Abstract
Ku is a heterodimeric protein with double-stranded DNA end-binding activity that operates in the process of nonhomologous end joining. Ku is thought to target the DNA-dependent protein kinase (DNA-PK) complex to the DNA and, when DNA bound, can interact and activate the DNA-PK catalytic subunit (DNA-PKcs). We have carried out a 3′ deletion analysis of Ku80, the larger subunit of Ku, and shown that the C-terminal 178 amino acid residues are dispensable for DNA end-binding activity but are required for efficient interaction of Ku with DNA-PKcs. Cells expressing Ku80 proteins that lack the terminal 178 residues have low DNA-PK activity, are radiation sensitive, and can recombine the signal junctions but not the coding junctions during V(D)J recombination. These cells have therefore acquired the phenotype of mouse SCID cells despite expressing DNA-PKcs protein, suggesting that an interaction between DNA-PKcs and Ku, involving the C-terminal region of Ku80, is required for DNA double-strand break rejoining and coding but not signal joint formation. To gain further insight into important domains in Ku80, we report a point mutational change in Ku80 in the defective xrs-2 cell line. This residue is conserved among species and lies outside of the previously reported Ku70-Ku80 interaction domain. The mutational change nonetheless abrogates the Ku70-Ku80 interaction and DNA end-binding activity.
DNA-dependent protein kinase (DNA-PK) is a complex comprising the heterodimeric Ku protein, which consists of subunits of 70 and 80 kDa (Ku70 and Ku80), and a large catalytic subunit, DNA-PKcs (7, 14). Ku has non-sequence-specific double-stranded DNA (dsDNA) end-binding activity and, when DNA bound, can interact with DNA-PKcs, enhancing its kinase activity. DNA-PK functions in a major pathway for rejoining DNA double-strand breaks (DSBs) in mammalian cells termed nonhomologous end joining (NHEJ) (for reviews, see references 18 and 23). This process is additionally used to recombine the site-specific DNA DSBs introduced during the process of V(D)J recombination. Cell lines defective in components of DNA-PK are both radiation sensitive, since the major lethal lesion induced by ionizing radiation is a DNA DSB, and defective in their ability to carry out V(D)J recombination (19). Compatible with these cellular studies is the finding that mice with disrupted components of DNA-PK exhibit a severe combined immunodeficiency (SCID) phenotype and display radiation sensitivity (1, 3, 11, 17, 31, 49). However, SCID mice, which are defective in DNA-PKcs, have a less severe V(D)J recombination defect than do Ku-defective mice and cell lines. Mutants with mutations in Ku are defective in recombining the two distinct types of junctions that arise during the process, namely, signal and coding joints. In contrast, signal joint formation can proceed largely unimpaired in mutants defective in DNA-PKcs, although the possibility remains that there is some residual kinase activity in the DNA-PKcs-defective mutants analyzed to date which is sufficient for signal joint but not coding-joint formation. This caveat notwithstanding, it appears that Ku has a function independent of its role as a component of DNA-PK (for reviews, see references 12, 23, and 43).
Neither Ku70 nor Ku80 cDNA has any obvious motifs (28, 37, 48). Ku80 appears to require association with Ku70 for DNA binding, although Ku70 has been reported to show Ku80-dependent and -independent DNA binding (45). Ku has been reported to have ATP-dependent helicase activity, but both Ku70 and Ku80 lack a helicase domain (44). Ku has been reported to have ATPase activity following autophosphorylation, but mutations in putative ATP-binding sites do not impair the ability of Ku70 or Ku80 to correct defective cell lines (4, 24, 39). Several approaches can provide insight into the identification of functional domains within Ku70 and Ku80. Multiple rodent cell lines defective in Ku80 have been reported, and identification of the causal mutational changes could potentially provide insight into functionally important domains. To date, however, such analysis has shown that either the mutants lack Ku80 expression or the mutations result in large deletions or truncations due to a changed reading frame (9, 29, 39). Although important in verifying that Ku80 is the gene defective in group 5 mutants, these studies have provided little insight into important functional domains. Other studies have examined the regions of Ku70 and Ku80 required for heterodimer formation by using the two-hybrid system and by analysis of in vitro-expressed cDNA fragments (5, 32, 46). The results show some discrepancies but, taken together, define a minimal domain of 28 amino acids from the central region of Ku80 (residues 449 to 477) that is required for Ku70-Ku80 interaction. Additionally, amino acid residues 334 to 449 of Ku80 appear to be required for DNA end-binding activity (46). Two studies have reported aberrant Ku80 proteins in human cell extracts that apparently lack the C terminus and have DNA end-binding but not DNA-PK activity, suggesting that the C-terminal region of Ku80 might be involved in interacting with DNA-PKcs (16, 30). Ku has been widely recognized as the DNA-binding component of DNA-PK that activates the catalytic subunit (DNA-PKcs) when DNA bound. The lack of DNA-PK activity in both Ku-defective and DNA-PKcs-defective cell lines has substantiated these in vitro findings (2, 10, 35). Recent studies, however, have challenged this view, showing that DNA-PKcs can bind to linear DNA fragments and function as a protein kinase in the absence of Ku and that Ku merely stimulates DNA-PK activity up to eightfold (47). Hammarsten and Chu (15) have also reported similar findings and concluded that Ku stabilizes the recruitment of DNA-PKcs to DNA ends and that a cooperative interaction between Ku, DNA-PKcs, and DNA efficiently activates the DNA-PKcs activity.
In this study, we have used two approaches to study Ku80 function. First, to gain further insight into the role of the Ku80 C terminus, we have examined C-terminally truncated Ku80 proteins for their ability to interact with Ku70, to bind dsDNA ends, and to interact with and activate DNA-PKcs. We examined the activity of the proteins generated following cotranslation with full-length Ku70 and, to gain insight into the significance of these in vitro findings, introduced truncated Ku80 cDNAs into Ku80-defective xrs-6 cells. Our results suggest that the C-terminal region of Ku80 is dispensable for interaction with Ku70 and for DNA end binding but is required for optimal DNA-PK activity. Cells lacking this region of Ku80 acquire the phenotype of cells defective in DNA-PKcs and are able to rejoin signal but not coding junctions during V(D)J recombination. We also report the identification of a point mutational change in Ku80-defective xrs-2 cells. This is the first Ku80 mutant harboring a point mutational change, and it identifies a site outside of the previously described Ku70-Ku80 minimal interaction domain that is nonetheless required for heterodimer formation.
MATERIALS AND METHODS
Cell culture, DNA transfections, and protoplast fusions.
The xrs cell lines were derived from the CHO-K1 cell line on the basis of their sensitivity to ionizing radiation (21). Cells were cultured in minimal essential medium (Gibco) supplemented with nonessential amino acids, penicillin, streptomycin, glutamine, and 10% fetal calf serum as described previously (21). Transfection was carried out by the Polybrene transfection method as described previously (22). Transfectants were selected by using 600 μg of G418 per ml alone or with 50 μg of zeocin per ml and subsequently maintained in 300 μg of G418 per ml alone or with 50 μg of zeocin per ml. Yeast artificial chromosomes (YACs) were transferred to rodent cells by the protoplast fusion protocol described previously (2). Isolation and preliminary characterization of the DNA-PKcs YAC have been described previously (2, 36). Experiments to assess survival in response to ionizing radiation were as described previously (21). V(D)J recombination assays were as described previously (40, 41).
