Abstract
The plant hormone auxin governs many aspects of normal plant growth and development. Auxin also plays an important role in plant–microbe interactions, including interactions between plant hosts and pathogenic microorganisms that cause disease. It is now well established that indole-3-acetic acid (IAA), the most well-studied form of auxin, promotes disease in many plant–pathogen interactions. Recent studies have shown that IAA can act both as a plant hormone that modulates host signaling and physiology to increase host susceptibility and as a microbial signal that directly impacts the pathogen to promote virulence, but large gaps in our understanding remain. In this article, we review recent studies on the roles that auxin plays during plant–pathogen interactions and discuss the virulence mechanisms that many plant pathogens have evolved to manipulate host auxin signaling and promote pathogenesis.
To colonize their plant hosts, pathogenic microbes must enter plant tissue, evade or suppress basal defense, obtain nutrients and water, and grow to high levels. In many cases, this ultimately results in the development of disease symptoms, such as abnormal plant growth or development and/or generation of chlorotic or necrotic lesions. Plant pathogens have evolved a variety of virulence strategies that allow them to achieve these ends. One common theme that has emerged from studies of plant–pathogen interactions is that pathogens often manipulate plant hormone signaling to promote disease, especially to suppress host defenses or to alter plant development.
The roles of plant hormone signaling in plant–pathogen interactions have been discussed in several recent reviews (Robert-Seilaniantz et al. 2011; Kazan and Lyons 2014; Ma and Ma 2016; Bürger and Chory 2019; Han and Kahmann 2019), so here we focus on summarizing studies that provide new insight into the roles that auxin and host auxin responses play during plant–pathogen interactions. One discovery that we find especially exciting is that, in addition to functioning as a plant hormone to regulate host physiology and defense, auxin can also act as a microbial signaling molecule that modulates pathogen gene expression.
Because of space limitations, we are not able to discuss the demonstrated and potential roles of auxins in beneficial interactions with mycorrhizal fungi, nodulating bacteria, or plant growth–promoting rhizobacteria (PGPR), nor in the establishment and function of the plant microbiome. We refer readers interested in these areas to reviews by others (Grunewald et al. 2009; Singh et al. 2019; Legein et al. 2020).
OVERVIEW OF PLANT–PATHOGEN INTERACTIONS
Many different microorganisms, including viruses, bacteria, fungi, oomycetes, and nematodes, can infect and cause disease on plants. Collectively, pathogenic organisms can colonize all plant tissues (e.g., leaves, fruits, seeds, vascular tissue, and roots) and give rise to a variety of different disease symptoms, such as chlorotic or necrotic lesions on aerial portions of the plant, wilting, altered growth habits (e.g., stunting or loss of apical dominance), and formation of tumor-like growths (e.g., galls, knots, and cankers) (Agrios 1997). The majority of pathogens, with the exception of viruses and vascular pathogens, colonize the intercellular space between plant cells. This space, known as the apoplast, is believed to be a relatively inhospitable environment for microbes, as it is relatively poor in water and nutrients (Beattie 2011; Xin et al. 2018). The apoplast also contains antimicrobial compounds that are either constitutively present or that are secreted into the extracellular space as part of an induced defense response (Heath 2000; Wang et al. 2007). Thus, to successfully colonize and grow to high levels within the apoplast, pathogens must be able to tolerate or detoxify these antimicrobial compounds, evade or suppress any additional host defenses, and further modify host tissue to render it a suitable place for pathogen growth.
Plant Defense Responses
Plants constantly come into contact with many different microorganisms in their environment, many of them potential pathogens. Despite this fact, it is important to note that plant disease is rare. This is due in part to the ability of plants to detect microbes in their proximity and induce basal defense responses that prevent most microbes from colonizing plant tissue. Recognition of potential pathogens is dependent upon plasma membrane–localized receptors, known as pattern recognition receptors (PRRs). These receptors have evolved to recognize conserved molecules commonly associated with microbes (microbial-associated molecular patterns [MAMPs]), such as flagellin peptide or extracellular polysaccharides (EPS), or molecules associated with tissue damage caused by microbial attack (DAMPs). Recognition of one or more of these molecules results in activation of defense responses, including production of reactive oxygen species (ROS), activation of mitogen-activated protein (MAP) kinase cascades, expression of defense-related genes, synthesis of antimicrobial compounds, and the accumulation of one or more defense hormones (Boller and Felix 2009; Bigeard et al. 2015; Boutrot and Zipfel 2017; Saijo et al. 2018).
The plant defense hormones salicylic acid (SA), jasmonates (JAs), and ethylene play central roles in protection against microbial attack. SA is important for defense against biotrophic and hemibiotrophic pathogens that infect living tissue. In contrast, JAs and ethylene activate defense responses that provide protection against necrotrophic organisms that rapidly kill plant cells and grow on dying or dead tissue. The regulation of plant defenses by these three hormones is not as simple as the previous statement may imply. In fact, the regulation of plant defense hormone signaling is complicated and involves intricate regulatory interactions in a complex signaling network (Katagiri and Tsuda 2010). Further, additional plant hormones including auxin, abscisic acid (ABA), gibberellins (GAs), cytokinins, and brassinolides (BRs) also play roles in plant–pathogen interactions, for example, by modulating the SA/JA/ethylene signaling network (Robert-Seilaniantz et al. 2011; Kazan and Lyons 2014; Kunkel and Harper 2018; Bürger and Chory 2019). A fascinating area of research is the investigation of mechanisms pathogens have evolved to take advantage of this intrinsic hormone-signaling network to promote disease. A major focus of this review is how pathogens manipulate auxin signaling.
