Abstract
The newly identified p53 homolog p73 can mimic the transcriptional activation function of p53. We investigated whether p73, like p53, participates in an autoregulatory feedback loop with MDM2. p73 bound to MDM2 both in vivo and in vitro. Wild-type but not mutant MDM2, expressed in human p53 null osteosarcoma Saos-2 cells, inhibited p73- and p53-dependent transcription driven by the MDM2 promoter-derived p53RE motif as measured in transient-transfection and chloramphenicol acetyltransferase assays and also inhibited p73-induced apoptosis in p53-null human lung adenocarcinoma H1299 cells. MDM2 did not promote the degradation of p73 but instead disrupted the interaction of p73, but not of p53, with p300/CBP by competing with p73 for binding to the p300/CBP N terminus. Both p73α and p73β stimulated the expression of the endogenous MDM2 protein. Hence, MDM2 is transcriptionally activated by p73 and, in turn, negatively regulates the function of this activator through a mechanism distinct from that used for p53 inactivation.
In response to various cellular stresses, the p53 tumor suppressor protein accumulates dramatically and becomes activated, subsequently inducing cell growth arrest or apoptosis and thereby preventing tumorigenesis (18, 29, 33). These cellular roles of p53 are achieved primarily through its ability to transcriptionally activate specific target genes (18, 29, 33). Many p53-responsive genes have been identified as important downstream targets in p53-dependent cell growth arrest and apoptosis. For instance, p21waf1, a cellular inhibitor of the cyclin-dependent kinase family (60), is mainly responsible for p53-induced G1 arrest (12, 13). Recently, the 14-3-3ς protein was reported to play a part in p53-mediated G2/M arrest (23). Moreover, several apoptotic inducers, including Bax-1 (41), and proteins involved in the generation of reactive oxygen species (49) have been implicated in p53-dependent apoptosis. Thus, the transcriptional activity of p53 is central to p53-mediated cell growth arrest and programmed cell death.
In accordance with the crucial importance of the transcriptional activity of p53 in cell growth regulation, this activity is also conserved among p53 homologs. The gene for a p53-like protein, p73, has been identified recently (28) and encodes of two spliced polypeptides, p73α and p73β. p73β (499 aa), which possesses a unique pentamer at its extreme C terminus, is 137 aa shorter than p73α (636 aa). Further analyses of this p53 homolog showed that not only its primary sequence (approximately 60% identity to p53 within the core DNA-binding domain and 29% identity within the N terminus) but also its function resembles that of p53. Like p53, p73 transactivates in vivo p53 target genes, such as those encoding p21waf1 and Bax1, and causes apoptosis and growth suppression (27, 28). Another group of p53 homologs, including p51 and p63 (47, 52, 57, 61), has been identified in humans, mice, and rats and has 55 to 65% homology to p53 in the central domain. These p53 homologs, like p73 and p53, can suppress cell growth, induce apoptosis, and transactivate several p53-responsive genes (47, 61), although it remains unclear whether they are also tumor suppressors. Moreover, two additional p53-like activities have been identified, p53CP from mice (4) and NBP from humans (62). They both bind to the p53-responsive DNA element (p53RE), and NBP also activates the p53RE-dependent transcription in vitro (62). Hence, these studies indicate that the transcriptional activity is well conserved among the p53 family members. However, they do not reveal whether the activities of the different family members are regulated by similar mechanisms.
One crucial p53 regulator is the cellular protein MDM2 (42), which is encoded by the mdm2 oncogene amplified on a mouse double-minute chromosome in the 3T3DM cell line (6). This gene is amplified or overexpressed in approximately one-third of human sarcomas (11, 43, 45). MDM2 can immortalize and, in cooperation with Ras, transform rat embryo fibroblasts (16). Furthermore, overexpression of this gene potentiates the tumorigenicity of NIH 3T3 cells (15). The tumorigenic potential of MDM2 is closely linked to its ability to inhibit the growth suppression function of p53. MDM2 binds to the p53 N-terminal transactivation domain (7, 46) and inhibits its transcriptional activity in cells (42). MDM2 can also stimulate the ubiquitination (24) and proteolytic degradation of p53 (22, 31), although it is also proposed that MDM2 down-regulates p53 by concealing its transactivation domain (7, 8, 38). Targeting of MDM2 by MDM2 antisense oligonucleotides (10), an MDM2-binding miniprotein (5), and the tumor suppressor p19arf (50, 64) activates the p53 pathway. The physiological relationship between p53 and MDM2 is further supported by studies of p53/mdm2 double-knockout mice in which the early embryonic death of mdm2 null mice was rescued by further deleting the p53 gene (25, 44). Interestingly, the mdm2 gene itself is transcriptionally activated by p53 (2, 58), thus forming a unique autoregulatory feedback loop for restraining p53 activity.
Although p73 mimics p53 in transcriptional activation and apoptotic induction (27, 28), it is not induced in response to DNA damage signals (27a, 28), suggesting that p73 may either respond to different cellular signals or use different mechanisms to function in cells. Our recent study (63) showed that p73 is positively regulated by p300/CBP, which can mediate transactivation by a variety of transcriptional factors (reviewed by Shikama et al. [53]) including p53 (1, 20, 34). p73 and p53, however, were found to bind to different domains of the coactivators in order to activate transcription (63). Here we describe the negative regulation of the functions of p73 by MDM2. Like p53, p73 stimulates transcription of the mdm2 gene. MDM2 associates with p73 both in vitro and in vivo. Wild-type but not p53-binding-defective mutant MDM2 inhibits p73-dependent transcription, as measured in reporter gene assays, as well as p73-induced apoptosis in p53-deficient human tumor-derived cells. Interestingly, MDM2 disrupts the interaction of p73 but not p53 with p300/CBP in vitro. Conversely, MDM2 can reduce the protein level of p53 but not p73 in vivo. These results demonstrate that MDM2 negatively regulates the transcriptional activity of p73 through a mechanism distinct from that used for p53 regulation.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: CBP, CREB-binding protein; SDS, sodium dodecyl sulfate; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, hemagglutinin; DAPI, 4′,6-diamidino-2-phenylindole; IP, immunoprecipitation; NGS, normal goat serum; MDM2, a protein encoded by gene amplified in mouse double-minute chromosome; NBP, non-p53 p53RE binding protein; p53CP, p53 competing protein; p53RE, p53-responsive DNA element; P11, phosphocellulose; PBS, phosphate-buffered saline; PMSF, phenylmethylsulfonyl fluoride; WB, Western blotting; aa, amino acids; CAT, chloramphenicol acetyltransferase; NP-40, Nonidet P-40; TdT, terminal deoxynucleotidyl transferase.
Plasmids and antibodies.
The BP100CAT reporter plasmid contained two copies of the p53RE motif derived from the MDM2 promoter as described previously (58). The p53RE motifs were subcloned into the region upstream of the adenovirus major late TATA box/TdT initiation motif-CAT gene of the parental reporter plasmid p1634CAT (58). The pGST-MDM2 fusion protein-expressing plasmids were constructed as described previously (7). The pGST-p73 fusion protein expression vectors were constructed as described previously (63). The pCDNA3-HA-p73α and pCDNA3-HA-p73β plasmids encoding HA-tagged simian p73α and p73β, respectively, were described previously (27). pCHDMG58A and V75A encoding the human MDM2 mutants, G58A and V75A, respectively, were gifts from Deborah Freedman and Arnold J. Levine (17). The human p53 cDNA in pGEX2T-p53 (a gift from Peter Howley, Harvard Medical School) was excised by digestion with BamHI and EcoRI and ligated into pCDNA3-HA that had been cut with these two enzymes to make pCDNA3-Hap53 (40). pCOC-mdm2X2, encoding full-length mouse MDM2, and pCOC-mdm2ΔXM, encoding a mouse MDM2 deletion mutant lacking the first 61 aa, used for the experiments in Fig. 1D and 4, were described previously (3). pCMV-hMDM2 and pCMV-hMDM2Δ (lacking aa 58 to 89) expression vectors, used for the experiments in Fig. 1A to C, 2, and 6, were described previously (8, 59). pCNA3-GFP encoding GFP was a gift from Y. Haupt (Hebrew University).