Analysis of DNA transfectants.
Between 6 and 15 clones were picked per transfection and analyzed by PCR for the presence of the construct. PCR was carried out in 25 μl containing 2.5 μl of 10× PCR buffer (100 mM Tris [pH 9], 500 mM KCl, 15 mM MgCl2, 0.1% gelatin, 1% Triton X-100), 1.25 μl of each deoxynucleoside triphosphate (2.5 mM), 20 ng of each primer, and 0.25 U of Taq polymerase (HT Biotechnology Ltd. and Advanced Biotechnologies). Clones transfected with Chinese hamster (CH) Ku80 cDNA were analyzed with primers AP34 and AP35 (39) or primers AP34 and SP6 (a vector-specific primer for SP6 promoter). Human (Hs) Ku80 cDNA was detected with primers Ku80 PN (5′GTC GGG GAA TAA GGC AGC TGT3′) (positions 9 to 29) and Ku80 PP (5′ACT TGG TTT GTC TTT GGG GGC3′) (positions 2136 to 2116) for full-length Ku80 or primers C (39) and D (5′TTT GTC TTT GGG GGC CAG AAA CTT3′) (positions 2130 to 2107) for the 3′ half of the cDNA. At least two PCR-positive clones from each transfection were analyzed further.
Construction of Ku80 constructs.
Full-length CH Ku80 cDNA was cloned into pcDNA3 as described previously (39). Full-length Hs Ku80 cDNA was PCR amplified with primers containing BamHI sites and cloned into the BamHI site of pcDNA3.1/HisC (Invitrogen). The deletion constructs were generated as described below. For Δ560, a termination codon at residue 560 followed by a XhoI site was created by site-directed mutagenesis. The C-terminal portion was then removed by XhoI digestion and religation. The remaining deletion constructs were generated by the introduction of two adjacent termination codons by site-directed mutagenesis at the residues indicated. The xrs-2 mutation was introduced into Ku80 cDNA by site-directed mutagenesis. All site-directed mutagenesis was carried out by the method of Kunkel et al. (25).
In vitro transcription and translation.
Full-length Hs Ku70 was subcloned into the pcDNA3 vector. A 0.5-μg portion of each Ku80 construct (or pcDNA3 vector alone) was coexpressed with 0.5 μg of pcDNA3 Hs Ku70 in a 25-μl transcription-translation system as specified by the manufacturer (Promega TNT T7 quick-coupled transcription/translation system). A 2.5-μl volume of each reaction mixture was used for Western blot analysis. Hs Ku80 proteins were detected with anti-Xpress antibody (Invitrogen) following 1:1,000 dilution in 1% milk. Ku70 was detected with Ku70-6 antibody diluted 1:5,000 in 5% milk. Coimmunoprecipitation was examined with 10 μl of TNT reaction product following overnight incubation at 4°C with Ku70-6. The antigen-antibody complexes were isolated following incubation with protein A-Sepharose and extensive washing. Immunoprecipitation of Ku70 was verified with anti-Ku70 antibody, N3H10, diluted 1:5,000 in 2% milk. Coimmunoprecipitation of Ku80 was detected with anti-Xpress antibody.
End-binding and DNA-PK assays.
DNA end-binding assays were carried out essentially as described previously (14, 41). In brief, extracts were prepared by a modification of the method of Scholer et al. (38) and were incubated with γ-32P-labelled ds oligonucleotide M1/M2 at room temperature for 30 min, and DNA-protein complexes were resolved on 4% polyacrylamide gels containing 5% glycerol. The modified DNA-PK assay in which the phosphorylated product is separated by polyacrylamide gel electrophoresis (PAGE) was carried out essentially as previously described (36).
Immunoblotting.
Whole-cell extracts (120 μg for hamster cell extracts and 60 μg for human cell extracts) were boiled in sodium dodecyl sulfate (SDS)-PAGE loading buffer, and separated by SDS-PAGE (8% polyacrylamide). Proteins were transferred to nitrocellulose by using a wet-blotting apparatus and blocked for 1 h to overnight at 4°C with 5% skim milk solution and 0.05% Tween. The primary antibodies were Ku80-4 and Ku70-6, which were raised against baculovirus-expressed Hs Ku80 and Ku70 proteins, respectively (Serotech). For the detection of hamster proteins, the Ku80-4 or Ku70-6 primary antibody was diluted 1:5,000 and 1:2,500, respectively, in 2% milk solution and incubated for 2 to 3 h at room temperature, the mixture was washed extensively in milk solution, and anti-rabbit immunoglobulin antibody (diluted 1:2,500) was added for 1 hr at room temperature. The filter was rewashed and developed with an ECL kit (Amersham). For the detection of human proteins, the Ku80-4 antibody was diluted 1:1,000 in 5% milk and the monoclonal anti-Ku70 antibody, N3H10, was diluted 1:5,000 in 2% milk.
cDNA synthesis and sequencing.
Poly(A)+ RNA was extracted from 5 × 107 cells with a Quickprep Micro mRNA Purification kit (Pharmacia Ltd.). Reverse transcription-PCR (RT-PCR) and sequencing of the Ku80 gene from xrs-2 cells were carried out with the primers and method described previously (39). To verify the sequence of mouse Ku80 cDNA, mouse Ku80 cDNA was amplified by RT-PCR with C57BL/6 testis RNA. Primers AP7 (5′GGA ACA AAT GAA ATA TAA AT3′) and D (3′TTT GTC TTT GGG AGC CAG GAA CTT3′) were used to amplify the relevant region. The products were cloned into a T vector and sequenced by standard procedures.
RESULTS
Analysis of Ku80 3′ deletion constructs.
To investigate the function of the C-terminal region of Ku80, a series of Ku80 cDNAs having 3′ deletions of different sizes were generated (Table 1). Some of the deletions were derived with CH Ku80 cDNA in the mammalian expression vector pcDNA3. Subsequently, additional deletions were generated with Hs Ku80 cDNA in a pcDNA3 vector containing an Xpress epitope tag. The former has the advantage that the full-length CH Ku80 cDNA fully corrects the Ku80-defective xrs-6 cells but has the disadvantage that the Hs Ku80 antibodies that cross-react with CH Ku80 do not recognize the C-terminally truncated proteins (the proteins show 80.3% homology at the amino acid level). The use of Hs cDNA constructs therefore enabled a wider range of antibodies as well as the anti-Xpress antibody to be used.
TABLE 1.
