Abstract
Single Particle Tracking (SPT) is a powerful class of tools for analyzing the dynamics of individual biological macromolecules moving inside living cells. The acquired data is typically in the form of a sequence of camera images that are then post-processed to reveal details about the motion. In this work, we develop a local time-varying estimation algorithm for estimating motion model parameters from the data considering nonlinear observations. Our approach uses several well-known existing tools, namely the Expectation Maximization (EM) algorithm combined with an Unscented Kalman filter (UKF) and an Unscented Rauch-Tung-Striebel smoother (URTSS), and applies them to the time-varying case through a sliding window methodology. Due to the shot noise characteristics of the photon generation process, this model uses a Poisson distribution to capture the measurement noise inherent in imaging. In order to apply our time-varying approach to the UKF, we first need to transform the measurements into a model with additive Gaussian noise. This is carried out using a variance stabilizing transform. Results from simulations show that our approach is successful in tracing time-varying diffusion constants at a range of physically relevant signal levels. We also discuss the initialization for the EM algorithm based on the available data.
I. Introduction
Single particle tracking (SPT) is an important class of techniques for studying the motion of single biological macromolecules. With its ability to localize particles with an accuracy far below the diffraction limit of light and the ability to track these particles across time, SPT continues to be an invaluable tool in understanding biology at the nanometer-scale. It has been applied to the study of a wide variety of molecules, including proteins [1], mRNA molecules [2], viruses [3], and more. While SPT encompasses many specific experimental techniques [4], [5], common to most of these is that measurements come in the form of CCD camera images which are analyzed to infer particle trajectories and to estimate parameters. Typically, these models are assumed time-invariant, but in reality parameters can vary according to local conditions or the actions of the cellular machinery.
There are several motion models relevant to biophysical applications, including diffusion, confined diffusion, directed motion, and combinations [6]. Given noisy observations of such a model (e.g. from producing a trajectory by localizing a fluorescent particle in each frame of an image sequence), the most common technique to estimate the motion model parameters is to fit the chosen model to the Mean Square Displacement (MSD) curve. This is simple and popular approach has been enormously successful in probing biomolecular dynamics [7]. However, the resulting estimates depend on choices such as the number of points to fit and the scheme does not account for many factors such as observation noise and motion blur arising from the camera shutter time [8].
Since the diffusion model is straightforward, it is amenable to analysis and to the use of optimal estimation. This was recognized and developed in [8]–[10] where it has been shown that such schemes outperform simple MSD. Using optimal techniques such as ML estimation places the problem on firm theoretical grounding, ensuring that the analysis method is both consistent and asymptotically efficient, and provides a rigorous understanding of the accuracy and performance of the estimator. A similar approach has also been developed for the analysis of confined diffusion [11].
In general, techniques for model parameter estimation assume a simple linear observation of the particle position corrupted by additive white Gaussian noise [12]. The actual data, however, are intensity measurements from a CCD camera that are well modeled as Poisson-distributed random variables with a rate depending on the true location of the particle and on experimental realities, including background intensity and the optics used. This already nonlinear model becomes even more complicated at low signal intensities (common to SPT data) where noise models specific to the type of camera being used become crucial [13], [14].
In SPT, especially at low SBR intensities, we thus face a nonlinear estimation problem. In [15], this nonlinear estimation problem (with time-invariant parameters) was solved by using Sequential Monte Carlo (SMC) techniques in the context of the EM algorithm. The approach is general, being able to handle nearly arbitrary nonlinearities in both the motion and observation models, at the cost of significant computational burden. Recently, in [9], two of the authors addressed the computational load challenge by replacing the particle-based methods by an Unscented Kalman filter (UKF) and Unscented Rauch-Tung-Striebel smoother (URTSS) [16], [17], which reduces the computational complexity drastically with very little compromise in performance.
In this work, we consider the time-varying setting and develop an estimation algorithm based on local likelihood estimation, focusing on diffusion with a nonlinear observation model. The idea of local likelihood is very old and is a natural development of the sliding window approach [18]. However, a thorough theoretical understanding of the method was not developed until the 1990s in the statistics literature under the name local least squares or local polynomial modelling [18], [19]. Despite these developments, the theory has not diffused widely outside the statistics literature and so a number of basic insights are still ignored in other application domains.