Pathogen Virulence Strategies
Given that plants are very effective at protecting themselves against generic environmental microbes, pathogenic organisms have had to evolve strategies and virulence factors to enable them to evade or suppress basal host defenses to colonize plant tissue. Once that is accomplished, they further modify the physiology of their hosts to render the plant tissue amenable for pathogen growth, such as stimulating water and nutrient release into the apoplastic space at the infection site. In the case of gall-forming pathogens such as Agrobacterium tumefaciens or cyst nematodes, this includes coercing the plant to form novel structures to house and feed the pathogen.
As mentioned above, many plant pathogens are extracellular and colonize the apoplast. Thus, to successfully modify the biology of plant cells at the infection site, these pathogens rely on secreted virulence factors. These secreted factors either remain in the apoplast and impact plant cells from outside or enter the plant cell cytosol to directly target host cell processes. Examples of apoplastic factors include cell wall degrading enzymes and EPS. Virulence factors that function inside plant cells include plant hormones and hormone analogs and fungal and oomycete effector proteins that can be translocated into plant cells, as well as bacterial effector proteins that are secreted directly into the host cell cytosol via a dedicated type III secretion system (T3SS) (Melotto and Kunkel 2013; Kunkel and Harper 2018; Han and Kahmann 2019). Pathogen effector proteins have been the focus of much exciting research in recent years, and several have been shown to target plant defense hormone signaling, and to promote disease susceptibility and symptom development. Several recent reviews provide more comprehensive summaries of type III secreted and RXLR effectors and their functions (Büttner 2016; Toruño et al. 2016; Kunkel and Harper 2018; Xin et al. 2018; Han and Kahmann 2019; Boevink et al. 2020).
As will be discussed in more detail below, many pathogens manipulate host auxin signaling to promote disease. Strategies to accomplish this include production of auxins, including indole-3-acetic acid (IAA) and phenyl acetic acid (PAA), inhibition of auxin signaling in interactions in which auxin contributes to resistance, and enhancing auxin signaling to promote pathogenesis in interactions in which auxin increases susceptibility (Ma and Ma 2016; Han and Kahmann 2019).
A BRIEF INTRODUCTION TO AUXIN SIGNALING
As the role of host auxin responses during plant–pathogen interactions is a main focus of this review, we provide a brief introduction to plant auxin signaling pathways. We refer the reader to the literature cited within for more details on this fascinating area of plant biology.
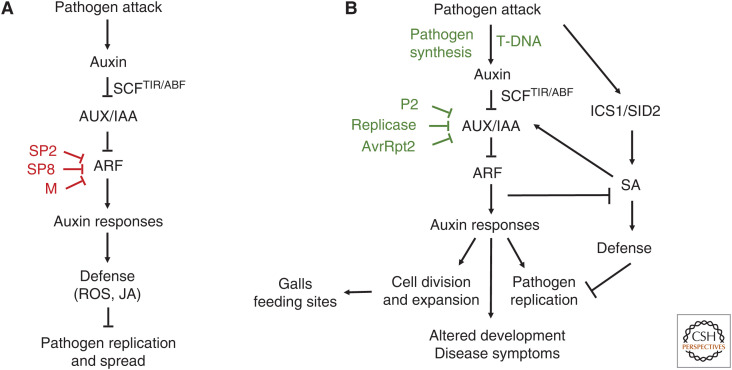
Auxin induces transcriptional changes in plant cells by promoting ubiquitin-mediated degradation of a family of AUX/IAA transcriptional repressor proteins. In the absence of auxin, the AUX/IAA proteins bind to and inhibit the function of transcription factors called auxin response factors (ARFs) to repress expression of auxin-responsive genes. When auxin levels increase in the cell, auxin binds to auxin sensor proteins (also called auxin coreceptors) belonging to the TRANSPORT INHIBITOR RESPONSE1/AUXIN F-BOX (TIR1/AFB) family of F-box proteins and promotes the interaction between TIR1/AFB proteins and AUX/IAA proteins. This results in the recruitment of the AUX/IAA proteins to the SCFTir ubiquitin protein ligase, ubiquitination, and degradation of the transcriptional repressors, activation of the ARFs, and expression of auxin-responsive genes (Fig. 1; Mockaitis and Estelle 2008; Lavy and Estelle 2016; Dubey et al. 2021).
Figure 1.