Analysis of Ku80 C-terminal deletion constructs
| Constructe | Species | In vitro translation analysis
|
Transfection into xrs-6 cells
|
Transfection into xrs-6/YAC fusion hybrids
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subunit interaction | EMSA | Ku70 (WB)b | Ku80 (WB) | EMSA | Kinase activity | IRb | Ku70 (WB) | Ku80 (WB) | EMSA | Kinase activity | IR | ||
| Ku80 | Hamster | ++ | ++ | ++ | ++ | ++ | + | R | ++ | ++ | ++ | +++ | R |
| Ku80 | Human | ++ | ++ | ++ | ++ | ++ | + | Partial R | ++ | ++ | ++ | +++ | Partial R |
| Δ560–732 | Hamster | NDab | ++ | ++ | NDa | ++ | ≈30%c | Partial R | ND | NDa | ++ | 40%c | R |
| Δ554–732 | Human | ++ | ++ | ++ | ++ | ++ | <5%d | S | ND | ++ | ++ | 23%d | S |
| Δ531–732 | Hamster | NDa | ± | − | NDa | − | <0.1% | S | ND | ND | ND | ND | ND |
| Δ510–732 | Human | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Δ474–732 | Hamster | NDa | − | − | NDa | − | <0.1% | S | ND | ND | ND | ND | ND |
CH-derived Ku80 truncated proteins were not recognized by the anti-Ku80 antibody, 80-4, and therefore could not be analyzed by Western blot analysis.
WB, Western blot; ND, not done; S, as sensitive to ionizing radiation (IR) as xrs-6 cells; R, as resistant to ionizing radiation as CHO-K1 cells.
Expressed relative to the activity seen in xrs-6 cells complemented with CH Ku80 cDNA.
Expressed relative to the activity seen in xrs-6 cells complemented with Hs Ku80 cDNA.
Constructs C-terminally deleted at positions 622, 591, and 563 were also analyzed in the in vitro transcription system. All the resulting truncated proteins were proficient in subunit interaction and in DNA end-binding.
(i) Analysis with in vitro-expressed proteins.
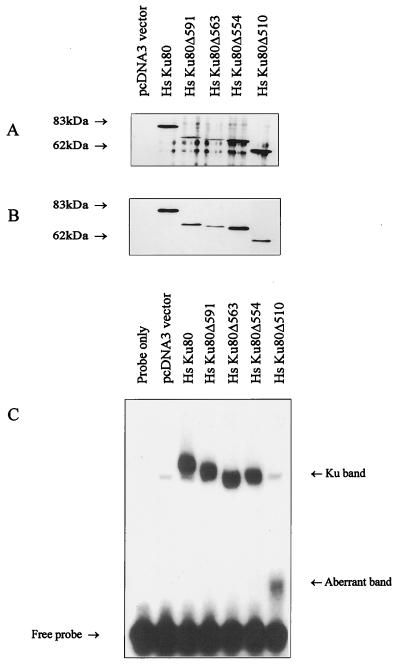
First, the truncated Ku80 proteins of human origin were examined for their ability to interact with Ku70 following their coexpression with full-length Hs Ku70 cDNA in an in vitro translation system with rabbit reticulocyte lysates. The expression of Ku80 protein was assessed by Western blotting with anti-Xpress antibody (Fig. 1A). To examine the Ku70-Ku80 interaction, Ku70 protein was immunoprecipitated with anti-Ku70 antibodies and coimmunoprecipitation of Ku80 was determined with anti-Xpress antibodies (Fig. 1B). In the absence of Ku80 cDNA in the translation system, no anti-Xpress cross-reacting protein was detectable (Fig. 1A, lane pcDNA3 vector). Ku80 truncated proteins lacking up to 178 amino acid residues interacted as efficiently with Ku70 as did the full-length protein, while Hs Ku80Δ510 (where Δ510 denotes deletion of C-terminal residues 510 to 732) displayed slightly reduced Ku70 interaction, as determined by the amount of Ku80 coimmunoprecipitated relative to that expressed (compare Fig. 1B with Fig. 1A). The same results were obtained by examining the ability of Ku80 antibodies to coimmunoprecipitate Ku70 protein (data not shown). The smaller size of the Ku80 protein verifies that the site-directed mutations generated yield the anticipated truncated proteins. Next, we determined whether the in vitro-translated proteins had DNA end-binding activity by examining them in an electrophoretic mobility shift assay (EMSA) (Table 1; Fig. 1C). A low background level of DNA end-binding activity was obtained in the absence of Ku70 and Ku80 cDNA or in the presence of either cDNA alone, probably due to Ku homologues present in the reticulocyte lysates (Fig. 1C). Full-length Hs Ku80 and deletions up to Hs Ku80Δ554 had similar DNA end-binding activity, while the protein with the larger truncation (Hs Ku80Δ510) gave no end-binding activity above background (Fig. 1C). Surprisingly, Hs Ku80Δ510 showed significant Ku70-Ku80 interaction but no residual DNA end-binding activity. However, EMSA with Hs Ku80Δ510 gave smaller, smeared band shift products (Fig. 1C), possibly suggesting an unstable Ku complex dissociating during the electrophoresis. Results consistent with this were also obtained with the truncated proteins of hamster origin, although the Ku70-Ku80 interaction could not be assessed due to the lack of cross-reacting Ku80 antibody (Table 1). CH Ku80Δ531 had impaired DNA end-binding activity and also showed a smaller, smeared band shift product like that observed with Hs Ku80Δ510. CH Ku80Δ474 had no end-binding activity above background. Collectively, these results show that the C-terminal 178 amino acid residues of Ku80 are dispensable for both Ku70-Ku80 interaction and DNA end-binding activity. Loss of additional sequences to residue 510 impinges upon both heterodimer formation and DNA end-binding activity.
FIG. 1.
Analysis of in vitro-expressed C-terminally truncated Ku80 proteins. (A) Western blot analysis of in vitro-expressed Ku80 proteins. A 2.5-μl volume of reticulocyte lysate, following coexpression of full-length Ku70 and pcDNA3 vector alone, full-length Hs Ku80 cDNA, and truncated Ku80 cDNAs as indicated, was used for Western blot analysis with the anti-Xpress antibody to detect Ku80 protein. (B) Coimmunoprecipitation of Ku80 and Ku70. A 10-μl volume of coexpressed Ku70/vector control (“pcDVA3 vector only” lane) or Ku70-Ku80 proteins as indicated were immunoprecipitated with the anti-Ku70 antibody, Ku70-6, and coimmunoprecipitation of Ku80 was examined with the anti-Xpress antibody. (C) DNA end-binding activity of in vitro-expressed proteins. Expressed proteins (2.5 μl) as indicated were used in an EMSA. The band labeled “Ku band” represents Ku-dependent dsDNA end-binding activity. The aberrant band represents a smeared product routinely observed when construct Hs Ku80Δ510 is used (see text for details). The mobility of the band shift products generally reflected the size of the Ku80 product, with the exception of Hs KuΔ563, which may be due to altered conformation of the heterodimer. Autoradiographic images were scanned on an AGFA StudioStar scanner with FotoLook32 software.
(ii) Analysis following transfection into xrs-6 cells.