There are, of course, alternatives to a local modeling approach. For example, there is significant literature on modeling data using a global polynomial fit. However, this method potentially needs a large number of components to yield a low bias [19], and, as a consequence, may introduce over-parametrization. The local approximation approach, in general, only requires a small number of parameters.
In this work, we restrict our attention to 2-D diffusion since: (i) diffusion is the workhorse of motion modeling in biophysical systems at the nanometer scale; (ii) there are many 2-D biophysical settings of interest, such as motion in plasma membranes; and (iii) this allows us to concentrate the discussion on a straightforward and concrete setting. As the corresponding dynamic model is linear with additive Gaussian noise, applying the UKF to the state update equations is straightforward. The observation model, however, involves Poisson distributed noise whose parameters depend upon the state. To apply the UKF, the model must be converted into one where the measurement noise is Gaussian. Based on prior work [9], we use the Anscombe transform.
One important aspect in the EM algorithm is the selection of the initial value of the parameter. It is usually the case that, in a real application, the user has prior information about the values of the parameters in their data based on domain knowledge. However, this information is not necessarily reliable or might reflect a bias of the experimenter, and thus it is important to provide a more formal approach to initialization. It is well-known that the selection of the initial estimate of the parameter of interest is critical in the performance of the EM algorithm, see e.g. [20]. Here, we investigate two initialization approaches. The first one is based on a warm start, which means we use the last value of the algorithm obtained in the previous window. The second approach is given by the spectral factorization (SF) [12] of an approximated time series to the SPT model. Results for different kind of signal-to-background levels (SBR) are then compared based on the two aforementioned initializations.
II. Problem formulation
A. Motion model
The model of 2-D anisotropic diffusion in discrete time is
| (1) |
where represents the location of the particle at time t and is a covariance matrix given by
| (2) |
Here Dx and Dy are independent diffusion coefficients and Δt is the time between frames of the image sequence.
B. Observation model
Because the single particle is smaller than the diffraction limit of light, the image on the camera is described by the point spread function (PSF) of the instrument. In 2-D (and in the focal plane), the PSF is well approximated by
| (3) |
where the particle is located at the origin, the pair (x, y) represents a position on the plane x-y at which the PSF is being evaluated, and σx and σy are given by
| (4) |
Here λ is the wavelength of the emitted light and NA is the numerical aperture of the objective lens being used [21]. This PSF is then imaged by the CCD camera.
Assuming segmentation has been done (a standard preprocessing step), the image acquired by the camera is composed of P pixels arranged into a square array. The pixel size is Δx by Δy with the actual dimensions determined by the physical size of the CCD elements on the camera and the optical magnification. At time step t. the expected photon intensity measured for the pth pixel is
where G denotes the peak intensity of the fluorescence and the integration bounds are over the given pixel.
In addition to the signal, there is always a background intensity rate arising from autofluorescence and out-of-focus fluorescence. While this background can vary across the sample, the region covered by the P pixels is small and it is assumed that Nbgd is a constant, uniform rate that is independently estimated (though it can be integrated into our inference problem in Sec. III). Combining these signals, the measured intensity in the pth pixel at time t is given by
| (5) |
where Poiss(·) represents a Poisson distribution.
C. Measurement model transformation
The UKF applies to nonlinear observation models with additive Gaussian noise [17]. The model in (5) can be transformed into such a form using a variance stabilizing transformation such as the Anscombe [22] or the Freeman and Tukey [23] transformation, or at sufficiently large intensities, it can be directly approximated by a Gaussian model [24]. These different approaches were compared in [9] where it was found that in general the Anscombe transform performs the best, especially at low signal levels and thus this is the approach we take here. Applying this transformation, the observation model (5) becomes
| (6) |
III. Inference problem
Our goal is to use a local likelihood approach to handle time-varying estimation. We begin with a single window where the problem reduces to the time-invariant case.
A. Time-invariant parameter estimation via EM
Consider the problem of identifying an unknown parameter for the nonlinear state space model
| (7) |
Our goal is to find an ML estimate of θ from the data . The ML estimates can be found through the optimization problem:
| (8) |
This optimization can only be solved in closed form in certain simple cases as pθ(YN) is typically intractable.