Host auxin signaling during plant–pathogen interactions. (A) Infection by some necrotrophic pathogens, such as Alternaria and Rhizoctonia, and some plant viruses stimulates auxin signaling, resulting in activation of defenses that inhibit pathogen replication and spread. The SP2 protein from rice stripe virus, the SP8 protein from Southern rice black-streaked dwarf virus, and the M protein from rice stripe mosaic virus interact with different regions of OsARF17 and modulate its function, thereby inhibiting activation of auxin-dependent defenses (Zhang et al. 2019, 2020). (B) Biotrophic pathogens, such as Pseudomonas spp., Agrobacterium tumefaciens, and Pantoea agglomerans, stimulate auxin signaling to promote disease. Stimulation of auxin signaling can occur at many points in the signaling process including auxin synthesis by the pathogen (Kunkel and Harper 2018). Enhanced auxin signaling can promote production of disease symptoms, development of galls and feeding sites, and/or suppress salicylic acid (SA)-mediated defenses. SA-mediated defense signaling and auxin signaling are mutually antagonistic (Wang et al. 2007; Cui et al. 2013). Several pathogens have evolved virulence factors to promote auxin signaling. The Pseudomonas syringae T3S-effector protein AvrRpt2 enhances auxin sensitivity by promoting degradation of AUX/IAA proteins (Cui et al. 2013). The 126/183 kDa replicase protein from tobacco mosaic virus inhibits PAP1/IAA16 function (Padmanabhan et al. 2006), and the P2 protein from rice dwarf virus targets OsIAA10 and prevents its degradation (Jin et al. 2016). Virulence factors shown in red inhibit auxin signaling. Virulence factors shown in green stimulate auxin responses. (SCF) Skp, Cullin, F-box ubiquitin protein ligase complex, (TIR/AFB) TRANSPORT INHIBITOR RESPONSE1/AUXIN F-BOX, F-box proteins (auxin coreceptor), (AUX/IAA) AUXIN/INDOLE ACETIC ACID (transcriptional repressors), (ARF) AUXIN RESPONSE FACTOR (transcription factor), (ROS) reactive oxygen species, (JAs) jasmonates, (ICS1/SID2) isochorismate synthase (SA synthesis), (SA) salicylic acid.
Investigation of the roles of auxin signaling during plant growth, development, and responses to environmental stimuli has been greatly facilitated by the availability of a variety of Arabidopsis thaliana mutants and transgenic lines that are impaired in various aspects of auxin signaling. For example, plants carrying the dominant axr2-1 allele, which encodes a mutant form of an AUX/IAA protein that is not ubiquitinated and degraded upon auxin treatment, can be used to examine the impact of impaired auxin responses on the process of interest (Timpte et al. 1994; Djami-Tchatchou et al. 2020). Likewise, a variety of higher-order mutants disrupted for various combinations of the TIR1/AFB auxin coreceptors can be used to examine the effect of disrupted auxin perception (Djami-Tchatchou et al. 2020). Several studies mentioned in this review take advantage of these types of genetic resources to investigate the roles of auxin signaling during plant–pathogen interactions.
THE ROLES OF AUXIN AS A PLANT-SIGNALING MOLECULE IN PLANT–PATHOGEN INTERACTIONS
The role of host auxin signaling during pathogen infection, and whether it promotes disease or defense, depends on the pathogen and the nature of the interaction. For example, activation of auxin signaling enhances resistance against some necrotrophic pathogens and plant viruses (Fig. 1A). In contrast, auxin signaling tends to promote disease in plants challenged with biotrophic or hemibiotrophic pathogens (Fig. 1B; Kazan and Manners 2009; Robert-Seilaniantz et al. 2011; Kazan and Lyons 2014; Ma and Ma 2016; Kunkel and Harper 2018; Bürger and Chory 2019).
Auxin Signaling and Promotion of Plant Defense
As mentioned above, the primary defense hormones are SA, JAs, and ethylene (ET). Thus, there are only a few reports in the literature of auxin and auxin signaling playing a role in activation of plant defense responses. For example, in interactions involving necrotrophic fungi (e.g., Botrytis, Rhizoctonia, and Alternaria), infection can result in elevated IAA levels, and application of exogenous auxin results in reduced disease susceptibility (Llorente et al. 2008; Mah et al. 2012; Qi et al. 2012; Qiao et al. 2020). Consistent with a role for auxin in defense in these interactions, disruption of auxin signaling due to mutations that stabilize AUX/IAA transcriptional repressors (e.g., axr mutants) or overexpression of small RNAs that negatively regulate expression of TIR1/AFB family auxin receptors, results in enhanced disease susceptibility (Llorente et al. 2008; Qi et al. 2012; Qiao et al. 2020). In contrast, overexpression of the TIR1/AFB family auxin coreceptor FBL55 in rice resulted in enhanced resistance to Rhizoctonia solani (Qiao et al. 2020). In most of the above studies, the mechanism through which auxin signaling results in increased resistance was not investigated. It seems reasonable to speculate that in many cases auxin signaling leads to activation of JA-mediated defenses, which are protective against necrotrophs (Fu and Wang 2011; Qi et al. 2012; Zhang et al. 2017). The observation that auxin signaling activates defense in these interactions raises the question of whether necrotrophic pathogens have evolved strategies to perturb auxin signaling to promote susceptibility, similar to what has been reported for their ability to interfere with JA signaling (Kazan and Lyons 2014; Zhang et al. 2017).
We could find evidence of only a few interactions in which resistance to infection by plant viruses is associated with activation of auxin-mediated defense signaling. In two recent studies, Zhang et al. (2019, 2020) discovered that several viral proteins can interfere with host auxin signaling by interacting with plant ARFs (Fig. 1A). The SP8 protein from Southern rice black-streaked dwarf virus (SRBSDV) is a transcriptional repressor that can interact with and inhibit the carboxy-terminal dimerization domain (CTD) of OsARF17, an activator ARF. Similarly, the M protein from rice stripe mosaic virus (RSMV) and the SP2 protein from rice stripe virus (RSV) interact with and inhibit OsARF17. Like SP8, the RSMV M protein also interacts with the CTD domain. However, the RSV SP2 protein interacts with the DNA-binding domain of OsARF17 and interferes with its ability to bind DNA (Zhang et al. 2020). Thus, three different viruses have evolved strategies to interfere with host auxin signaling, presumably to promote virulence, by targeting different functions of the same host-signaling component. It is interesting to contemplate what might make OsARF17 such a popular target. A question for the future is whether this particular activator ARF has a critical function in auxin-mediated defenses against viruses and possibly also against other pathogens.