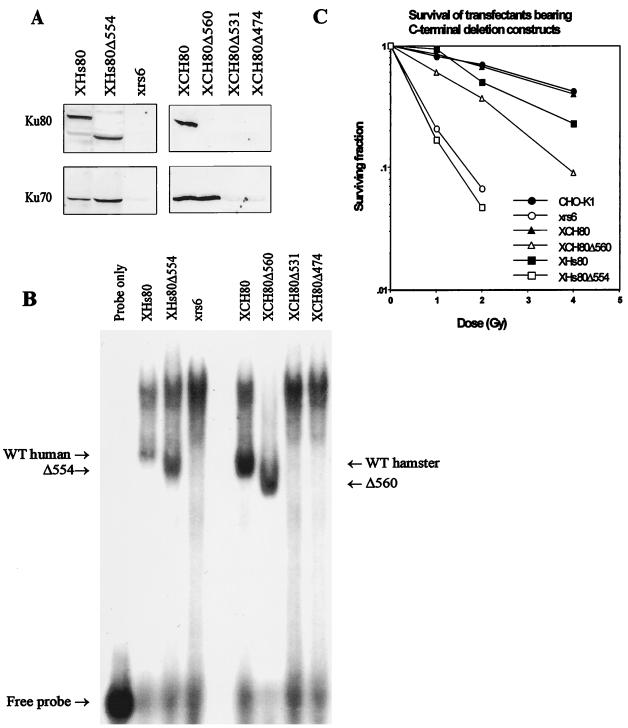
To gain insight into the function of the Ku80 C-terminal region in vivo, a subset of the 3′-deleted cDNAs were introduced into Ku80-defective xrs-6 cells by DNA transfection (Table 1). xrs-6 cells have no residual Ku-dependent DNA end-binding or DNA-PK activity due to the presence of an inactivating missplicing mutation close to the N terminus of the Ku80 open reading frame (29, 39). Individual stable transfectants were selected with the neomycin marker present in pcDNA3 and screened by PCR with primers that amplify the Ku80 cDNA construct used. A minimum of two transfectants from each transfection were analyzed for the presence of Ku70 and Ku80 protein by Western blot analysis, for DNA end binding and DNA-PK activity. The clones derived from these transfections are named to reflect the construct used: e.g., XCH80Δ560 represents a stable transformed clone of xrs-6 with hamster Ku80Δ560. Normal levels of truncated Ku80 protein are expressed in XHs80Δ554 cells (Fig. 2A). Expression of Ku80 protein in clones derived with the other constructs could not be assessed since the truncated CH proteins do not cross-react with the anti-Ku80 antibodies. xrs-6 cells, as well as Ku80 knockout cell lines, have little or no Ku70 protein, demonstrating that Ku70 is unstable in the absence of Ku80 (6, 31, 39, 41). The faint residual signal seen in Western blot analysis of xrs-6 cells with anti-Ku70 antibodies could be due to residual Ku70 protein in the absence of Ku80 or to some other cross-reacting protein (Fig. 2A). The recovery of Ku70 protein in the transfected clones assessed by Western blot analysis was therefore used to monitor the interaction of Ku80 with Ku70 in vivo. Ku70 protein was clearly stabilized in XCH80Δ560 and XHs80Δ554 cells (Fig. 2A). Thus, it is likely that Ku80 is expressed in XCH80Δ560 clones, although it is not seen in the Western blots due to lack of cross-reactivity of the Ku80 antibodies. When the constructs with larger deletions were used, only low residual Ku70 protein levels were seen, comparable to that observed in xrs-6 cells (Fig. 2A). Taken together, these results suggest that the Hs80Δ554 and CH80Δ560 truncated proteins are stably expressed and interact efficiently with Ku70 in vivo. The lack of Ku70 protein in clones bearing the larger deletions probably reflects the reduced ability of the truncated Ku80 proteins to interact with Ku70, although we have not verified that Ku80 is expressed in these clones. The DNA end-binding activity of the clones is closely correlated with the stabilization of Ku70 protein (Fig. 2B). Thus, XHs80Δ554 and XCH80Δ560 had wild-type levels of DNA end-binding activity, while the two larger deletions resulted in loss of such activity.
FIG. 2.
Analysis of xrs-6 transfectants expressing C-terminally truncated Ku80 proteins. (A) Western blot analysis of xrs-6 transfectants with anti-Ku70 and anti-Ku80 antibodies. Protein (120 μg) in whole-cell extracts from cells expressing CH Ku80 and protein (60 μg) in whole-cell extract from cells expressing Hs Ku80 were subjected to Western blotting with antibodies Ku80-4 and Ku70-6. The lack of Ku80 protein in XCH80Δ560, XCH80Δ531, and XCH80Δ474 is due to the inability of the Ku antibody to recognize the truncated CH proteins. The presence of Ku70 protein with XCH80Δ560 indicates heterodimer formation and hence Ku80 expression. (B) EMSA of extracts from xrs-6 transfectants. Whole-cell extracts (30 μg) from the xrs-6 transfectants indicated were subjected to EMSA. Cold circular DNA was not added as competitor in these assays, resulting in the presence of a heavy band of non-Ku-specific DNA binding. The Ku-specific bands are labeled. The human samples were analyzed on a different gel from the hamster samples, and so a direct comparison of the mobilities cannot be made. However, the band shift involving Hs Ku80 routinely has a lower mobility than that involving CH Ku80, as also seen in Fig. 4 and as observed previously (41). WT, wild type. (C) Survival of transfectants following exposure to ionizing radiation. Cells were exposed to the indicated dose of ionizing radiation, and survival was estimated after 7 days of incubation. Autoradiographic images were scanned on an AGFA StudioStar scanner with FotoLook32 software.
To measure DNA-PK activity, DNA-binding proteins were first microfractionated from whole-cell extracts by using DNA-cellulose beads and then assayed for kinase activity with a p53-derived peptide as substrate (10). The phosphorylated product was separated by polyacrylamide gel electrophoresis and quantified by scanning. This procedure considerably enhanced the sensitivity of the assay by separating DNA-PK independent, nonspecific phosphorylated products from the p53 peptide (36). Only clones with DNA end-binding activity were examined for kinase activity. Phosphorylation of the p53 peptide with extracts of xrs-6 cells was not significantly above background (the signal obtained in the absence of p53 peptide). Clones expressing full-length CH Ku80 have levels of activity comparable to that obtained with CHO-K1 cells, which is three- to fivefold above background (data not shown). The activity obtained with clones expressing Hs Ku80 cDNA was approximately half of that obtained with wild-type cells (data not shown). XHs80Δ554 and XCH80Δ560 clones reproducibly had decreased DNA-PK activity compared to that of clones expressing the corresponding full-length Ku80 protein, which by scanning was estimated to be 4% (XHs80Δ554) and 28% (XCH80Δ560) of the signal obtained with clones expressing full-length Ku80 of the same species (data not shown). However, although the decrease in kinase activity was reproducible, the quantitative estimation of the decrease is highly inaccurate due to lack of sensitivity of the kinase assay in the rodent cells. These limitations notwithstanding, these results raise the possibility that clones with normal DNA end-binding activity have reduced kinase activity and suggest that the C-terminal region of Ku80 is required for kinase activation.