An alternative way to find the estimate that optimizes (8) is by using the EM algorithm [20]. EM defines a hidden (or latent) variable and iteratively optimizes the log-likelihood function l(θ) = pθ(YN) through a new function, ,
| (9) |
The calculation of is called the Expectation (E)-step at the ith iteration. It has been shown [20] that any choice of such that also increases the original likelihood. Thus, the E-step is followed by a Maximization (M)-step to produce the next estimate,
| (10) |
Notice that (9) can be decomposed as, see e.g. [25]
| (11) |
where
| (12a) |
| (12b) |
| (12c) |
In the SPT context, the unknowns are the diffusion constants, captured in the (unknown) variance Q. Hence, our E-step is given by (ignoring constant terms):
| (13) |
where | · | and tr(·) indicate the determinant and trace operators, respectively. The M-step is simply obtained by setting the derivative of with respect to Q−1 to zero and solving to obtain:
This depends on the smoothed distributions p(xt|YN) and p(xt, xt−1|YN). If the underlying model in (III-A) is linear with Gaussian noise then these distributions are easily obtained [26] using the Kalman Filter. For nonlinear systems, an analytical solution cannot be found. Therefore, either an approximation or numerical approach must be used. Here we take an approximation approach and apply the UKF and URTSS. Finally, from (2), our estimates are given by:
| (14) |
B. Time-varying likelihood
In this section, we describe an algorithm to trace time-varying parameters associated with the general model in (1), using as a base the algorithm in section III-A. The idea is to pose a time-varying likelihood which is time-invariant in a window of a nominated length. The likelihood is then optimized in this window and the window then shifted.
The local likelihood is defined as:
| (15) |
where is usually called the kernel, h is the window length, t is a point within the window, usually the middle point, and k indicates any point within the window. Different values for h can be used, as well as different kernels, and the most appropriate selection is a nontrivial issue. One common choice is the Epanechnikov kernel, see e.g [19], which is used throughout this work.
Due to the nonlinear measurements of SPT, obtaining the likelihood function is intractable and we use the auxiliary function of the EM algorithm as an approximation of the likelihood, extending it to the time-varying setting through the local approach. We then consider the following equation:
| (16) |
From (11), we can write
| (17) |
As noted above, in the SPT setting, the parameter θt only appears in I2 so that the time-varying E-step is
| (18) |
To optimize this time-varying function, we use a similar procedure as in the time-invariant case, that is, we set the derivative of Qt with respect to θt to zero and solve. The resulting estimates optimize the (windowed) likelihood within window h. The estimation algorithm continues when the time points are shifted by one unit and finishes when the last data point is included in the window. Of course, the shifting can be done by more than one unit to reduce computation, see e.g. [19].
To summarize, the local likelihood approach introduces a time-varying E-step, denoted by , optimized within a window h. This, in turn, will optimize the log-likelihood function within the same window, to create the local likelihood approach. For our SPT problem, discarding the constant terms and the parameters for the initial conditions, we obtain
| (19) |
with
| (20) |
| (21) |
| (22) |
The values for , Pk|h, and Pk,k−1|h are the smoothed state, variance, and covariance, which are found using the UKF and URTSS, for details see [9].
The M-step is completed by taking the derivative of with respect to and setting it to zero, obtaining as a result:
| (23) |
Remark 1. To focus on our time-varying approach, we use the naïve version of EM. However, we note that there is a robust version for the M-step, see e.g. [27], based on the Cholesky factorization, with improved robustness.
C. Convergence
It is known that the EM algorithm converges to a stationary point of the likelihood function (not necessarily the global maximum) so long as [28]. We have shown ( [29] that the local likelihood does increase at each step. Then, since our algorithm is based on EM, it inherits its convergence properties. Of course, this also implies we cannot guarantee convergence to the true value because the EM algorithm itself only yields the global maximum under certain special cases [28, ch.3].
D. Initialization of the EM algorithm
One crucial aspect to consider is the initial estimate for the EM algorithm. If the initial value of the estimate is too far from the actual value, and the log-likelihood has multiple stationary points, then the EM algorithm might not converge to the true value of the parameters, see e.g. [28, p.33]. Focusing on the x-axis, we rewrite (1) as
| (24a) |
| (24b) |
Through a small abuse of notation, the output in (24b), yk, is no longer the camera image a time k but the position of the particle in the x-axis. This trajectory can be estimated from the camera images by using a variety of methods with one a fit of the image intensities to a Gaussian model being the most common [30]. We now consider the model in (24) to calculate our initial estimate for Q. We rewrite (24) as yk+1 − yk = wk + (z − 1)vk, where z is the shift operator. Our initial estimate of Q, namely , is given by [12]
| (25) |
where σϵ and c are the parameters of an equivalent MA(1) model of the form yk − yk−1 = (z − c)ϵk, , that are estimated using data from (24b) using, for example, Prediction Error Methods (PEM).