Consistent with the hypothesis that auxin signaling promotes antiviral defense, studies involving both overexpression lines and CRISPR/cas9 mutants demonstrated that auxin signaling plays a critical role in defense against both SRBSDV and the related RBSDV (Zhang et al. 2019, 2020). These studies also showed that increased susceptibility in transgenic rice lines with reduced auxin signaling was correlated with a reduction in JA signaling and ROS in infected plants, suggesting that auxin signaling defends against viruses through activation of JA-dependent defenses and accumulation of ROS (Zhang et al. 2019).
Auxin Signaling and Promotion of Disease Susceptibility
There are many examples of host auxin signaling enhancing disease susceptibility. For example, application of exogenous auxins promote susceptibility to a number of biotrophic and hemibiotrophic pathogens, including the bacterial pathogens Pseudomonas syringae and Xanthomonas oryzae (Navarro et al. 2006; Chen et al. 2007; Wang et al. 2007; Fu et al. 2011), the fungal pathogen Magnaporthe grisea (Fu et al. 2011), and the oomycete Phytophthora parasitica (Evangelisti et al. 2013). Additionally, A. thaliana lines with elevated levels of free IAA exhibit increased susceptibility to P. syringae (Mutka et al. 2013; Djami-Tchatchou et al. 2020) and plant lines with impaired auxin perception or signaling (e.g., axr2/3 or tir1/afb mutants) exhibit reduced susceptibility to several biotrophs and hemibiotrophs (Wang et al. 2007; Kidd et al. 2011; Fousia et al. 2018; French et al. 2018; Djami-Tchatchou et al. 2020).
Perhaps the most compelling evidence that auxin signaling promotes disease is the growing number of reports of pathogens that stimulate host auxin signaling to promote pathogenesis and disease development. These microbes use a variety of strategies, including synthesis of auxin (Patten et al. 2013; Duca et al. 2014; Kunkel and Harper 2018) and deployment of pathogen virulence proteins that act within host cells to enhance host auxin signaling (Chen et al. 2007; Kazan and Lyons 2014; Ludwig-Müller 2015; Ma and Ma 2016).
There are several mechanisms by which host auxin signaling promotes disease, and they fall into two general categories: (1) manipulation of host auxin biology (e.g., signaling, homeostasis, and/or transport) to directly promote disease development, and (2) suppression of host defenses by taking advantage of regulatory “cross talk” inherent in hormone signaling networks (Fig. 1B; Robert-Seilaniantz et al. 2011; Kazan and Lyons 2014; Ma and Ma 2016; Kunkel and Harper 2018). Some examples from the first category have been observed in viral pathogens (Fig. 1B). Infection by plant viruses often results in changes in hormone homeostasis that results in disease symptoms, including severe stunting, altered shoot architecture, and tissue chlorosis. These symptoms are commonly reflected in the names of the viruses, such as rice dwarf virus (RDV) and tobacco mosaic virus (TMV). Several viral pathogens have evolved mechanisms to perturb or reprogram auxin signaling as a strategy to promote disease development (Islam et al. 2019). For example, the TMV 126/183 kDa replicase protein interacts with several AUX/IAA proteins to prevent their localization to the nucleus (Padmanabhan et al. 2006), leading to changes in auxin-responsive gene expression and development of disease symptoms (Padmanabhan et al. 2005). Likewise, RDV modifies host auxin signaling through the action of its P2 protein, which interacts with the region II degron domain of the rice OsIAA10 transcriptional repressor. This interferes with the normal interaction between OsIAA10 and OsTIR1 and prevents degradation of OsIAA10 upon auxin binding. Consistent with a role for OsIAA10 in RDV infection, transgenic rice plants expressing a stabilized OsIAA10 variant exhibited phenotypes reminiscent of RDV-infected plants, even in the absence of virus, and plants expressing reduced levels of OsIAA10 were less susceptible to RDV infection (Jin et al. 2016).
The virulence strategies of gall-forming pathogens such as A. tumefaciens, Pseudomonas savastanoi, Pantoea agglomerans (formerly Erwinia herbicola), and parasitic nematodes involve manipulation of host auxin physiology to directly stimulate abnormal plant cell division and expansion, to form tumor-like structures to shelter and feed the pathogen (Fig. 1B). In these interactions, the pathogens either make auxin themselves (P. savastanoi, P. agglomerans) (Patten et al. 2013; Duca et al. 2014), stimulate plant cells at the site of infection to synthesize auxin (A. tumefaciens) (Thomashow et al. 1986; Mashiguchi et al. 2019), or redirect auxin flow within the plant so that auxin accumulates at the feeding site and stimulates cell division or syncytium development (nematodes) (Grunewald et al. 2009; Kyndt et al. 2016). Presumably, nutrient flow into the infected host tissue is also altered to support the infection site, but this is not well understood.