The ability of the Ku80 deletion constructs to complement the radiation sensitivity of xrs-6 cells was also examined (Fig. 2C). XCH80Δ560 clones that retained approximately one-third of the kinase activity found in control cells had elevated γ-ray survival compared with xrs-6 cells or cells transfected with an empty vector but less than that observed with wild-type CH Ku80 cDNA or parental cells. In contrast, the larger deletions were unable to complement the radiation sensitivity of xrs-6 cells. Clones expressing Hs Ku80Δ554 cDNA resemble cell lines defective in DNA-PKcs (e.g., the mouse SCID cell line) in having DNA end-binding activity but lacking DNA-PK activity (2, 13). DNA-PKcs-defective cell lines (V-3, irs20, and the mouse SCID line) differ from Ku80-defective lines in the nature of their V(D)J recombination defect (26, 27, 34, 40, 42). Ku80-defective mutants have major defects in both signal joint and coding-joint formation, whereas the DNA-PKcs-defective lines can rejoin signal sequences with only a slightly impaired frequency. Additionally, while signal junctions are normally accurate in control cells, those formed in DNA-PKcs-defective lines have an elevated frequency of deletions. We therefore examined the ability of the XHs80Δ554 cells to carry out signal joint and coding-joint formation. XHs80Δ554 cells significantly recovered the frequency of signal joint formation to 50% of the level seen in clones expressing full-length Hs Ku80, although the defect in coding-joint formation remained (Table 2). The accuracy of signal joint formation also increased, although some junctions had deletions. Therefore, cells expressing this Ku80 deletion construct have the V(D)J recombination phenotype of cells lacking DNA-PKcs, even though they do express DNA-PKcs protein. XCH80Δ560 clones showed considerable recovery of coding-joint formation, suggesting that their higher level of DNA-PK activity is sufficient to carry out coding joint formation effectively. This is consistent with their elevated survival (Fig. 2C).
TABLE 2.
Analysis of signal joint and coding-joint formation by transient V(D)J recombination assaya
| Cell line | PJH200 (signal joints)
|
PJH290 (coding joints)
|
|||
|---|---|---|---|---|---|
| No. Ampr/no. Chlr | % recombinedb | Fidelity (%) | No. Ampr/no. Chlr | % recombinedb | |
| XHs80 | 220/7,650 | 2.9 | 100 | 292/17,400 | 1.7 |
| XHs80Δ554 | 27/1,770 | 1.5 | 93 | 4/5,220 | 0.07 |
| XCH80 | 120/3,275 | 3.2 | 100 | 116/3,670 | 3.2 |
| XCHΔ560 | 471/13,950 | 3.4 | 100 | 148/14,100 | 1.1 |
| xrs-6 | 20/15,000 | 0.15 | 0 | 2/3,150 | 0.06 |
Mouse SCID and V-3 cells have approximately a 50-fold defect in coding-joint formation and a 2-fold defect in signal joint formation. SCID and V-3 cells rejoin signal joints with 60 to 80% fidelity (34, 40).
The percent correct RS joints screened by ApaLI digestion of the PCR product by using the recombination substrate pJH200. The results represent the mean of at least three independent transfection experiments.
Overexpression of DNA-PKcs in xrs-6 cells.
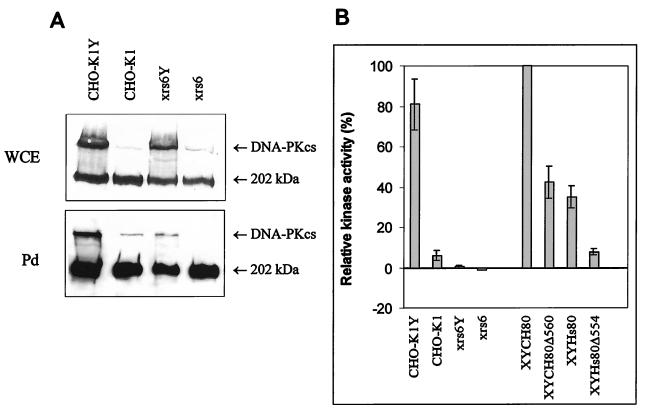
Rodent cells have approximately 50-fold-lower levels of DNA-PK proteins than do human cells. While human cells have high levels of DNA-PK activity, we routinely observed levels of DNA-PK activity in parental CHO-K1 cells only three- to fivefold above background. This made it difficult to monitor accurately the decrease in kinase activity in transfectants expressing the Ku80 deletion constructs. Previously, we have shown that rodent cells harboring a YAC expressing Hs DNA-PKcs express human levels of the protein and have high DNA-PK activity compared with wild-type rodent cells (2, 36). This suggests that DNA-PKcs is rate limiting for DNA-PK activity and provided a tool to overexpress DNA-PKcs and enhance the sensitivity of the DNA-PK assay in the rodent cells. First, we examined the effect of overexpression of DNA-PKcs in xrs-6 cells by introducing YACs encoding Hs DNA-PKcs into CHO-K1 and xrs-6 cells by protoplast fusion. As previously reported, fusion clones (designated CHO-K1Y or xrs-6Y) derived from both CHO-K1 and xrs-6 expressed the higher levels of DNA-PKcs expected (36) (Fig. 3A). Cell extracts from the fusion clones were next examined for DNA-PK activity. While CHO-K1Y clones had approximately 20- to 30-fold-higher levels of activity than control CHO-K1 cells, the DNA-PK activity in xrs-6 cells expressing DNA-PKcs YACs (xrs-6Y) was only marginally increased (Fig. 3B). It is also notable that xrs-6Y clones expressing full-length CH Ku80 protein had DNA-PK activity 50- to 100-fold above that present in xrs-6Y clones, demonstrating the significant increase in kinase activity when Ku is present (Fig. 3B). The overexpression of DNA-PKcs in these clones did not affect the phenotype of either CHO-K1 or xrs-6 cells and did not affect their level of radiosensitivity (36). The high levels of DNA-PKcs in these fusion clones, however, allowed us to examine the effect of Ku on the ability of DNA-PKcs to bind to DNA. To assess DNA-PKcs/DNA binding, half the microfractionated DNA-binding proteins obtained during the DNA-PK assay were boiled to remove the bound proteins and examined for the presence of DNA-PKcs by Western blotting (Fig. 3A, Pd). High levels of DNA-PKcs were recovered from extracts of CHO-K1Y cells in this pull-down assay, and lower but measurable levels were obtained from control CHO-K1 cells expressing only endogenous DNA-PKcs (Fig. 3A). No detectable DNA-PKcs was recovered from xrs-6 cells in this pull-down assay, even though DNA-PKcs was present in the whole-cell extract (Fig. 3A), but low levels have been recovered in other experiments (data not shown). The level of DNA-PKcs recovered from the pull-down fraction by using extracts from xrs-6Y cells was variable but was always markedly lower than that recovered from CHO-K1Y extracts (Fig. 3B). However, despite the recovery of DNA-PKcs from extracts of xrs-6Y cells, only low DNA-PK activity was observed (Fig. 3B). The response to radiation and the V(D)J recombination phenotypes of xrs-6 and CHO-K1 cells were unaffected by the presence of the DNA-PKcs YAC (for data on the survival of xrs-6 cells, see Fig. 4C). Taken together, these results show that Ku significantly enhances the ability of DNA-PKcs to bind to DNA and to function as a DNA-dependent protein kinase.
FIG. 3.