FIG. 1.
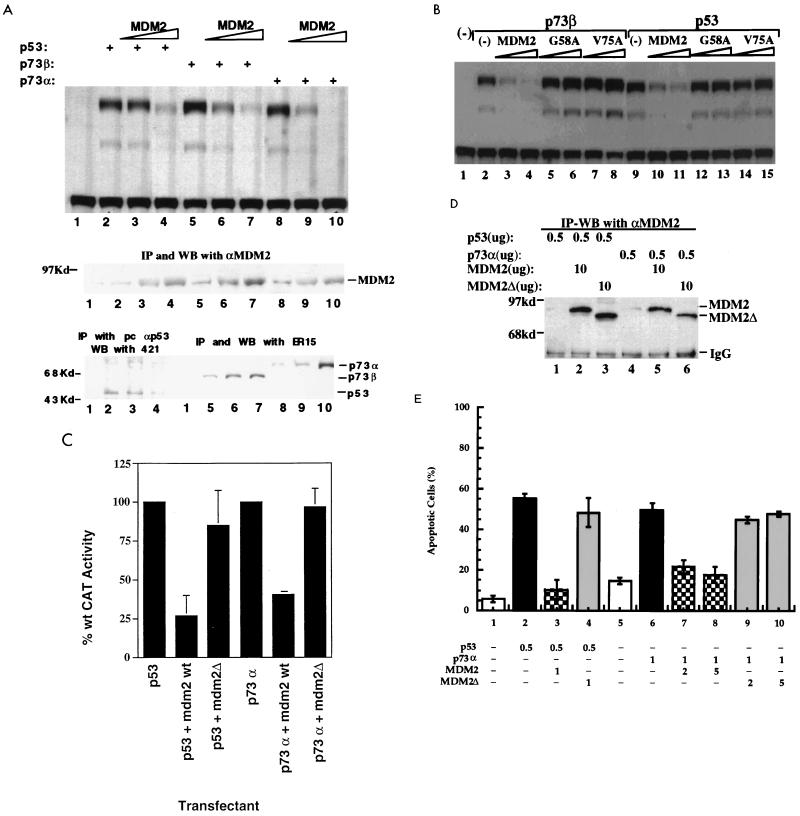
Wild-type but not mutant MDM2 inhibits the transcription mediated by p73. (A) Overexpression of MDM2 leads to decrease of p73-dependent transcription as measured by CAT activity (top panel). Saos-2 cells were transfected with expression vectors for p53 (200 ng), p73α (200 ng), and p73β (200 ng) and increasing amounts (0, 1.5 and 3.0 μg) of MDM2-expressing vectors as indicated at the top. The CAT assay was conducted with a CAT reporter plasmid driven by two copies of the p53RE motif derived from the MDM2 promoter as described in Materials and Methods. Expression of MDM2 (middle panel) and p53 and p73α/β (bottom panel) under the same conditions was detected by co-IP followed by WB with cell lysates of triplicate plates for each lane. The antibodies used for co-IP-WB are indicated above the panels. (B and C) MDM2 mutants fail to inhibit p73-dependent transcription in vivo. An experiment identical to that in panel A was carried out, except that two MDM2 mutants, G58A (replacement of Gly 58 by Ala) and V75A (replacement of Val 75 by Ala), were also included. Saos-2 cells were transfected with plasmids encoding HA-p53 or HA-p73α (500 ng each) in the presence or absence of plasmids encoding either wild-type (wt) or mutant hMDM2Δ (10 μg each), as indicated, along with a p53-responsive CAT reporter plasmid (4 μg) containing two p53-binding sites derived from the ribosomal gene cluster and a cytomegalovirus-driven β-galactosidase reporter plasmid (4 μg). Sufficient pBSK was added to bring the total to 24 μg. MDM2Δ lacks residues 58 to 89 (7). The raw CAT activity was corrected for β-galactosidase activity to normalize for differences in transfection efficiency. To facilitate comparison, the activities of p53 and p73 in the absence of MDM2 were both set to 100%. Error bars indicate the standard error of the mean. (D) IP-WB analysis of MDM2 and MDM2Δ expression in transfected Saos-2 cells. The same transient transfection as that in the experiment in panel C was conducted to detect MDM2 expression levels. 2A10 was used for IP, and polyclonal MDM2 antibodies were used for WB. IgG, immunoglobulin G. (E) MDM2 inhibits p73- and p53-induced apoptosis. H1299 cells (3 × 105/60-mm dish) were transfected with plasmids encoding proteins indicated at the bottom. The amounts used per transfection are indicated (in micrograms) in the lower part of the figure. Mouse MDM2 and N-terminally deleted MDM2 (MDM2Δ) were used in this experiment. A GFP expression plasmid (50 ng) was included in all the combinations. Transfected dishes were scored by fluorescence microscopy for the appearance of cells with distinct apoptotic morphology (rounding and shrinkage), 40 or 32 h after transfection for p53 or p73α, respectively. Only GFP-positive cells were counted. The percentage of apoptotic cells shown in each column represents the mean of three independent transfections; standard deviations are indicated. The total amount of DNA was kept constant by including an appropriate amount of empty vector DNA. Columns 1 and 5 are the vector-only controls for p53 (2 μg of vector DNA) and p73α (6 μg of vector DNA), respectively.
FIG. 4.
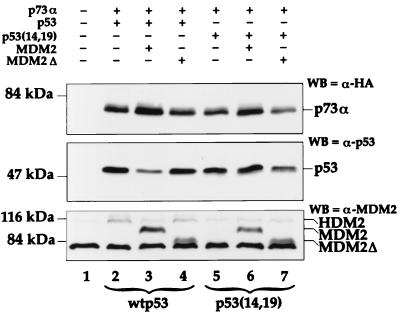
MDM2 does not induce rapid proteolytic degradation of p73. H1299 cells (4 × 105/60-mm dish) were transfected with the indicated combinations of the following expression plasmids (total DNA, 3 μg): pCDNA3 vector (3 μg/dish; lanes 1), HA-tagged p73α (0.5 μg/dish; lanes 2 to 7), wild-type human p53 (0.5 μg/dish; lanes 2 to 4), human p53 mutant p53L14N,F19S [p53(14,19), 0.5 μg/dish; lanes 5 to 7], full-length mouse MDM2 (2 μg/dish; lanes 3 and 6), or N-terminally deleted mouse MDM2 (MDM2Δ, 2 μg/dish; lanes 4 and 7). At 28 h posttransfection, the cells were harvested and extracted in protein sample buffer. WB was performed with either a monoclonal anti-HA antibody (HA.11 [Babco, Berkeley, Calif.]; WB=α-HA), a mixture of the p53-specific monoclonal antibodies Pab 1801 and Pab 421 (WB=α-p53), or 2A10 (WB=α-MDM2). The endogenous human MDM2 protein of the H1299 cells, migrating slower than the mouse MDM2, is also indicated (HDM2). The fast-migrating bands below the MDM2Δ are nonspecific signals detected by anti-MDM2 antibodies in the bottom panel.