IV. Demonstration and Analysis
We generate data according to the optical parameters and other fixed constants shown in Table I. These were chosen to replicate experimental settings found in many SPT experiments. Notice that we produce a step-like change in the diffusion constant at time tc = 200, from 1 × 10−3 to 2×10−3 μm2/s. For space reasons, results are presented only for a single axis. Note that in general, real SPT experiments are often data-poor as the biological samples can typically only be viewed for short periods of time. In our simulated data, then, we have considered a somewhat short total of 400 images. Here, and in the sequel, we select the tuning parameters for the UKF as follows: [α κ β]T = [1 0 2]T. For details about these parameters, see e.g. [9].
TABLE I.
Parameter settings
| Symbol | Parameter | Values |
|---|---|---|
| Δt | Image period (discrete time step) | 100 ms |
| T | Number of images per dataset | 400 |
| tc | time of change | 200s |
| P | Number of pixels per squared image | 25 |
| Diffusion coeff. in x direction before tc | 0.001 μm2/s | |
| Diffusion coeff. in x direction after tc | 0.002 μm2/s | |
| Diffusion coeff. in y direction before tc | 0.001 μm2/s | |
| Diffusion coeff. in y direction after tc | 0.002 μm2/s | |
| Δx | Length of unit pixel | 100 nm |
| Δy | Width of unit pixel | 100 nm |
| λ | Emission wavelength | 540 nm |
| NA | Numerical aperture | 1.2 |
A. Demonstration of the effect of kernel K(v)
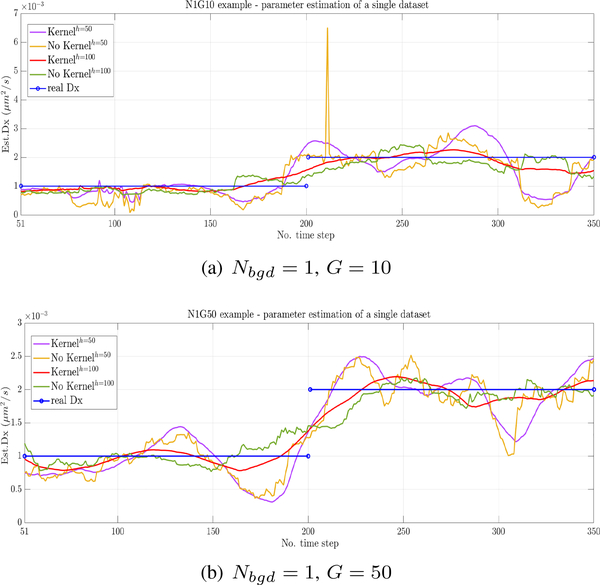
We first demonstrate the effect of the inclusion of a Epanechnikov kernel function K(v), for two different peak intensities G = {10, 50} with 10 being a relatively weak signal and 30 a relatively strong one. Fig. 1 shows five lines corresponding to two different window sizes each with and without a kernel is used or not, and the real value of Dx. As expected, the kernel smoothing effect is stronger with shorter windows and with lower signal level as in Fig. 1(a) compared to Fig. 1(b). These results thus indicate that a kernel is an important element of the estimation process.
Fig. 1.
Effect of the kernel with window sizes of 50 (purple, with kernel; yellow, without kernel) and 100 (red, with kernel; green, without kernel). (a) Low SBR: Ngbd = 1, G = 10. (b) High SBR: Ngbd = 1, G = 50.