Interaction of DNA-PKcs with DNA and DNA-PK activity in the absence of Ku. (A) Recovery of DNA-PKcs from DNA cellulose beads. (Top) Protein (50 μg) from whole-cell extracts (WCE) of CHO-K1 and xrs-6 cells either expressing DNA-PKcs YACs (lanes CHO-K1Y and xrs6Y) or not carrying DNA-PKcs YACs (lanes CHO-K1 and xrs6) was subjected to Western blotting with the monoclonal anti-DNA-PKcs antibody, 18-2. (Bottom) DNA-binding proteins from 150 or 300 μg of protein from whole-cell extracts were microfractionated with DNA-cellulose beads and then analyzed for DNA-PKcs by Western blotting. Pd represents extracts microfractionated for DNA-binding proteins by the pull-down assay with DNA-cellulose beads. The lighter upper band is DNA-PKcs, and the heavy lower band is a 220-kDa protein routinely seen when anti-DNA-PKcs antibodies are used. Autoradiographic images were scanned on an AGFA StudioStar scanner with FotoLook32 software. (B) Kinase activity. DNA-PK activity was assayed following a pull-down assay of DNA-binding proteins with DNA-cellulose beads. The substrate is a p53-derived peptide, which was analyzed for phosphorylation by PAGE. The intensity of the p53 band was assessed by phosphorimager scanning. The activity is expressed relative to that obtained with xrs-6/YAC cells complemented with full-length CH Ku80 cDNA. No activity above background was obtained with xrs-6 cells. The activity obtained with xrs-6/YAC fusion hybrids was low but reproducibly above background levels. CHOK1-Y and xrs-6Y represent CHO-K1 and xrs-6 cells expressing the YAC encoding DNA-PKcs, respectively; XYCH80, XYCH80Δ560, XYHs80, and XYHs80Δ554 are xrs-6/YAC fusion hybrids expressing CH Ku80, CH Ku80Δ560, Hs Ku80, and Hs Ku80Δ554 constructs, respectively.
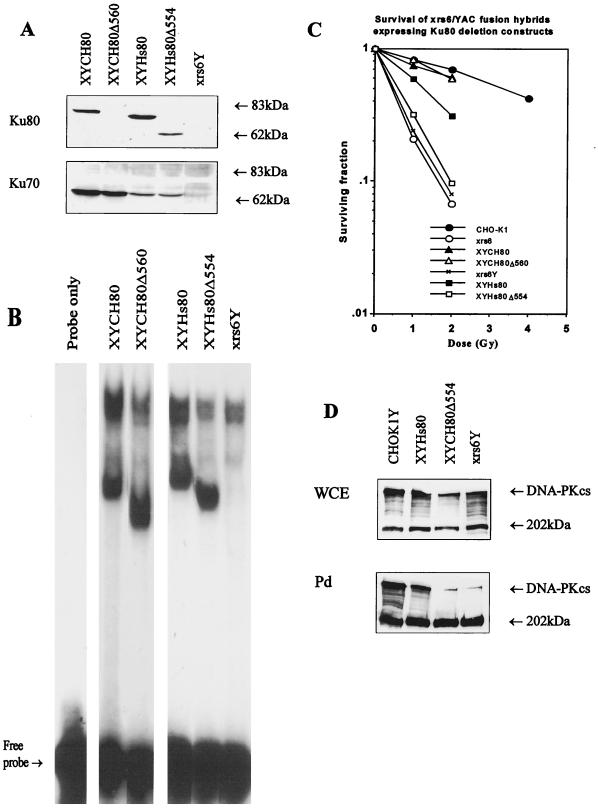
FIG. 4.
Analysis of extracts of xrs-6Y clones. (A) Protein (50 μg) from whole-cell extracts was separated by SDS-PAGE through 8% polyacrylamide gels and subjected to Western blotting with the Ku70 and Ku80 antibodies, N3H10 and Ku80-4. (B) EMSA analysis. Whole-cell extracts (30 μg) from the xrs-6Y clones indicated were subjected to EMSA. (C) Survival of xrs-6Y clones in response to ionizing radiation. (D) Recovery of DNA-PKcs from DNA-cellulose beads. (Top) Protein (15 μg) from whole-cell extracts (WCE) of CHO-K1Y cells, xrs-6Y cells expressing full-length Hs Ku80, xrs-6Y cells expressing CH Ku80Δ554, and xrs-6Y cells was subjected to Western blotting with anti-DNA-PKcs antibody, 18-2. (Bottom) DNA-binding proteins derived from 150 μg of protein from the same whole-cell extracts were first microfractionated with DNA-cellulose beads and then analyzed for DNA-PKcs with antibody 18-2. Pd represents extracts microfractionated for DNA-binding proteins by pull-down assays with DNA-cellulose beads. Autoradiographic images were scanned on an AGFA StudioStar scanner with FotoLook32 software.
Transfection of Ku80 C-terminal deletion constructs into DNA-PKcs-overexpressing xrs-6 cells.
The low level of DNA-PKcs activity in rodent cells limited our ability to assess the magnitude of the defect in DNA-PKcs interaction resulting from the C-terminal truncations of Ku80. We were also unable to assess efficiently whether the Ku80 deletion constructs impaired the ability of DNA-PKcs to interact with DNA. We therefore introduced the two most informative 3′ deletion constructs (Hs Ku80Δ554 and CH Ku80Δ560) and full-length Hs or CH Ku80 cDNA into xrs-6Y cells, yielding transformants designated XYHs80Δ554, XYCH80Δ560, XYHs80, and XYCH80, respectively. First, the constructs were subcloned into a pZeoSV vector, since the YACs encoded the neomycin selectable marker, and subsequently they were transferred to xrs-6Y cells by DNA transfection and selection for resistance to zeocin. All the clones expressed a truncated Ku80 protein, and Ku70 protein was stabilized, indicating efficient Ku70-Ku80 interaction (Fig. 4A; not visible for CH80Δ560 due to the inability of Ku80 antibody to cross-react with rodent protein). Cell extracts from these clones also had DNA end-binding activity with the predicted altered mobility (Fig. 4B). xrs-6Y cells expressing full-length CH Ku80 protein had DNA-PK activity 20- to 50-fold above that obtained with CHO-K1 cell extracts and similar to that found in CHO-K1Y cells (Fig. 3B) and human cells (36). In contrast, DNA-PK activity was decreased in clones expressing the 3′ deletion constructs, although some residual activity was detectable, which was lower with the larger deletion (Fig. 3B; Table 1). Although XYHs80Δ554 has markedly lower kinase activity than XYCH80Δ560 did, this is more likely to be due to the different species origin of the proteins than to a specific functional significance of the extra 6 amino acids deleted. An additional deletion construct (Hs Ku80Δ543) was introduced into xrs-6Y cells, and the transfectants derived had an almost identical phenotype to XYHs80Δ554 cells, with 6% of the parental kinase activity (data not shown). Thus, deletion of an additional 11 amino acid residues does not decrease the residual kinase activity. These results substantiated those obtained with the xrs-6 parental cells and verified that the C-terminal region of Ku is required for optimal DNA-PK activity. To assess whether DNA-PKcs can interact with DNA in the presence of the Ku80 truncated proteins, we used Western blotting to examine the microfractionated DNA-bound proteins obtained in XYHs80Δ554 cells for DNA-PKcs levels. The level of DNA-PKcs protein recovered from extracts of XYHs80Δ554 cells was markedly lower than that recovered from xrs-6Y cells expressing full-length Ku80 protein and similar to that obtained with xrs-6Y cells (Fig. 4D). These results show that the major impact of the C-terminal Ku80 deletions is to impair DNA-PKcs binding to DNA. Decreased but measurable kinase activity was detected with these extracts, however. The survival of these clones after exposure to γ-irradiation was also examined. While XCH80Δ560 cells were partially complemented for radiation sensitivity (Fig. 2C), XYCH80Δ560 cells had a level of resistance close to that of parental CHO-K1 cells (Fig. 4C). In contrast, expression of the Hs Ku80Δ554 protein resulted in only minor complementation of the radiation sensitivity of the xrs-6Y cells.