The discovery that auxin can also promote disease caused by biotrophic pathogens such as P. syringae, that do not result in dramatic changes in plant growth or development, was surprising at first. Insight into the mode of auxin action in these interactions was provided by the observation that auxin signaling and SA signaling can be mutually antagonistic (Fig. 1B; Wang et al. 2007). Additional evidence for cross talk between auxin and SA signaling has come from more recent studies demonstrating that the virulence-promoting effect of enhanced auxin signaling involves down-regulation of SA-mediated defenses (McClerklin et al. 2018; Djami-Tchatchou et al. 2020). Further, the observation that A. thaliana auxin signaling mutants express elevated SA-mediated defenses and that the reduced disease susceptibility phenotype of these mutant plants can be genetically rescued by alterations in SA synthesis or accumulation further support the hypothesis that auxin promotes disease by suppressing SA-mediated defenses (Wang et al. 2007; Fousia et al. 2018; Djami-Tchatchou et al. 2020). However, as will be discussed below, there is growing evidence that auxin can also promote disease susceptibility via one or more additional SA-independent mechanism (Chen et al. 2004; Kidd et al. 2011; Mutka et al. 2013; Djami-Tchatchou et al. 2020).
AUXIN AS A MICROBIAL SIGNAL
In addition to promoting pathogenesis by manipulating host auxin signaling, auxin may act directly to promote virulence of several pathogens. There is growing evidence that IAA acts as a molecular signal, both in plant-associated and non-plant-associated microbes, to regulate gene expression (Spaepen and Vanderleyden 2011; Kunkel and Harper 2018). However, the majority of what is known to date about the mechanisms used by microorganisms to sense and respond to IAA comes from studies of non-plant-associated bacteria.
IAA Regulates Genes Involved in Stress Tolerance, Metabolism, and Antibiotic Production
There are several examples in which IAA dramatically alters gene expression in non-plant-associated bacteria. For instance, in Escherichia coli, IAA regulates genes involved in central metabolism, which is believed to be why IAA-treated E. coli cells exhibited increased tolerance to several environmental stresses, including osmotic, heat and cold shock, and oxidative stress (Bianco et al. 2006a,b). Likewise, Donati et al. observed induction of genes involved in stress responses in Bradyrhizobium japonicum in response to IAA (Donati et al. 2013).
Several bacteria also have the ability to use IAA as a carbon source. In many of these organisms, IAA acts as a signal to up-regulate IAA catabolism genes, which are encoded in an IAA catabolic (iac or iaa) operon responsible for aerobic breakdown of IAA (Laird et al. 2020). This operon has been identified in several diverse bacterial species, including Pseudomonas putida, Enterobacter soli, Acinetobacter baumannii, Paraburkholderia phytofirmans, Caballeronia glathei, Aromatoleum evansii, and Aromatoleum aromaticum (Laird et al. 2020).
In A. baumannii, regulation of the iac operon by IAA is conferred by IacR, a MarR-type transcriptional regulator that negatively regulates the iac operon. In the absence of IAA, IacR binds the promoter of the iac operon. IAA is hypothesized to bind to IacR and release it from the iac promoter, thereby allowing RNA polymerase access to the promoter to initiate transcription (Shu et al. 2015). Similarly, in E. soli, IAA derepresses the iac operon via a MarR-type transcriptional repressor (Greenhut et al. 2018). Donoso et al. also identified an iac gene cluster in the PGPR P. phytofirmans strain PsJN. The iac operon in this strain has a different gene organization and lacks the iacR MarR family repressor. Instead, the PsJN genome carries other genes adjacent to the iac operon, including genes encoding a LysR-type regulator and a putative two-component signal transduction system, composed of iacS and iacR1. Expression of these regulatory genes is induced by IAA and is required for IAA-induced transcription from the iac promoter (Donoso et al. 2017). This is the only sensory system that has been identified in bacteria in which IAA does not directly target a transcription factor and instead may initiate a large-scale physiological response downstream of a two-component system. It will be interesting to learn whether other microbes use a similar system to sense and respond to IAA.
Adapting to a New Environment
An additional example of auxin-regulated gene expression has been observed in the soil bacterium Serratia plymuthica (Matilla et al. 2018), which produces a wide range of antibiotics including andrimid. Production of andrimid is regulated by AdmX, a transcriptional activator with an amino-terminal DNA-binding domain and a LysR-type ligand-binding domain. AdmX has been shown to bind IAA, which may change its conformation. The proposed change in conformation upon IAA binding is thought to reduce AdmX's ability to bind DNA, thereby disabling its transcriptional activation function. Thus, IAA lowers andrimid production in this strain. The biological relevance of reducing andrimid production is unclear, but one possibility is that this results in a physiological or metabolic change that enables S. plymuthica to acclimate to a new environment.
In several plant-associated bacteria, IAA may act as a signal of a new environment. Two examples of bacteria that show dramatic changes in gene expression in response to IAA are Azospirillum brasilense (Vande Broek et al. 2005; Van Puyvelde et al. 2011) and Rhizobium etli (Spaepen et al. 2009). In A. brasilense, IAA activates a positive feedback loop in which IAA biosynthesis genes are up-regulated (Vande Broek et al. 2005). Van Puyvelde et al. (2011) then showed extensive transcriptional responses to IAA by analyzing gene-expression patterns in auxin biosynthesis mutants. Interestingly, in wild-type A. brasilense, exposure to IAA altered expression of genes encoding transport proteins, cell surface proteins, and proteins involved in type VI secretion. These observations support the hypothesis that IAA may stimulate bacteria to alter gene expression to increase their competitiveness in the rhizosphere. Spaepen et al. observed that in R. etli, genes involved in motility and attachment are differentially regulated by IAA. Because these genes are believed to be important for symbiosis with plants, the authors hypothesized that IAA plays a role in promoting R. etli plant interactions (Spaepen et al. 2009).