B. Demonstration of window sizes
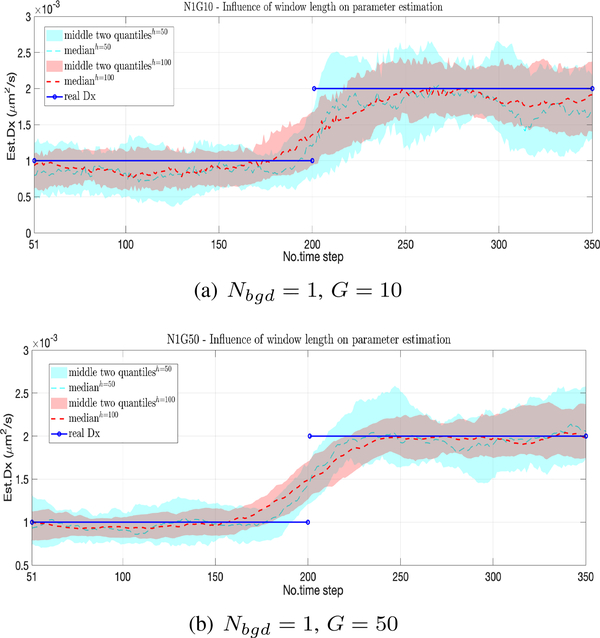
We now fix the kernel to the Epanechnikov form and look at the effect of the different window sizes. The results for h = 50,100, at two different SBR levels, running 50 different simulation trials, are shown in Fig. 2 with the median indicated by the center lines and the middle two quantiles by the shaded area. Color corresponds to window size with 50 in blue and 100 in red. As expected, the smaller window size leads to a quicker response but larger variance. Comparing the results in Fig. 2(a) and (b), we note an increased variance in the estimate at lower SBR.
Fig. 2.
Effect of window length h=50 (light blue) and h=100 (red) with median across 50 trials indicated by dashed lines and the center two quantiles by the shaded region. (a) Low SBR: Nbgd = 1, G = 10. (b) High SBR: Nbgd = 1, G = 50.
Table II shows the RMSE (over 50 runs) for different window lengths and signal intensities. The table indicates improvement in terms of accuracy for longer window lengths but with diminishing returns as the window increases. Note that while here the window length is chosen empirically, there are data-driven methods available, e.g. the Steins Unbiased Risk Estimator (SURE) [31], to select good window sizes.
TABLE II.
Root mean square error (RMSE) for different window lengths and signal intensity (G)
| window h | (RMSE ± std) ×10−3 G=50 | (RMSE ± std) ×10−3 G=10 |
|---|---|---|
| 50 | 0.8006 ± 0.1408 | 0.9197 ± 0.2752 |
| 75 | 0.7616 ± 0.1406 | 0.8179 ± 0.2626 |
| 100 | 0.7375 ± 0.1386 | 0.7622 ± 0.2547 |
| 125 | 0.7144 ± 0.1347 | 0.7227 ± 0.2515 |
C. Effect of initial estimate for the EM algorithm
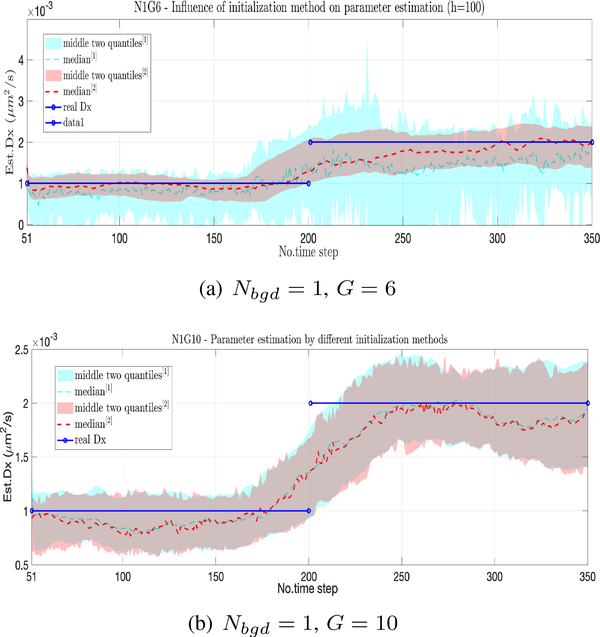
Next, we study the effect of using the initialization given by the SF decomposition versus a warm start, comparing again at low and high SBR. The results at a background rate of Nbgd = 1 and a signal intensity of G = 6, shown in Fig. 3(a), indicate that in this regime, initialization using SF fails to give a reasonable estimate while the warm start still leads to reasonable values. This can be explained in part by the fact that the SF decomposition uses an observation model of position plus noise, generated from using Gaussian fits to each image to localize the particle. This fitting becomes unreliable when the SBR drops too low. The warm start, on the other hand, does not rely on such estimates and uses only the full, nonlinear model. At higher SBR, we observe from Fig. 3(b) that both the warm start and the initialization given by the SF give almost identical results. From this we conclude that the SF decomposition is most useful to initialize the first window only with the warm start being used in all subsequent windows.