The monoclonal anti-p73 antibody ER15 recognizes both p73α and p73β, as described previously (40). Monoclonal anti-p300 antibodies against the C-terminal domain of p300 were purchased from Upstate Biotechnology. Polyclonal anti-p53 antibodies were purchased from Santa Cruz. The polyclonal anti-MDM2 antibody was a gift from Arnold J. Levine. The anti-p53 antibody PAb421 and the anti-MDM2 antibody 2A10 were produced from cell clones generously offered by David Lane and Arnold J. Levine.
Buffers.
The following buffers were used for the experiments in Fig. 2–4. Lysis buffer consisted of 50 mM Tris-HCl (pH 8.0), 0.5% NP-40, 5 mM EDTA, 150 mM NaCl, and 1 mM PMSF. SNNTE is composed of 50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 1% NP-40, 500 mM NaCl, and 5% sucrose. RIPA is composed of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1% (wt/vol) sodium deoxycholate. Buffer B consists of 50 mM Tris-HCl (pH 7.9), 10% glycerol, 100 mM NaCl, 0.1 mM EDTA, 0.2 mM PMSF, 1 mM dithiothreitol, and 0.1% NP-40. Buffer C 100 (BC100) consists of 20 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM PMSF, 1 mM dithiothreitol, and 0.25 μg of pepstatin A per ml.
FIG. 2.
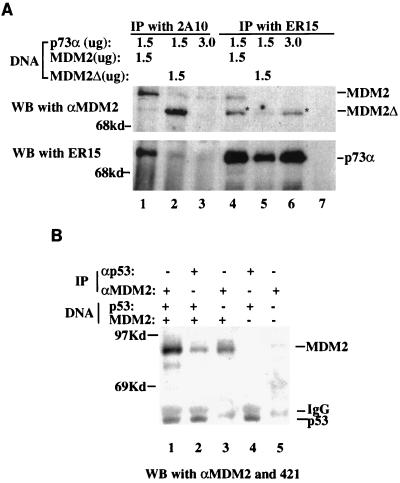
p73 binds to MDM2 in vivo. (A) Coimmunoprecipitation of p73 with MDM2. (10)1 cells (5 × 105/60-mm dish) were transfected with plasmids encoding HA-p73α or no insert, in the presence or absence of plasmids encoding either wild-type or mutant hMDM2Δ, as indicated at the top. hMDM2Δ lacks residues 58 to 89 (7). In this experiment, 1.5 μg of the relevant plasmids was used, and the parental pCMV vector was added to bring the total amount of plasmid DNA to 3 μg for each transfection. The antibodies used for IP and WB are indicated. Each lane presents a result from cell extracts of three dishes. αMDM2 denotes the polyclonal anti-MDM2 antibody. The strong enhanced chemiluminescence signals of p73α were due to a longer exposure (20 min) of the membrane when blotting with ER15. Because the same membrane was probed first with ER15 and then with anti-MDM2 antibodies, the slight bands comigrating with MDM2Δ in lanes 1, 4, and 5 were the remaining signals of p73α, since this band also appeared in lane 6, where only the p73α plasmid was added, but not in lane 7, where p73α was not included. Owing to the comigration of p73α with MDM2Δ, the results for MDM2 and p73α are presented separately. (B) Co-IP of p53 with MDM2. The same assay was performed, except that MDM2 (1.5 μg) and/or p53 (1.5 μg) expression plasmids were used. The parental vector was used to bring the total amount of plasmids to 3 μg where necessary. Anti-MDM2 antibodies used for IP and WB were 2A10 and polyclonal anti-MDM2 antibodies, respectively. Each lane is representative of the results from three dishes of cells. IgG, immunoglobulin G.
Cell culture.
Human osteosarcoma Saos-2, lung adenocarcinoma H1299, and murine embryonic fibroblast (10)1 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 50 U of penicillin per ml, and 0.1 mg of streptomycin per ml at 37°C in a 5% CO2 atmosphere. These cells are p53 deficient and contain no detectable p73, as analyzed by IP-WB (see Fig. 1A and 2) or WB (see Fig. 4).
Purification of recombinant MDM2 and Flag-p300.
Recombinant MDM2 and Flag-p300 proteins were purified from baculovirus-infected Sf9 insect cells by immunoaffinity chromatography with anti-MDM2 and anti-Flag antibodies as described previously (14, 54). Baculovirus encoding Flag-p300 was a kind gift from Hongwu Chen (Salk Institute).
Cotransfection, co-IP, and WB.
Saos-2 or (10)1 cells (4 × 105/100-mm plate) were transfected with the parental pCDNA3, pCDNA3-MDM2, or pCDNA3-MDM2Δ lacking aa 58 to 89 and/or with HA-p73α expression plasmids (the amounts of the plasmids used are indicated in the legend to Fig. 2) by using calcium phosphate or Lipofectamine (GIBCO). Transfected cells were harvested 48 h posttransfection for IP-WB as described below.
Cell lysates were prepared as described previously (39). Lysate containing 400 μg of proteins was precleaned with 30 μl of protein A-agarose (50% slurry) and then incubated at 4°C for 3 h with fresh protein A-beads (35 μl) in the presence of 2 μg of monoclonal antibodies specific for MDM2, p73, or p53. The beads were washed vigorously once with lysis buffer, twice with SNNTE, and once with RIPA and loaded directly onto an SDS–10% polyacrylamide gel. The immunoprecipitated proteins were detected by WB, as described (39, 63).
In vitro co-IP assays.
Increasing amounts of MDM2 purified from Sf9 cells were preincubated in ice for 15 min with in vitro-translated [35S]Met-labeled p73α, p73β, and p53, as indicated in the legend to Fig. 5, prior to addition of Flag-p300 purified from Sf9 cells. After a 40-min incubation at room temperature, the mixtures were blended with IP cocktails containing protein A-beads and anti-Flag antibodies and incubated for another 2 h at 4°C. After rigorous washing as described above, the bound p73 and p53 proteins were analyzed on an SDS-containing gel and detected by autoradiography after being transferred to a nitrocellulose membrane. This membrane was then probed with anti-p300 or anti-MDM2 antibodies for detecting p300 or MDM2 as indicated in the legend to Fig. 5.
FIG. 5.