Bacterial plant pathogens have also evolved the ability to use IAA as a signal to modulate gene expression and virulence mechanisms. One well-known pathogen that relies on its own ability to alter host auxin biology is A. tumefaciens, the causal agent of crown gall disease. This pathogen expresses virulence (vir) genes during pathogenesis that are ultimately responsible for genetic transformation of plant cells so that they will abnormally produce auxins and cytokinins and stimulate the formation of galls at the site of infection (Thomashow et al. 1986; Mashiguchi et al. 2019). After sufficient accumulation of IAA in the infected tissue and thus pathogen vir genes are presumably no longer required, the high level of IAA inhibits their expression (Liu and Nester 2006). A large number of additional non-vir genes are also regulated by IAA, suggesting IAA may also regulate processes important for bacterial persistence after gall formation (Yuan et al. 2008).
Plant pathogens in the Pseudomonas genus also respond to IAA by altering gene expression. Experiments to investigate the possibility that IAA regulates virulence gene expression in P. syringae strain PtoDC3000 were prompted by studies suggesting that auxin promotes disease development via a mechanism independent of suppressing SA-mediated host defenses (Chen et al. 2004; Mutka et al. 2013). In a recent study, Djami-Tchatchou et al. (2020) demonstrated that IAA regulates expression of known virulence genes, including genes involved in T3S, in PtoDC3000 growing in culture. However, rather than promoting expression of T3SS-related genes, as was initially expected given that IAA promotes pathogenesis, they observed that T3SS-related genes were down-regulated in response to IAA. However, other virulence genes, such as tvrR, which encodes a TetR family transcription factor, were up-regulated in response to IAA. Similar effects of IAA on expression of these virulence genes were also observed in planta. These findings led to the hypothesis that IAA acts as a signaling molecule that coordinates expression of virulence genes required during different stages of pathogenesis (Djami-Tchatchou et al. 2020). IAA has also been shown to down-regulate expression of T3SS genes in P. savastanoi (Aragon et al. 2014; Ryu 2015). Currently, we do not know how P. syrinage and other plant pathogens sense and respond to auxin. Elucidating the mechanisms involved will greatly enhance our understanding of the roles of IAA as a pathogen signal during infection.
CONCLUDING REMARKS
It is evident from the examples discussed above that auxin plays several different roles during plant–pathogen interactions. It should come as no surprise that one of its primary roles is as a plant hormone, and that alterations in host auxin signaling and physiology can either result in enhanced defense or increased disease, depending on the pathogen. Perhaps more surprising are the more recent discoveries that auxin also acts as a microbial signaling molecule that can directly impact pathogen biology, for example, by modulating virulence gene expression. These findings have opened up an exciting new area of research focused on elucidating the mechanisms by which pathogens sense and respond to IAA.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agrios GN. 1997. Plant pathology, 4th ed.Academic, San Diego. [Google Scholar]
- Aragon IM, Perez-Martinez I, Moreno-Perez A, Cerezo M, Ramos C. 2014. New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol Lett 356: 184–192. 10.1111/1574-6968.12413 [DOI] [PubMed] [Google Scholar]
- Beattie GA. 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol 49: 533–555. 10.1146/annurev-phyto-073009-114436 [DOI] [PubMed] [Google Scholar]
- Bianco C, Imperlini E, Calogero R, Senatore B, Amoresano A, Carpentieri A, Pucci P, Defez R. 2006a. Indole-3-acetic acid improves Escherichia coli’s defences to stress. Arch Microbiol 185: 373–382. 10.1007/s00203-006-0103-y [DOI] [PubMed] [Google Scholar]
- Bianco C, Imperlini E, Calogero R, Senatore B, Pucci P, Defez R. 2006b. Indole-3-acetic acid regulates the central metabolic pathways in Escherichia coli. Microbiolog (Reading) 152: 2421–2431. 10.1099/mic.0.28765-0 [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539. 10.1016/j.molp.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Boevink PC, Birch PRJ, Turnbull D, Whisson SC. 2020. Devastating intimacy: the cell biology of plant–Phytophthora interactions. New Phytol 228: 445–458. 10.1111/nph.16650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C. 2017. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55: 257–286. 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- Bürger M, Chory J. 2019. Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26: 163–172. 10.1016/j.chom.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. 2016. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol Rev 40: 894–937. 10.1093/femsre/fuw026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN. 2004. The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J 37: 494–504. 10.1111/j.1365-313X.2003.01984.x [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. 2007. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci 104: 20131–20136. 10.1073/pnas.0704901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, Coaker G, Kunkel B, He P, Shan L. 2013. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol 162: 1018–1029. 10.1104/pp.113.219659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djami-Tchatchou AT, Harrison GA, Harper CP, Wang R, Prigge MJ, Estelle M, Kunkel BN. 2020. Dual role of auxin in regulating plant defense and bacterial virulence gene expression during Pseudomonas syringae PtoDC3000 pathogenesis. Mol Plant Microbe Interact 33: 1059–1071. 