Fig. 3.
Effect of initialization algorithm, comparing a warm start (blue) against an SF-approximation (red). Median across 50 trials indicated by dashed lines and the center two quantiles by the shaded regions. (a) Nbgd = 1, G = 6. (b) Nbgd = 1, G = 10.
D. Longer slides
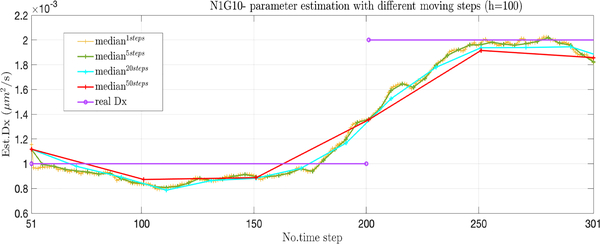
The proposed scheme involves non-trivial calculations as the EM needs to be run across the entire set of data in each window. While these computations can be performed off-line, implying that the computational complexity is not a major concern, it is often a matter of convenience to a user for estimates to be performed as quick as possible. One simple approach for reducing the overall computation time is to simply shift the window by more than one data point each time. In Fig. 4, we show the results of running the time-varying approach, stepping forward by 1, 5, 20, and 50 steps. Somewhat surprisingly, there is little loss in accuracy, though of course there is loss of sensitivity to multiple, rapid changes in parameter values with larger shifts.
Fig. 4.
Comparison of different shift sizes with shifts of 1 (yellow), 5 (green), 20 (blue), and 50 (red). The true parameter is shown in purple.
V. Conclusions
In this paper we introduced an algorithm for time-varying parameter estimation in single particle tracking where we have a complex, nonlinear measurement model. The proposed algorithm considers the use of previously developed tools, namely the EM algorithm combined with a UKF and the URTSS, in a local approach. Using physically accurate simulations, we explored the effect of using a kernel in the window, in the window lengths, and in the initialization method for the EM algorithm, all at two different SBRs. We also considered the effect of increasing shifts in the window as a means of reducing the computational load of the analysis technique.
Acknowledgement
This work was supported in part by the NIH through 1R01GM117039-01A1.
References
- [1].Simson R, Sheets ED, and Jacobson K, “Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis,” Biophysical Journal, vol. 69, no. 3, pp. 989–993, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Park HY, Buxbaum AR, and Singer RH, “Single mRNA tracking in live cells,” in Methods in Enzymology, 2010, vol. 472, pp. 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, and Helenius A, “Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes,” Proc. National Academy of Sciences, vol. 102, no. 42, pp. 15110–15115, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].von Diezmann A, Shechtman Y, and Moerner WE, “Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking,” Chemical Reviews, vol. 117, no. 11, pp. 7244–7275, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma Y, Wang X, Liu H, Wei L, and Xiao L, “Recent advances in optical microscopic methods for single-particle tracking in biological samples,” Analytical and Bioanalytical Chemistry, pp. 1–19, 2019. [DOI] [PubMed] [Google Scholar]
- [6].Monnier N, Guo S-M, Mori M, He J, Lénárt P, and Bathe M, “Bayesian approach to MSD-based analysis of particle motion in live cells,” Biophysical Journal, vol. 103, no. 3, pp. 616–626, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manzo C and Garcia-Parajo MF, “A review of progress in single particle tracking: from methods to biophysical insights,” Reports on Progress in Physics, vol. 78, no. 12, p. 124601, 2015. [DOI] [PubMed] [Google Scholar]
- [8].Berglund AJ, “Statistics of camera-based single-particle tracking,” Physical Review E, vol. 82, no. 1, p. 011917, 2010. [DOI] [PubMed] [Google Scholar]