Identification of a point mutation in Ku80-deficient xrs-2 cells.
xrs-2 is a Ku80-defective mutant that displays slightly less radiation sensitivity compared to other xrs mutants (21). Like other xrs mutants, xrs-2 lacks Ku70 and Ku80 proteins, detectable dsDNA end binding, and DNA-PK activity, but the Ku80 transcript levels are normal (data not shown). Sequence analysis of Ku80 cDNA from xrs-2 cells by RT-PCR identified a C-to-T base change at position 1229 that results in the substitution of a leucine for a proline at residue 410. Although a proline at this position is conserved between human, hamster, Caenorhabditis elegans, and Saccharomyces cerevisiae Ku80 proteins, the GenBank sequence (U68181) for mouse Ku80 showed a leucine at this position. We sequenced this region from mouse Ku80 cDNA and found, contrary to the published sequence, a proline at residue 410. This amino acid residue is therefore conserved between species, enhancing the likelihood that the mutational change identified in xrs-2 at this residue represents the causal inactivating mutation.
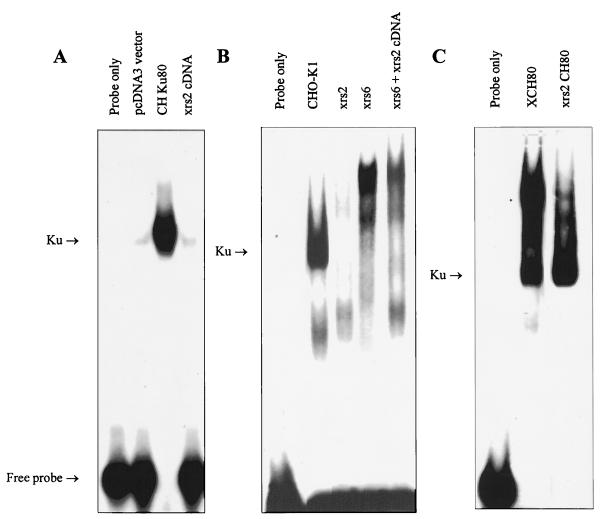
To verify the significance of this mutational change, we generated the mutation in CH Ku80 cDNA by site-directed mutagenesis and coexpressed the mutated cDNA (designated xrs-2 cDNA) with full-length Ku70 in the reticulocyte lysate in vitro translation system. The xrs-2 Ku80 protein generated cross-reacted with anti-Ku80 antibodies but was unable to coimmunoprecipitate Ku70 (data not shown), and the coexpressed proteins lacked DNA end-binding activity (Fig. 5A). xrs-2 cDNA was also introduced into xrs-6 mutants by DNA transfection, and individual clones were analyzed for radiation sensitivity, for DNA end binding, and for the presence of Ku70 and Ku80 protein by Western blotting (data not shown). The level of radiation sensitivity varied between clones generated from three independent transfection experiments, but none of the clones had detectable Ku70 protein or DNA end-binding activity (Fig. 5B shows the DNA end-binding activity). Ku80 protein was also undetectable in the majority of clones. We also showed that wild-type CH Ku80 cDNA fully corrected xrs-2 cells (Fig. 5C shows the DNA end-binding activity). Taken together, these results support our contention from the sequence analysis that the likely causal mutation in xrs-2 cells is a proline-to-leucine change at position 410. This impairs the ability of Ku80 to interact with Ku70 and thus abolishes DNA end-binding activity. Potentially, however, some interaction remains in vivo, allowing some level of complementation. However, the aberrant Ku complex appears to be unstable following our extraction procedure.
FIG. 5.
Examination of the effects of the xrs-2 mutation on DNA end binding. (A) DNA end-binding activity of in vitro expressed xrs-2 Ku80 protein. Ku80 cDNA harboring the xrs-2 mutation was coexpressed with Hs Ku70 in vitro and analyzed by EMSA. Coexpression of Hs Ku70 and pcDNA3 vector, coexpression of Hs Ku70 and CH Ku80, and coexpression of Hs Ku70 and Ku80 cDNA harboring the xrs-2 mutation are shown in the three right-hand lanes. (B) DNA end-binding activity of cells expressing the mutant xrs-2 Ku80 protein. The middle three lanes contain extracts of CHO-K1, xrs-2, and xrs-6 cells, respectively; the right-hand lane represents extracts from xrs-6 cells expressing mutant Ku80 protein carrying the P-to-L mutation identified in xrs-2 cells. (C) Restoration of DNA end-binding activity in xrs-2 cells expressing CH Ku80 cDNA. The two right-hand lanes contain xrs-6 and xrs-2 cells expressing wild-type CH Ku80, respectively. Autoradiographic images were scanned on an AGFA StudioStar scanner with FotoLook32 software.
DISCUSSION
In this and a previous study we have used two approaches to gain insight into the function of the Ku protein (39). One approach involves the identification of mutations in Ku80-defective rodent cells, and the other involves a structure-function analysis of Ku80 by site-directed mutagenesis. Others have also carried out a structural analysis of Ku80 and identified domains required for interaction with Ku70 and for DNA end-binding activity (5, 32, 46). Here we describe the analysis of C-terminally truncated Ku80 proteins following coexpression with Ku70 in vitro and following expression in Ku80-deficient xrs-6 cells. Our results, summarized in Fig. 6, show that the C-terminal region of Ku80 is involved in an interaction with DNA-PKcs. To facilitate the examination of this interaction, we introduced human YACs encoding DNA-PKcs into xrs-6 cells. Rodent cell-YAC fusion hybrids express levels of DNA-PKcs similar to that found in human cells and, in the presence of full-length Ku80 protein, have high DNA-PK activity (36). The introduction of the 3′-deleted Ku80 cDNAs into these DNA-PKcs-overexpressing cells therefore increased our ability to monitor the interaction between the truncated Ku80 proteins and DNA-PKcs. The results we have obtained in vivo are generally in agreement with our in vitro findings. Taken together, our results show that the terminal 178 amino acid residues are dispensable for interaction with Ku70 and for DNA end-binding activity. In contrast, amino acid residues from 474 to 531 are required for DNA end-binding activity. The loss of DNA end-binding activity is probably due to reduced Ku70-Ku80 interaction as demonstrated for Hs Ku80Δ510. Unfortunately, this could not be verified with the constructs generated with CH Ku80 cDNA since our Ku80 antibodies do not cross-react with the truncated CH proteins. The results of previous studies have defined a minimal region of 28 amino acids from 449 to 477 required for heterodimerization, although this minimal region alone has not been shown to be sufficient for Ku70-Ku80 interaction (5, 32, 46). Therefore, some aspects of our results differ from these previous studies. First, although the C-terminal 178 amino acid residues are dispensable for heterodimerization, loss of further amino acids to residue 531 impairs interaction. Additionally, we show that changing residue 410 from a proline to a leucine impairs heterodimerization. Thus, in contrast to previous results, we show that the presence of the minimal region of 28 amino acids from 449 to 477 is not sufficient for Ku70-Ku80 interaction. A likely explanation is that the larger protein context of our truncated or mutated proteins may hinder protein-protein interactions if the correct protein conformation is not achieved, a feature that may not arise in the analysis of short protein fragments. In a separate collaborative study in which mutational changes have been constructed at various sites in full-length Ku80, we have obtained further evidence that changes at multiple sites in the N-terminal two-thirds of Ku80 can impinge upon Ku70-Ku80 heterodimerization (39a). Furthermore, there are many sites in Ku70 at which mutations abolish heterodimerization (24).
