [Google Scholar]
- 56.Kim T; Zhou R; Mir M; Babacan SD; Carney PS; Goddard LL; Popescu G White-light diffraction tomography of unlabelled live cells. Nat. Photonics 2014, 8, 256–263. [Google Scholar]
- 57.Horstmeyer R; Chung J; Ou X; Zheng G; Yang C Diffraction tomography with Fourier ptychography. Optica 2016, 3, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cotte Y; Toy F; Jourdain P; Pavillon N; Boss D; Magistretti P; Marquet P; Depeursinge C Marker-free phase nanoscopy. Nat. Photonics 2013, 7, 113–117. [Google Scholar]
- 59.Izatt JA; Hee MR; Owen GM; Swanson EA; Fujimoto JG Optical coherence microscopy in scattering media. Opt. Lett 1994, 19, 590–592. [DOI] [PubMed] [Google Scholar]
- 60.Beaurepaire E; Boccara AC; Lebec M; Blanchot L; Saint-Jalmes H 1998. Full-field optical coherence microscopy. Opt. Lett 1998, 23, 244–246. [DOI] [PubMed] [Google Scholar]
- 61.Žurauskas M; Iyer RR; Boppart SA Simultaneous 4-phase-shifted full-field optical coherence microscopy. Biomed. Opt. Express 2021, 12, 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh WY; Bouma BE; Iftimia N; Yun SH; Yelin R; Tearney GJ Ultrahigh-resolution full-field optical coherence microscopy using InGaAs camera. Opt. Express 2006, 14, 726–735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