10.1094/MPMI-02-20-0047-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati AJ, Lee HI, Leveau JH, Chang WS. 2013. Effects of indole-3-acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum. PLoS ONE 8: e76559. 10.1371/journal.pone.0076559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso R, Leiva-Novoa P, Zúñiga A, Timmermann T, Recabarren-Gajardo G, González B. 2017. Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium Paraburkholderia phytofirmans PsJN. Appl Environ Microbiol 83: e01991-16. 10.1128/AEM.01991-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dubey SM, Calixte Serre NB, Oulehlová D, Vittal P, Fendrych M. 2021. No time for transcription—rapid auxin responses in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca D, Lorv J, Patten CL, Rose D, Glick BR. 2014. Indole-3-acetic acid in plant-microbe interactions. Antonie van Leeuwenhoek 106: 85–125. 10.1007/s10482-013-0095-y [DOI] [PubMed] [Google Scholar]
- Evangelisti E, Govetto B, Minet-Kebdani N, Kuhn ML, Attard A, Ponchet M, Panabières F, Gourgues M. 2013. The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol 199: 476–489. 10.1111/nph.12270 [DOI] [PubMed] [Google Scholar]
- Fousia S, Tsafouros A, Roussos PA, Tjamos SE. 2018. Increased resistance to Verticillium dahliae in Arabidopsis plants defective in auxin signalling. Plant Pathol 67: 1749–1757. 10.1111/ppa.12881 [DOI] [Google Scholar]
- French E, Kim BS, Rivera-Zuluaga K, Iyer-Pascuzzi AS. 2018. Whole root transcriptomic analysis suggests a role for auxin pathways in resistance to Ralstonia solanacearum in tomato. Mol Plant Microbe Interact 31: 432–444. 10.1094/MPMI-08-17-0209-R [DOI] [PubMed] [Google Scholar]
- Fu J, Wang S. 2011. Insights into auxin signaling in plant–pathogen interactions. Front Plant Sci 2: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S. 2011. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol 155: 589–602. 10.1104/pp.110.163774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhut IV, Slezak BL, Leveau JHJ. 2018. Iac gene expression in the indole-3-acetic acid-degrading soil bacterium Enterobacter soli LF7. Appl Environ Microbiol 84: e01057-18. 10.1128/AEM.01057-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, van Isterdael G, Beeckman T, Gheysen G, Mathesius U. 2009. Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562. 10.1105/tpc.109.069617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Kahmann R. 2019. Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front Plant Sci 10: 822. 10.3389/fpls.2019.00822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M. 2000. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 3: 315–319. 10.1016/S1369-5266(00)00087-X [DOI] [PubMed] [Google Scholar]
- Islam W, Naveed H, Zaynab M, Huang ZQ, Chen HYH. 2019. Plant defense against virus diseases; growth hormones in highlights. Plant Signal Behav 14: 1596719. 10.1080/15592324.2019.1596719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Qin QQ, Wang Y, Pu YY, Liu LF, Wen X, Ji SY, Wu JG, Wei CH, Ding B, et al. 2016. Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. Plos Pathog 12: e1005847. 10.1371/journal.ppat.1005847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Tsuda K. 2010. Understanding the plant immune system. Mol Plant Microbe Interact 23: 1531–1536. 10.1094/MPMI-04-10-0099 [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R. 2014. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309. 10.1105/tpc.114.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2009. Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci 14: 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, Thatcher LF, Aitken EA, Schenk PM, Manners JM, Kazan K. 2011. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol Plant Microbe Interact 24: 733–748. 10.1094/MPMI-08-10-0194 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Harper CP. 2018. The roles of auxin during interactions between bacterial plant pathogens and their hosts. J Exp Bot 69: 245–254. 10.1093/jxb/erx447 [DOI] [PubMed] [Google Scholar]
- Kyndt T, Goverse A, Haegeman A, Warmerdam S, Wanjau C, Jahani M, Engler G, de Almeida Engler J, Gheysen G. 2016. Redirection of auxin flow in Arabidopsis thaliana roots after infection by root-knot nematodes. J Exp Bot 67: 4559–4570. 10.1093/jxb/erw230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird TS, Flores N, Leveau JHJ. 2020. Bacterial catabolism of indole-3-acetic acid. Appl Microbiol Biotechnol 104: 9535–9550. 10.1007/s00253-020-10938-9 [DOI] [PubMed] [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legein M, Smets W, Vandenheuvel D, Eilers T, Muyshondt B, Prinsen E, Samson R, Lebeer S. 2020. Modes of action of microbial biocontrol in the phyllosphere. Front Microbiol 11: 1619. 10.3389/fmicb.2020.01619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Nester EW. 2006. Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc Natl Acad Sci 103: 4658–4662. 10.1073/pnas.0600366103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodráguez C, Jordá L, Parker J, Molina A. 2008. Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant 1: 496–509. 10.1093/mp/ssn025 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. 2015. Bacteria and fungi controlling plant growth by manipulating auxin: balance between development and defense. J Plant Physiol 172: 4–12. 10.1016/j.jplph.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Ma KW, Ma WB. 2016. Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol Biol 91: 713–725. 10.1007/s11103-016-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah KM, Uppalapati SR, Tang YH, Allen S, Shuai B. 2012. Gene expression profiling of Macrophomina phaseolina infected Medicago truncatula roots reveals a role for auxin in plant tolerance against the charcoal rot pathogen. Physiol Mol Plant Pathol 79: 21–30. [Google Scholar]