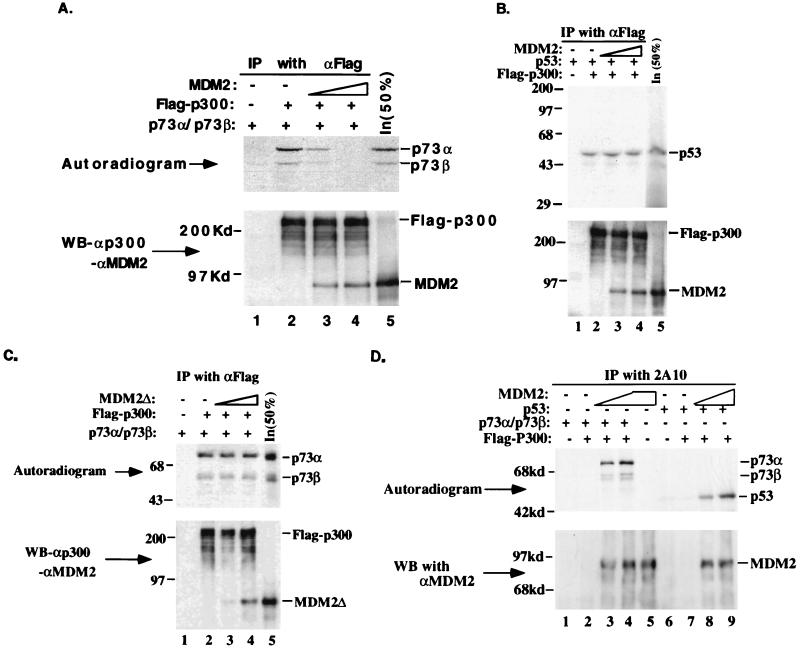
MDM2 blocks p73- but not p53-p300/CBP interactions. (A) MDM2 disrupts the p73-p300 interaction in an in vitro co-IP/protein competition assay. A 4-μl volume of in vitro-translated [35S]Met-labeled p73α and p73β, and 275 ng of the purified Flag-p300 protein were used in the assay. In lanes 3 and 4, 200 and 400 ng of MDM2 was added, respectively. The anti-Flag antibody used for IP is indicated at the top, and inputs (50%) of p73α and p73β as well as MDM2 (200 ng) are shown in lane 5. αp300 and αMDM2 indicate polyclonal anti-p300 and anti-MDM2 antibodies, respectively. The top panel is an autoradiogram of the same membrane for the WB in the bottom panel (the same is true for panels B to D). (B) MDM2 does not interfere with the p53-p300/CBP interaction. Experimental details were as for panel A and the corresponding test, except that 4 μl of in vitro-translated [35S]Met-labeled p53 was used. In lane 5, 50% of the p53 or MDM2 input was directly loaded. (C) The MDM2 mutant MDM2Δ does not interfere with the p73-p300 interaction. The same experiment was conducted, except that the N-terminally deleted mutant MDM2Δ was used as a competitor. For all these WB results, the strong signals were due to a longer exposure (15 min) to X-ray film. (D) Detecting p73- or p53-MDM2 complexes in the same experimental settings as those in panels A and B. The same IP-autoradiography-WB analysis was performed as described above, except the monoclonal anti-MDM2 antibody 2A10 was used for IP. The bound p73 or p53 was detected by autoradiography, and MDM2 was detected by WB with polyclonal anti-MDM2 antibodies, as indicated. The amount of p73, p53, MDM2, or p300 used in this analysis was the same as that in panels A to C.
Cotransfection and CAT assay.
Transfection of H1299 and Saos-2 cells was carried out as described above. For 60-mm plates, 4 μg of DNA was used (for the experiments in Fig. 1A and B), and the amounts of plasmids used in Fig. 1C are indicated in the legend. At 48 h after transfection, the transfected cells were harvested for CAT assays as described previously (26, 27, 58).
Cotransfection and immunofluorescent staining.
H1299 cells at 30% confluency were transfected with plasmids encoding p53, p73α, p73β, or no insert, by using Lipofectamine. The expression of MDM2 was detected by immunofluorescent staining with monoclonal anti-MDM2 2A9 antibody (7). Briefly, 24 h after transfection, cells were fixed for 20 min at room temperature with 4% paraformaldehyde in PBS. Then they were washed in PBS twice for 5 min to completely remove fixative, blocked with PBS–10% NGS for 20 min, and incubated with 2A9 at 1/100 dilution in PBS–10% NGS for 3 h. The slides were then washed with PBS–0.1% Triton X-100 three times for 5 min and incubated with a 1/400 dilution of fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G in PBS–10% NGS for 1 h. Following three 5 min washes with PBS–0.1% Triton X-100, the slides were incubated for 2 min with PBS–0.1% Triton X-100–0.1 μg of DAPI per ml. They were then mounted in the presence of anti-fade solution.
GST fusion protein association assay.
The fusion proteins were expressed in Escherichia coli and purified on a glutathione-Sepharose 12B column. Protein-protein association assays were conducted as described previously (37). A 10-μl volume of in vitro-translated 35S-labeled p73α, p73β, or MDM2 (TNT; Promega) was incubated with either the GST-MDM2 (for p73) or GST-p73 (for MDM2) fusion protein (400 ng) coupled to glutathione-Sepharose beads (50-μl total reaction volume). At 1 h after incubation at room temperature, the mixtures were washed once in buffer B, twice in SNNTE, and once in RIPA. The bound proteins were analyzed on an SDS–10% polyacrylamide gel and detected by autoradiography.
Apoptotic analysis.
H1299 cells (3 × 105/60-mm dish) were transfected with a plasmid encoding GFP together with combinations of expression plasmids (see Fig. 1E). Cultures were analyzed under a fluorescence microscope, and transfected cells were identified by the presence of green fluorescence. Apoptotic cells were identified by their rounded morphology, in contrast to the spread-out morphology of nonapoptotic H1299 cells. Apoptotic cells were counted and presented as a percentage of the total population of fluorescent cells (see Fig. 1E).
RESULTS
Wild-type but not inactive mutant MDM2 inhibits p73-dependent transcription in vivo.
To elucidate the relationship between MDM2 and p73, we first determined whether overexpression of MDM2 affects p73-dependent transcription in cells. Human p53-deficient osteogenic sarcoma Saos-2 cells were transfected with equal amounts (200 ng) of p53, p73α, and p73β expression vectors, together with increasing amounts (1.5 and 3.0 μg) of MDM2 expression vector, in the presence of a CAT reporter plasmid. This reporter plasmid was under the control of p53RE, derived from the MDM2 promoter (58). Consistent with previous studies (27, 28, 58, 63), each of these p53 family members stimulated CAT activity in a manner dependent upon the p53RE sequence (Fig. 1A, lanes 2, 5 and 8) (63), since these proteins were unable to do so when the p53RE-null CAT reporter plasmid was used (data not shown). In the presence of increasing levels of MDM2 (lanes 3, 4, 6, 7, 9, and 10 of the middle panel) as detected by IP-WB, the CAT activity in each case decreased significantly in a dose-dependent fashion (lanes 2 to 10 of top panel). Consistent with previous studies (22, 31), p53 levels were correspondingly reduced (lanes 3 and 4 of the bottom panel). In striking contrast, the levels of both p73α and p73β increased with increasing MDM2 levels (lanes 6, 7, 9, and 10 of the bottom panel). These results suggest that MDM2 can target both p53 and p73, inhibiting the transcriptional function of these proteins within cells, probably through distinct mechanisms, as described in more detail later.
Several MDM2 mutants with a single point mutation at the N terminus are inactive in repressing p53 transcriptional activity (17). Two of the mutants, G58A and V75A, were tested for their effect on the transcriptional activity of p73. As expected, these mutants failed to inhibit p53-dependent transcription (Fig. 1B, lanes 12 to 15). Similarly, G58A and V75A did not impact the activity of p73 (lanes 5 to 8), suggesting that these two residues in the N-terminal domain of MDM2 may also be critical for the inhibitory effect of MDM2 on p73. Consistent with this result, an MDM2 mutant lacking part of the N-terminal p53-binding domain (aa 58 to 89 deleted) was also unable to affect p73- and p53-dependent transcription (Fig. 1C). The different effects of the wild-type and mutant MDM2 proteins on transcription were not due to differences in expressed protein levels, since they all were expressed comparably in transfected cells (Fig. 1D and data not shown). Taken together, these results indicate that wild-type but not inactive mutant MDM2 can repress transcription mediated by p73, as has been previously described for p53 (7, 17, 46).