- [9].Lin Y and Andersson SB, “Simultaneous localization and parameter estimation for single particle tracking via sigma points based EM,” in 58th IEEE Conf. on Decision and Control (CDC), 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Michalet X and Berglund AJ, “Optimal diffusion coefficient estimation in single-particle tracking,” Physical Review E, vol. 85, no. 6, p. 061916, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Calderon CP, “Motion blur filtering: A statistical approach for extracting confinement forces and diffusivity from a single blurred trajectory,” Physical Review E, vol. 93, no. 5, p. 053303, 2016. [DOI] [PubMed] [Google Scholar]
- [12].Godoy BI, Lin Y, and Andersson SB, “A 2-step algorithm for the estimation of time-varying single particle tracking models using maximum likelihood,” in Asian Control Conf. (ASCC), 2019, pp. 1078–1083. [PMC free article] [PubMed] [Google Scholar]
- [13].Krull A, Steinborn A, Ananthanarayanan V, Ramunno-Johnson D, Petersohn U, and Tolic-Nørrelykke IM, “A divide and conqueŕ strategy for the maximum likelihood localization of low intensity objects,” Optics Express, vol. 22, no. 1, pp. 210–228, 2014. [DOI] [PubMed] [Google Scholar]
- [14].Lin R, Clowsley AH, Jayasinghe ID, Baddeley D, and Soeller C, “Algorithmic corrections for localization microscopy with sCMOS cameras-characterisation of a computationally efficient localization approach,” Optics Express, vol. 25, no. 10, pp. 11701–11716, 2017. [DOI] [PubMed] [Google Scholar]
- [15].Ashley TT and Andersson SB, “Method for simultaneous localization and parameter estimation in particle tracking experiments,” Physical Review E, vol. 92, no. 5, p. 052707, 2015. [DOI] [PubMed] [Google Scholar]
- [16].Van Der Merwe R et al. , “Sigma-point Kalman filters for probabilistic inference in dynamic state-space models,” Ph.D. dissertation, OGI School of Science & Engineering at OHSU, 2004. [Google Scholar]
- [17].Särkkä S, Bayesian filtering and smoothing. Cambridge Univ. Press, 2013. [Google Scholar]
- [18].Loader C, Local Regression and Likelihood. Springer, 1999. [Google Scholar]
- [19].Fan J and Gijbels I, Local Polynomial Modelling and its Applications. Chapman& Hall, CRC, 1996. [Google Scholar]
- [20].Dempster AP, Laird NM, and Rubin DB, “Maximum likelihood from incomplete data via the EM algorithm,” Journal of the Royal Statistical Society. Series B, vol. 39, no. 1, pp. 1–38, 1977. [Google Scholar]
- [21].Zhang B, Zerubia J, and Olivo-Marin J-C, “Gaussian approximations of fluorescence microscope point-spread function models,” Applied Optics, vol. 46, no. 10, pp. 1819–1829, 2007. [DOI] [PubMed] [Google Scholar]
- [22].Anscombe FJ, “The transformation of poisson, binomial and negative-binomial data,” Biometrika, vol. 35, pp. 246–254, 1948. [Google Scholar]
- [23].Freeman MF and Tukey JW, “Transformations related to the angular and the square root,” Ann. Math. Statist, vol. 21, no. 4, pp. 607–611, 1950. [Google Scholar]
- [24].Gnedenko BV, Theory of probability. Routledge, 2017. [Google Scholar]
- [25].Schön T, Wills A, and Ninness B, “System identification of nonlinear state-space models,” Automatica, vol. 47, no. 1, pp. 39–49, 2011. [Google Scholar]
- [26].Gibson S and Ninness B, “Robust maximum-likelihood estimation of multivariable dynamic systems,” Automatica, vol. 41, no. 10, pp. 1667–1682, 2005. [Google Scholar]
- [27].——, “Robust maximum-likelihood estimation of multivariable dynamic systems,” Automatica, vol. 41, no. 10, pp. 1667–1682, 2005. [Google Scholar]
- [28].McLachlan GJ and Krishnan T, The EM algorithm and its extensions, 2nd ed. John Wiley& Sons, 2008. [Google Scholar]
- [29].Godoy BI, Vickers N, Lin Y, and Andersson SB, “Estimation of general time-varying single particle tracking linear models using local likelihood,” in European Control Conf. (ECC), 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Thompson RE, Larson DR, and Webb WW, “Precise nanometer analysis for individual fluorescent probes,” Biophysical Journal, vol. 82, no. 5, pp. 2775–2783, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Long CJ, Brown EN, Triantafyllou C, Wald LL, and Solo V, “Nonstationary noise estimation in functional MRI,” Neuroimage, vol. 28, pp. 890–093, 2005. [DOI] [PubMed] [Google Scholar]