- Mashiguchi K, Hisano H, Takeda-Kamiya N, Takebayashi Y, Ariizumi T, Gao YB, Ezura H, Sato K, Zhao Y, Hayashi K, et al. 2019. Agrobacterium tumefaciens enhances biosynthesis of two distinct auxins in the formation of crown galls. Plant Cell Physiol 60: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Daddaoua A, Chini A, Morel B, Krell T. 2018. An auxin controls bacterial antibiotics production. Nucleic Acids Res 46: 11229–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM, Kunkel BN. 2018. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog 14: e1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Kunkel BN. 2013. Virulence strategies of plant pathogenic bacteria. In The prokaryotes (ed. Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F). Springer, Berlin, Heidelberg. 10.1007/978-3-642-30141-4_62 [DOI] [Google Scholar]
- Mockaitis K, Estelle M. 2008. Auxin receptors and plant development: a new signaling paradigm. Ann Rev Cell Dev Biol 24: 55–80. [DOI] [PubMed] [Google Scholar]
- Mutka AM, Fawley S, Tsao T, Kunkel BN. 2013. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J 74: 746–754. 10.1111/tpj.12157 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. 2005. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J Virol 79: 2549–2558. 10.1128/JVI.79.4.2549-2558.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Shiferaw H, Culver JN. 2006. The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol Plant Microbe Interact 19: 864–873. 10.1094/MPMI-19-0864 [DOI] [PubMed] [Google Scholar]
- Patten CL, Blakney AJ, Coulson TJ. 2013. Activity, distribution, and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit Rev Microbiol 39: 395–415. 10.3109/1040841X.2012.716819 [DOI] [PubMed] [Google Scholar]
- Qi LL, Yan J, Li YN, Jiang HL, Sun JQ, Chen Q, Li HX, Chu JF, Yan CY, Sun XH, et al. 2012. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol 195: 872–882. 10.1111/j.1469-8137.2012.04208.x [DOI] [PubMed] [Google Scholar]
- Qiao LL, Zheng LY, Sheng C, Zhao HW, Jin HL, Niu DD. 2020. Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J 102: 948–964. 10.1111/tpj.14677 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu Rev Phytopathol 49: 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Ryu CM. 2015. Against friend and foe: type 6 effectors in plant-associated bacteria. J Microbiol 53: 201–208. 10.1007/s12275-015-5055-y [DOI] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S. 2018. Pattern recognition receptors and signaling in plant–microbe interactions. Plant J 93: 592–613. 10.1111/tpj.13808 [DOI] [PubMed] [Google Scholar]
- Shu HY, Lin LC, Lin TK, Chen HP, Yang HH, Peng KC, Lin GH. 2015. Transcriptional regulation of the iac locus from Acinetobacter baumannii by the phytohormone indole-3-acetic acid. Antonie van Leeuwenhoek 107: 1237–1247. 10.1007/s10482-015-0417-3 [DOI] [PubMed] [Google Scholar]
- Singh D, Raina TK, Kumar A, Singh J, Prasad R. 2019. Plant microbiome: a reservoir of novel genes and metabolites. Plant Gene 18: 100177. 10.1016/j.plgene.2019.100177 [DOI] [Google Scholar]
- Spaepen S, Vanderleyden J. 2011. Auxin and plant–microbe interactions. Cold Spring Harb Perspect Biol 3: a001438. 10.1101/cshperspect.a001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Das F, Luyten E, Michiels J, Vanderleyden J. 2009. Indole-3-acetic acid-regulated genes in Rhizobium etli CNPAF512. FEMS Microbiol Lett 291: 195–200. 10.1111/j.1574-6968.2008.01453.x [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Hugly S, Buchholz WG, Thomashow LS. 1986. Molecular basis for the auxin-independent phenotype of crown gall tumor tissues. Science 231: 616–618. 10.1126/science.3511528 [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. 1994. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249. 10.1093/genetics/138.4.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G. 2016. Plant–pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Ann Rev Phytopathol 54: 419–441. 10.1146/annurev-phyto-080615-100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Broek A, Gysegom P, Ona O, Hendrickx N, Prinsen E, Van Impe J, Vanderleyden J. 2005. Transcriptional analysis of the Azospirillum brasilense indole-3-pyruvate decarboxylase gene and identification of a cis-acting sequence involved in auxin responsive expression. Mol Plant Microbe Interact 18: 311–323. 10.1094/MPMI-18-0311 [DOI] [PubMed] [Google Scholar]
- Van Puyvelde S, Cloots L, Engelen K, Das F, Marchal K, Vanderleyden J, Spaepen S. 2011. Transcriptome analysis of the rhizosphere bacterium Azospirillum brasilense reveals an extensive auxin response. Microb Ecol 61: 723–728. 10.1007/s00248-011-9819-6 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. 2007. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Xin XF, Kvitko B, He SY. 2018. Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16: 316–328. 10.1038/nrmicro.2018.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW. 2008. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and γ-amino butyric acid reveals signalling cross-talk and Agrobacterium–plant co-evolution. Cell Microbiol 10: 2339–2354. 10.1111/j.1462-5822.2008.01215.x [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. 2017. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot 68: 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Tan XX, Li LL, He YQ, Hong GJ, Li JM, Lin L, Cheng Y, Yan F, Chen JP, et al. 2019. Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol Plant Pathol 20: 1093–1104. 10.1111/mpp.12814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Li LL, He YQ, Qin QQ, Chen CH, Wei ZY, Tan XX, Xie KL, Zhang RF, Hong GJ, et al. 2020. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc Natl Acad Sci 117: 9112–9121. 10.1073/pnas.1918254117 [DOI] [PMC free article] [PubMed] [Google Scholar]