MDM2 alleviates p73-induced apoptosis.
p73 can induce apoptosis (27). To ask whether down-regulating p73-dependent transcription by MDM2 could abrogate this cellular response, p53-null human lung adenocarcinoma H1299 cells were transfected transiently with various combinations of expression vectors encoding p53, p73α, MDM2, MDM2Δ, or GFP, as described in Materials and Methods and in the legend to Fig. 1E. As expected (9, 21), wild-type but not N-terminally deleted mutant MDM2 inhibited p53-induced apoptosis (Fig. 1E, compare columns 3 and 4). As reported previously (27), p73 also induced apoptosis in transfected H1299 cells (column 6). In keeping with the inhibitory effect of MDM2 on p73-dependent transcription (Fig. 1B and C), wild-type (columns 7 and 8) but not mutant (columns 9 and 10) MDM2 significantly inhibited the apoptotic effect of p73. Thus, this inhibition requires an intact N-terminal domain of MDM2, as with p53 (columns 3 and 4). Of note, while the inhibitory effect of MDM2 on p73-induced apoptosis was significant, it was not as complete as that seen for p53 (compare column 3 to columns 7 and 8). This may be accounted for by the observation that, in contrast with p53, the p73 level was elevated in the presence of MDM2 (Fig. 1A; also see Fig. 2A and 4). Alternatively, p73 may induce apoptosis partly through a nontranscriptional pathway; therefore, concealing the p300-binding domain of p73 (see Fig. 5) and subsequently inhibiting its transcription (Fig. 1) may not lead to a complete inhibition of this more complicated cellular process by MDM2. Taken together, these results demonstrate that MDM2 can inhibit p73-dependent transcription and alleviate p73-induced apoptosis and that the N-terminal domain of this oncoprotein is crucial for these negative regulatory effects.
Association of p73 with MDM2 in vivo.
To test whether the inhibitory effect of MDM2 on p73-dependent transcription and apoptosis is accomplished through its direct interaction with p73 in cells, we performed an IP-WB analysis of murine p53 null embryonic fibroblast (10)1 cells transfected with HA-p73α in the presence of plasmids encoding either wild-type or N-terminally deleted MDM2 lacking aa 58 to 89. Proteins were coimmunoprecipitated with the monoclonal anti-p73 antibody ER15 and anti-MDM2 antibody 2A10, respectively, followed by WB with ER15 and polyclonal anti-MDM2 antibodies. As shown in Fig. 2A, p73α was coimmunoprecipitated by the monoclonal anti-MDM2 antibody when coexpressed with wild-type MDM2 (lane 1). Reciprocally, MDM2 was coimmunoprecipitated by the monoclonal anti-p73 antibody (lane 4), which recognizes the C-terminal domain of p73 (40). In agreement with the above observation that the MDM2 N-terminal mutants failed to inhibit p73-dependent transcription and apoptosis in vivo (Fig. 1), the p73-MDM2 interaction was not detected by the same sets of antibodies when p73α was coexpressed with the N-terminally deleted mutant MDM2Δ (lanes 2 and 5). The faint bands comigrating with wild-type MDM2 and p73α in lane 2 were probably due to the interaction of p73α with the endogenous MDM2 protein induced by this transcriptional factor (see Fig. 6), since this was also seen in lane 3, where only p73α was included, but not in a blank control (lane 7). Consistent with the previous study (7, 8), the p53-MDM2 interaction was expectedly detected by co-IP-WB analysis with either the anti-MDM2 or anti-p53 antibodies when p53 was co-expressed with MDM2 (Fig. 2B). Equal amounts of p53- and MDM2-expression plasmids were used in this specific experiment for the purpose of an efficient co-IP analysis. The p73-MDM2 binding was also observed with plasmids encoding p73β (data not shown) or when Saos-2 cells were used (27a). Taken together with the data discussed above (Fig. 1), it can be concluded that MDM2 binds to p73 within cells and subsequently inhibits p73-mediated transcription and apoptosis.
FIG. 6.
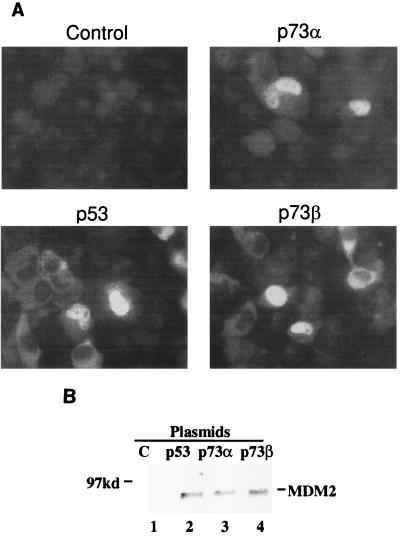
p73α and p73β, like p53, induce the expression of the endogenous MDM2 protein in transfected H1299 cells. (A) Immunofluorescent staining of the endogenous MDM2. Cells were transiently transfected with p73 or p53 expression plasmids and then subjected to immunofluorescent staining with the anti-MDM2 monoclonal antibody 2A9 as described in Materials and Methods. The vectors used in the experiment are indicated at the top of each panel. Similar results were obtained in two independent experiments. (B). IP-WB analysis of the endogenous MDM2. H1299 cells (106/100-mm dish) were transfected with plasmids (15 μg/transfection) encoding p53, p73α, and p73β, respectively. MDM2 was detected by IP with 2A10 followed by WB with polyclonal anti-MDM2 antibodies. Each lane is representative of the result from three dishes of cells. Lane 1 contains the vector only.
p73 binds to the N-terminal domain of MDM2.
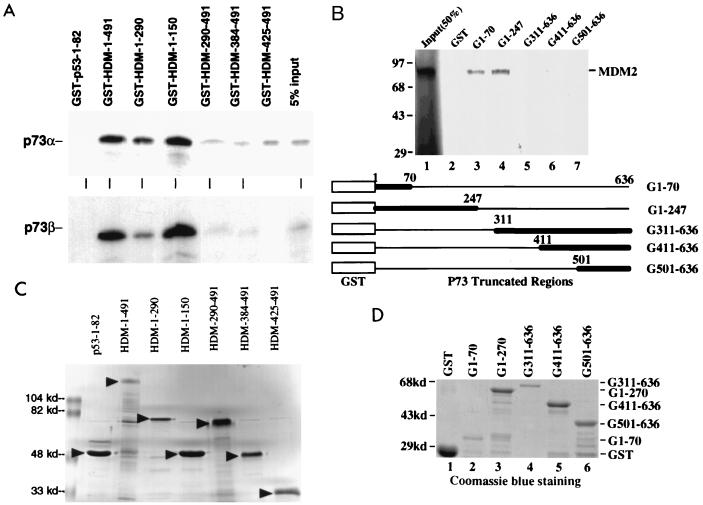
The above result suggests that the N terminus of MDM2 may be important for the MDM2-p73 association and functional relationship. To map unequivocally the p73-binding site on the MDM2 protein, a GST-protein association assay was conducted. In vitro translated [35S]Met-labeled p73α and p73β proteins were incubated with immobilized GST fusion proteins containing different portions of MDM2 or a p53 N-terminal region (Fig. 3C). After vigorous washing, the bound p73 proteins were subjected to SDS-polyacrylamide gel electrophoresis and detected by autoradiography (Fig. 3A). Both p73α and p73β bound to the GST–wild-type MDM2 protein (lane GST-HDM-1-491) as well as to the MDM2 N-terminal domain (aa 1 to 150) (lane GST-HDM-1-150). This binding was specific to the MDM2 N-terminal domain, since binding of p73 to three C-terminal fragments of MDM2 was barely detectable (lanes GST-HDM-290-491 to GST-HDM-425-491). Also, as a control, the GST-p53 N-terminal region did not retain any p73 (lane GST-p53-1-82). Hence, these results demonstrate that in agreement with our in vivo data (Fig. 2), both p73α and p73β bind to the MDM2 N terminus.
FIG. 3.
Mapping the protein interaction domains for MDM2 and p73. (A) Both p73α and p73β bind to the N terminus of MDM2. The indicated GST-MDM2 fusion proteins, immobilized on Sepharose, were incubated with 10 μl of in vitro-translated 35S-labeled p73α and p73β at room temperature for 1 h. Bound proteins were analyzed as described in Materials and Methods. As a control, 5% of the input was directly loaded on the last lane. Binding between GST-HDM-425–491 and p73β was not done due to the redundancy (see lanes for GST-290-491 and GST-384-491). (B) MDM2 binds to the N-terminal domain of p73 (aa 1 to 70). A set of GST pull-down experiments similar to those in panel A were performed, except that GST-p73 deletion fusion proteins (indicated at the top) were used in this case. In lane 1, 50% of MDM2 input (35S labeled) was loaded directly. The GST-p73 truncation fusion proteins are schematically drawn at the bottom of the panel, and their expression is shown in panel D. (C) Expression of GST-MDM2 fusion proteins used in panel A. GST-MDM2 fusion proteins (10 μl; 50% slurry) were loaded directly onto an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue. (D) Expression of GST-p73 fusion proteins used in panel B. GST- or GST-p73 fusion protein-containing beads (10 μl; 50% slurry) were directly analyzed on an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue. The amounts of these proteins used in panels A and B were normalized by comparing the intensity of the bands with BSA standards; 400 ng of each of these proteins was used for GST-pull down assays.
MDM2 binds to the N-terminal domain of p73.
The fact that MDM2 targets the p53 N-terminal transactivation domain (7) and that this domain has 29% amino acid identity to that of p73 (28) suggested that MDM2 may also contact the p73 N terminus. To define the MDM2-binding site on p73, we generated a set of p73 deletion mutants fused with GST, as shown in Fig. 3D. A similar GST pull-down assay to that described above was carried out, except that GST-p73 fusion proteins and in vitro-translated [35S]Met-labeled wild-type MDM2 were used. As shown in Fig. 3B, MDM2 bound to both the N-terminal fragments of p73 (lanes 3 and 4) but not to three C-terminal regions (lanes 5 to 7). The interactions of MDM2 with the fragments of p73 from aa 1 to 70 and 1 to 247 were approximately equivalent, suggesting that the N-terminal domain from aa 1 to 70 is the MDM2-binding site. As a control, GST alone did not retain any MDM2 (lane 2). Thus, this result, similar to the MDM2-p53 interaction (7), indicates that MDM2 targets the N-terminal region of p73. Since p300/CBP also contacts the same N-terminal domain of p73 and stimulates the transcriptional activity of this transcriptional factor (63), it is likely that MDM2 inhibits the function of p73 by competing with it for p300/CBP.
MDM2 does not reduce p73 protein levels.
To further examine the effect of MDM2 on p73 protein expression, H1299 cells were transiently transfected with a mixture of p73α and either wild-type p53 or the mutant p53(14,19) in the presence or absence of different forms of mouse MDM2. This mutant p53, carrying mutations of L14N and F19S, was used as a control because it does not bind to MDM2 but retains partial transcriptional activity (35). In these cells, transfected MDM2 leads to a marked reduction in steady-state p53 levels (22) (Fig. 4, middle panel, compare lane 3 with lanes 2 and 4). The p53 reduction was caused specifically by wild-type MDM2, since it did not occur when the p53-binding-defective MDM2 mutant MDM2Δ (lane 4) was used. This reduction also requires a direct contact with p53, since it spared the MDM2-binding-defective p53(14,19) mutant (middle panel, lanes 5 to 7). Of note, MDM2-mediated p53 degradation is dose dependent (references 22 and 31 and data not shown); this explains why it was easily detected when excess MDM2 expression plasmid was used in transient-transfection experiments (Fig. 1 and 4 and data not shown) but not when equal amounts of p53 and MDM2 plasmid DNA were used (Fig. 2B).
In contrast, in the same transfected cell culture, coexpression of full-length MDM2 did not cause a reduction in the steady-state level of p73; rather, it actually resulted in an increase in the level of p73 protein (Fig. 4, top panel, compare lanes 3 and 2), regardless of the presence or absence of wild-type p53 (lanes 3 and 6). This is consistent with the result in Fig. 1A. This increase was even visible when equal amounts of MDM2 and p73 plasmids were used (Fig. 2A, compare lanes 4 and 5; of note, twice as much p73 plasmid was added in lane 6 as in lanes 4 and 5). By contrast, an N-terminally deleted mutant of MDM2, which is unable to bind to p73 (Fig. 2 and 3), failed to increase the steady-state p73 level (Fig. 4, compare lanes 4 and 7 with lanes 2 and 5 of the top panel). Hence, MDM2 cannot promote the degradation of p73. The observed increase in the level of p73 implies that p73 may be degraded through a mechanism that does not require MDM2 but is somehow inhibited by excess MDM2. Alternatively, p73 might exert a detrimental effect on the transfected cells, leading to selective loss of p73-expressing cells. If MDM2 can alleviate this deleterious effect (Fig. 1D), efficient p73 expression might be maintained in the culture, resulting in an apparent increase in the overall levels of transfected p73. The observations that MDM2 inhibits p73-dependent transcription and apoptosis (Fig. 1) without reducing the protein level of this transcriptional activator (Fig. 1 and 4A) suggest that MDM2 regulates p73 through a mechanism distinct from that used for p53.
MDM2 disrupts the interaction of p73 but not of p53 with p300/CBP.
Given that both MDM2 (Fig. 3B) and p300/CBP (63) bind to the N terminus of p73 and that p300/CBP can mediate p73-dependent transcription in vivo (63), we tested whether MDM2 could compete with p300 for binding to p73. This was assessed by using in vitro co-IP/protein competition assays, with p53 serving as a control. The MDM2 and Flag-p300 proteins were purified from baculovirus expression systems by immunoaffinity chromatography with anti-MDM2 and anti-Flag antibodies, respectively (14, 54). Increasing amounts of MDM2 protein were preincubated for 15 min on ice with in vitro-translated [35S]Met-labeled p73α, p73β, or p53 prior to addition of equal amounts (275 ng) of Flag-p300. Following further incubation and subsequent IP with anti-Flag antibodies, bound proteins were analyzed by IP and SDS-polyacrylamide gel electrophoresis and detected by autoradiography after the proteins were transferred to a nitrocellulose membrane (Fig. 5A to C, top panels), and the bound p300 and MDM2 were monitored by WB with polyclonal anti-p300 and anti-MDM2 antibodies after IP (Fig. 5A to C, bottom panels). As shown in Fig. 5, both p73α and p73β (Fig. 5A and C, lanes 2) and p53 (Fig. 5B, lane 2) were coimmunoprecipitated by the anti-Flag antibody, confirming the interaction of p73 and p53 with p300 in vitro. The p73-p300 interaction was gradually eliminated by increasing the amounts of wild-type (Fig. 5A, compare lane 2 with lanes 3 and 4) but not N-terminally deleted (Fig. 5C, lanes 2 to 4) MDM2 protein. 400 ng of MDM2 (lane 4) completely blocked the interaction of p300 with either p73α or p73β. Under the same experimental conditions, MDM2-p73α or MDM2-p73β complexes were detected by IP followed by autoradiography with anti-MDM2 antibodies (Fig. 5D, lanes 2 to 4, corresponding to Fig. 5A, lanes 2 to 4), indicating that MDM2 can compete with p300 for p73. In contrast, the same amount of MDM2 was unable to interfere with the p53-p300 association (Fig. 5B, compare lane 2 with lanes 3 and 4), although MDM2 also formed a complex with p53 as detected by IP with anti-MDM2 antibodies followed by autoradiography (Fig. 5D, lanes 7 to 9, corresponding to Fig. 5B, lanes 2 to 4). This may be explained by the observation that p53 can bind to both the N (19, 63) and C (20, 63) termini of p300/CBP while p73 and MDM2 bind exclusively to the N termini of these coactivators (reference 63 and data not shown). The possible ternary complex containing p53, MDM2, and p300 has been shown to promote p53 degradation in vivo (19). Consistent with the observation that p300 associated with the MDM2 aa 110 to 220 fragment (19), MDM2Δ, lacking the region from aa 58 to 89, was also found to bind to p300 (Fig. 5C, lanes 3 and 4). However, this mutant was unable to compete with p73 (Fig. 5C), suggesting that the interactions of p73 with MDM2 and p300 are mutually exclusive. Thus, concealing the p300-binding domain of p73 by MDM2 appears to be one possible mechanism underlying the inhibitory effect of MDM2 on the transcription function of p73.
p73 stimulates the expression of MDM2 in vivo.
MDM2 has been documented to function in cells as a natural p53 inhibitor in an autoregulatory feedback loop (22, 31, 58). The finding that MDM2 also inhibits p73-dependent transcription and apoptosis in vivo (Fig. 1) raises the possibility that MDM2 acts as a feedback regulator for p73 as well if p73 can induce MDM2 expression. To test this hypothesis, expression of the endogenous MDM2 protein was examined by introducing exogenous p73α, p73β, p53 (a positive control) and parental vector (a negative control) plasmids, respectively, into H1299 cells, followed by immunofluorescence staining for MDM2. Basal expression levels of MDM2 were low in these p53-null cells. As expected, MDM2-specific immunofluorescence was hardly visible in the vector-transfected cells (Fig. 6A, Control). In contrast, introduction of both p73α and p73β into the cells stimulated significant expression of the endogenous MDM2 protein (Fig. 6A, p73α and p73β). This is consistent with previous reports showing that p73 transactivates the expression of two other p53 target genes, those encoding p21 and Bax-1 (27, 28). As expected, p53 induced MDM2 expression (Fig. 6A, p53). The induction of endogenous MDM2 by p73 and p53 was also confirmed by IP-WB with anti-MDM2 antibodies (Fig. 6B). Hence, MDM2 is also a p73 target gene. p73 and p53 can thus transactivate the expression of MDM2 and share the MDM2 autoregulatory feedback loop.
DISCUSSION
The MDM2 negative regulation feedback loop (2, 42, 58; see references 36, 43, and 48 for reviews) has been considered unique for p53 regulation. The present study indicates that this loop is also used by the p53 analog p73. Supporting this notion are several lines of evidence. First, p73α or p73β, when overexpressed in p53-deficient H1299 cells, induced expression of the endogenous MDM2 protein (Fig. 6). Second, p73 associated with MDM2 in vivo, as shown by a transient-transfection/co-IP-WB analysis (Fig. 2). This interaction was confirmed by in vitro IP (Fig. 5) and GST-protein association assays (Fig. 3). Similar to p53 (7, 46), the N-terminus of p73 bound to the MDM2 N terminus. Finally, this interaction correlated with inhibition of p73-dependent transcription, as measured in reporter gene assays (Fig. 1A to C), and with abrogation of p73-induced apoptosis (Fig. 1E). Together, these results imply that expression of MDM2 can be markedly stimulated by p73, leading to inhibition of p73-mediated transcription and apoptosis and thereby forming an MDM2 feedback loop for p73 regulation.
p53 binds to and utilizes one group of coactivators, p300 and CBP, for its transcriptional activation (1, 20, 34). In a parallel study, we found a synergy between p300/CBP and p73 in activation of the p53RE-containing MDM2 promoter (63). However, p73 and p53 use different domains of these coactivators for their transcriptional functions (63). This difference may be correlated with the distinct mechanisms by which MDM2 negatively regulates the transcriptional functions of p73 and p53, as discussed below.
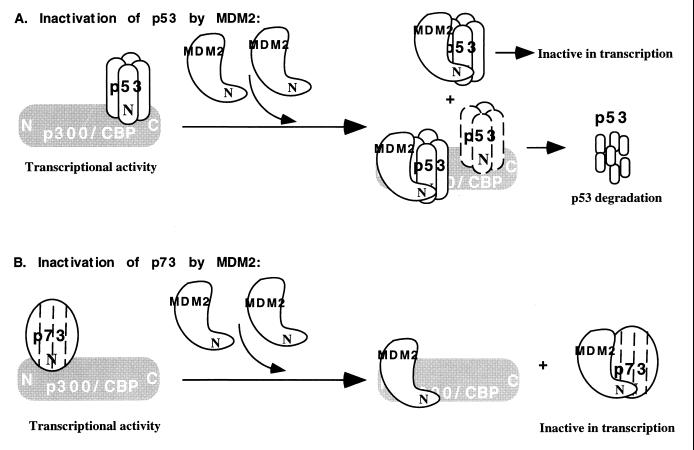
Although p73 and p53 share the MDM2 autoregulatory feedback loop, the underlying molecular mechanisms appear to be distinct for these two members of the p53 family, as schematically presented in Fig. 7. In addition to possibly blocking the interaction of p53 with the components of the RNA polymerase II transcriptional machinery (37, 38, 55, 56) and/or p300/CBP, MDM2 can enhance the rapid proteolytic degradation of p53 (5, 22, 31), probably through the ubiquitin-proteasome pathway (24). Direct binding between the N termini of p53 and MDM2 is critical for p53 degradation (5, 19, 22, 31) (Fig. 4), and the p53 C terminus also plays a role in regulating its sensitivity to the MDM2-mediated proteasome pathway (24, 32). This pathway contributes to the inhibitory regulation of p53 by MDM2 in cells. By contrast, p73 steady-state levels were not reduced by coexpression of MDM2 (Fig. 1A and 4). Instead, MDM2 disrupted the interaction of p73 with p300 (Fig. 5A) but was without effect on the p53-p300 complex in vitro (Fig. 5B). This is probably due to the competition of MDM2 with p73, but not with p53, for binding to p300, since MDM2 and p73 both interact exclusively with the same N-terminal domain of p300/CBP (Fig. 3 and data not shown) whereas p53 binds to either the N or C terminus of the coactivators (19, 63). Consistent with this in vitro observation, the association of p300 with MDM2 and p53 was detected in cells by a glycerol gradient sedimentation analysis (19). Coexpression and association of MDM2 with p300 was found to further stimulate p53 degradation (19). This cannot occur to p73, since associations of p73 with MDM2 and with p300/CBP appears to be mutually exclusive (Fig. 3B and 5A) and overexpression of MDM2 virtually leads to p73 stabilization (Fig. 1A and 4). Although MDM2 probably cooperates with p300 in degradation of p53 (19), MDM2 does not appear to influence the steady-state level of the coactivator (63a). In summary, these results suggest that MDM2 represses the transcriptional function of p73, probably by interrupting its contact with p300/CBP but not through a proteolytic pathway. MDM2 has recently been proposed to inhibit the transcription of p53 by binding to components, such as TFIIE and TBP, of the RNA polymerase II machinery (56). Whether this is also an alternate mechanism for regulating the function of p73 remains to be addressed.
FIG. 7.
Schematic presentation showing distinct mechanisms by which MDM2 inactivates p53 (A) and p73 (B). p53 with dashed lines in panel A indicates that this protein may or may not be in the ternary complex. See Discussion for details. This model does not address the suggested alternative mechanism for p53 inactivation by MDM2, namely, that MDM2 interferes with the binding of other transcription factors, e.g., hTAFII31 and hTAFII70, to the N-terminal transcription domain of p53 (37, 38, 55). Also, binding of p53 to the N terminus of p300/CBP (19, 63) in the absence of MDM2 is not shown.
It is perplexing why MDM2 adopts different mechanisms to down-regulate the functions of these two p53 family members. On the one hand, this may reflect diverse protein-protein interactions among members of this family and their partners. For example, while the interaction between p73 and p300/CBP appears to involve a single protein-protein interface formed by the N termini of these proteins, the binding of p53 to these coactivators involves multiple contacts. Thus, concealing the binding domain of the coactivators from transcriptional activators is sufficient for MDM2 to suppress the transcription activity of p73 but not of p53. Therefore, MDM2 has evolved an additional mechanism to inactivate p53 through ubiquitin-mediated proteolytic degradation. On the other hand, it is possible that the different mechanisms for MDM2-mediated down-regulation of p53 or p73 are correlated with their distinct cellular roles. The major role of p53 is to serve as a cellular gatekeeper (33). Thus, it responds to cellular stress at both the protein and activity levels, subsequently triggering apoptosis or growth arrest (18, 29, 33). The dramatically increased p53 protein might be detrimental to cells if it is not tightly controlled or quickly removed. This tight regulation of p53 is executed by MDM2, perhaps largely by accelerating the proteolytic degradation of p53. By contrast, although its biological role is unclear, the level of p73 is not responsive to stress signals (28). Hence, it might be unnecessary for MDM2 to adopt a destructive strategy to control p73 function.
In spite of their different responses to DNA damage signals (18, 28, 29, 33), both p73 and p53 appear to turn on a rather similar set of downstream genes (27, 28) (Fig. 5), both induce apoptosis (27) (Fig. 1D), and both are down-regulated by the same cellular protein MDM2 (Fig. 1). Even though p73 itself may not act as a tumor suppressor (30, 40), these remarkable similarities between the two proteins suggest new directions for developing novel therapies. For instance, it may be reasonable to activate the p73 protein by targeting MDM2 in tumors that contain no or altered p53 but significant levels of p73. It is possible that MDM2 regulates p73 in a cell-specific manner, since the p73 protein was detected only in a limited subset of cells or tissues (30, 40, 63a). Thus, a systematic investigation of the p73-MDM2 and p73-p53 relationship in cells, along with elucidation of the p73 signalling pathway, not only will be crucial for a better understanding of the cellular role of this protein but also will be instructive for possible anti-cancer drug discovery.
ACKNOWLEDGMENTS
We thank Deborah Freedman, Lin Wu, Angie Tereskey, Arnold J. Levine, Kristen Walker, Hongwu Chen, Ronald Evans, Roland Kwok, James Lundblad, Richard Goodman, David Livingston, Zhi-Xiong Xiao, and David Lane for anti-MDM2 antibodies, MDM2 mutants, CBP plasmids, anti-Flag antibodies, Flag-p300-expressing baculovirus, p300 plasmids, MDM2 plasmids, and the Pab421 cell clone. We thank Richard Goodman, Matt Thayer, and Buddy Ullman for stimulating discussion and critically reading the manuscript.
W. G. Kaelin, Jr., is an Assistant Investigator of the Howard Hughes Medical Institute. M. Oren was supported by the Israel Science Foundation, Israel Academy of Sciences and Humanities—Centers of Excellence Program. J. Chen was supported by an NIH grant. This work was supported partly by grants to H. Lu from the American Cancer Society (RPG-98-191-01-CBE), NIH (RO1 CA 79721-01), Oregon Cancer Center, Medical Research Foundation of Oregon, and Oregon Division of American Cancer Society.
REFERENCES
- 1.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 2.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 4.Bian J, Sun Y. P53CP, a putative p53 competing protein, that specifically binds to the consensus p53 DNA binding sites: a third member of the p53 family? Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottger A, Bottger V, Sparks A, Liu W L, Howard S F, Lane D P. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 6.Cahilly-Snyder L, Yang-Feng T, Francke U, George D L. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somatic Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Marechal V, Levine A J. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Lin J, Levine A J. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Wu X, Lin J, Levine A J. Mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Agrawal S, Zhou W, Zhang R, Chen J. Synergistic activation of p53 by inhibition of mdm2 expression and DNA damage. Proc Natl Acad Sci USA. 1998;95:195–200. doi: 10.1073/pnas.95.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordon Cardo C, Latres E, Drobnjak M, Oliva M R, Pollack D, Woodruff J M, Marechal V, Chen J, Brennan M F, Levine A J. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 12.Dulic V, Kaufmann W K, Lees S J, Tisty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine A J. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 15.Fakharzadeh S S, Trusko S P, George D L. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finlay C A. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman D A, Epstein C B, Roth J C, Levine A J. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta Gene Struct Expression. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 19.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z, Kumar S, Howley P M, Livingston D M. P300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 21.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 22.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H, Lengauer C, Polyak K, He T, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3ς is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 24.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 25.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 26.Jost C A, Ginsberg D, Kaelin W J. A conserved region of unknown function participates in the recognition of E2F family members by the adenovirus E4 ORF 6/7 protein. Virology. 1996;220:78–90. doi: 10.1006/viro.1996.0288. [DOI] [PubMed] [Google Scholar]
- 27.Jost C A, Marin M C, Kaelin W G. p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 27a.Jost, C. A., and W. G. Kaelin. Unpublished data.
- 28.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J, Valent A, Minty A, Chalon P, Lelias J, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 29.Ko J L, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 30.Kovalev S, Marchenko N, Swendeman S, LaQuaglia M, Moll U M. Expression level, allelic origin, and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ. 1998;9:897–903. [PubMed] [Google Scholar]
- 31.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 32.Kubbutat M H, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 34.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Chen J, Elenbass B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to MDM2 and the adenovirus 5 E1B 55-kd protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 36.Lozano G, de Montes O L. MDM2 function. Biochim Biophys Acta Rev Cancer. 1998;1377:M53–M57. doi: 10.1016/s0304-419x(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 37.Lu H, Levine A J. Human TAF-31 is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H, Lin J, Chen J, Levine A J. Regulation of p53 transcriptional function: TAFs and MDM2. Harvey Lect. 1996;90:81–93. [PubMed] [Google Scholar]
- 39.Lu H, Taya Y, Ikeda M, Levine A J. Phosphorylation of p53 at serine 389 is responsive uniquely to UV- but not to gamma- and etoposide-induced DNA damage. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marin M C, Jost C A, Irwin M S, DeCaprio J A, Caput D, Kaelin W G. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 42.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 43.Momand J, Zambetti G P. Mdm-2: “big brother” of p53. J Cell Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 44.Oca Luna R M, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 45.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 46.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 47.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimira Y, Nakagawara A, Obinata M, Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 48.Piette J, Neel H, Marechal V. Mdm2: keeping p53 under control. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- 49.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 50.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 51.Prabhu N S, Somasundaram K, Satyamoorthy K, Herlyn M, El-Deiry W S. p73beta, unlike p53, suppresses growth and induces apoptosis of human papillomavirus E6-expressing cancer cells. Int J Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 52.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 53.Shikama N, Lyon L, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 54.Tharakan J. Immunoaffinity purification. In: Malik V S, Lillehoj E P, editors. Antibody techniques. New York, N.Y: Academic Press, Inc.; 1994. pp. 327–340. [Google Scholar]
- 55.Thut C J, Chen J L, Klemin R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 56.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- 58.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 59.Xiao Z, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 60.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. P21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 61.Yang A, Kaghad M, Wang Y, Gillett E, Fleming M D, Dotsch V, Andrews N C, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 62.Zeng X Y, Levine A J, Lu H. Non-p53 p53RE binding protein, a human transcription factor functionally analogous to p53. Proc Natl Acad Sci USA. 1998;95:6681–6686. doi: 10.1073/pnas.95.12.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng, X. Y., A. Miller, W. Yuan, R. Kwok, R. Goodman, and H. Lu. p73 and p53 activate transcription through interaction with different domains of p300/CBP. Submitted for publication.
- 63a.Zeng, X. Y., and H. Lu. Unpublished data.
- 64.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]