FIG. 6.
Summary of results with deletion constructs analyzed. The solid box represents the minimal Ku70-Ku80 interaction domain previously identified (32); P→L at position 410 represents the mutation in xrs-2 cells; ND, not done; S and R, sensitive and resistant to ionizing radiation, respectively; Ku70/80, heterodimerization; DB, DNA end-binding activity; DNA-PK, DNA-PK activity.
Previously it has been reported that DNA-PKcs has Ku-independent DNA-binding and kinase activity, and Ku was believed merely to stabilize DNA-PKcs/DNA binding, enhancing the kinase activity no more than eightfold (15, 47). Here, using xrs-6 cells that overexpress DNA-PKcs, we show that Ku can stimulate DNA-PK activity 50- to 100-fold. Our deletion analysis shows that the C-terminal region of Ku80 is required for interaction with DNA-PKcs and stimulation of the kinase activity. Thus, Ku80 proteins truncated at positions 554 and 560 produce a Ku heterodimer efficient for DNA end binding but with decreased ability to stimulate DNA-PK kinase activity. XYHs80Δ554 clones were not complemented for γ-ray sensitivity and were defective in coding-joint formation but were able to recombine signal junctions. These cells have thus acquired the phenotype of mouse SCID cells. Although we cannot verify that the sole function of the C-terminal region of Ku80 required for DSB rejoining is the recruitment and/or activation of DNA-PKcs, the fact that cells lacking this region of Ku80 but having high DNA-PKcs protein acquire the phenotype of cells lacking DNA-PKcs is highly supportive of this notion. The decreased kinase activity of cells expressing Ku80 truncated proteins could arise because the C-terminal region of Ku80 facilitates or stabilizes binding of DNA-PKcs to DNA and/or because it activates DNA-PKcs catalytic activity, for example by inducing a conformational change in the protein. The presence of the C-terminal region of Ku80 markedly increased the recovery of DNA-PKcs from DNA cellulose beads, showing that Ku enhances DNA-PKcs binding to DNA. While the recovery of DNA-PKcs protein from DNA cellulose beads was similar in extracts expressing the Ku80Δ554 protein and those lacking Ku (Fig. 4D, lanes XYCH80Δ554 and xrs6Y), greater residual kinase activity was detected when the truncated protein was present (Fig. 3B). This is consistent with the notion that Ku enhances DNA-PKcs/DNA binding and activates DNA-PKcs catalytic activity and that the truncated proteins retain some residual DNA-PKcs activation activity. However, lack of sensitivity of this analysis precludes a firm conclusion about the relative contribution of these two potential aspects of Ku80 function. These limitations notwithstanding, we conclude that the C terminus of Ku80 interacts with DNA-PKcs, enhancing its ability to bind to DNA and to function as a DNA-dependent protein kinase.
Our results also provide evidence for Ku-independent DNA-PKcs/DNA binding and kinase activity. Since rodent cells have levels of DNA-PK activity only three- to fivefold above background, it was not possible previously to assess whether Ku80-defective mutants retain low DNA-PK activity. When cell extracts that lack Ku80 protein but have high DNA-PKcs activity (xrs-6/YAC fusion cells) are used, the level of DNA-PKcs protein recovered from DNA cellulose beads is similar to that found in wild-type hamster cells. This may represent Ku-independent DNA-PKcs binding to DNA but may also result from nonspecific recovery due to the elevated DNA-PKcs activity in these extracts. However, these same Ku-defective extracts have low but detectable DNA-PK activity, indicative of Ku-independent kinase activity. However, although the mutation in xrs-6 cells inactivates the Ku80 protein, we cannot exclude the possibility that there is some expression from a methylation-silenced allele that we have shown to be present in these cells and capable of being activated (20, 39). Furthermore, we cannot assess whether the low activity has functional significance, since such cells lack Ku DNA end-binding activity. Taken together, however, our results support the model proposed by Hammarsten and Chu (15), that Ku stabilizes an interaction between DNA-PKcs and DNA and that this interaction serves to stimulate DNA-PK activity.
Although XCH80Δ560 and XYCH80Δ560 clones retain residual kinase activity and were partially complemented for γ-ray sensitivity, no complementation was observed with the larger truncated protein (Hs Ku80Δ554), even though, in the DNA-PKcs-overexpressing background, the cells retained some kinase activity and indeed had DNA-PK activity greater than that observed in CHO-K1 cells. In this context, it should be noted that the in vitro kinase assay uses a p53 peptide substrate and may not represent the constraints imposed upon the physiologically relevant in vivo substrate. The ability to phosphorylate the relevant in vivo substrate may therefore be more strongly impaired than is evident from the in vitro kinase assay.
We and others have observed the presence of aberrant Ku80 proteins in some human cell extracts (references 16 and 30 and our unpublished observations). These smaller Ku80 proteins were similar in size to our Hs Ku80Δ554 protein, did not cross-react with a Ku80 antibody specific for a C-terminal epitope, and retained DNA end binding but had decreased kinase activity. One particular extract had 90% smaller Ku80 protein and barely detectable DNA-PK activity (our unpublished data). Proteolytic degradation of Ku80 has been previously reported (33), and, in our hands at least, the aberrant, smaller Ku80 product appears to arise during extraction. Therefore, for example, when cells that appeared to have a smaller Ku80 protein were lysed in SDS and immediately subjected to Western blotting, a normal-sized Ku80 protein was obtained. Our results would therefore caution that the variant Ku80 proteins may not represent the in vivo Ku80 state and may not be related to radiation sensitivity. Nonetheless, these results support our findings with defined Ku80 truncated proteins and raise the possibility that such modifications of Ku have some function in the regulation of the NHEJ process.
The C-terminal region of Ku80 is less highly conserved between species than is the rest of the Ku80 cDNA. For example, the S. cerevisiae Ku80 cDNA is 102 amino acid residues shorter than its human homologue and most of these nonconserved residues are located in the C-terminal region. It is possibly significant, therefore, that a DNA-PKcs homologue has not been detected in yeast. In contrast, there is greater conservation between the C. elegans and mammalian sequences in this region, raising the possibility that these cells will have a functionally homologous DNA-PKcs protein. It has also been reported that Ku70 and Ku80 are evolutionarily related (8). Significantly, Ku70 lacks the C-terminal region present in Ku80, raising the possibility that only Ku80 functions in interactions with DNA-PKcs. Further work, including additional mutational analysis of Ku80 and Ku70, is required to characterize this interaction further.
A second strategy for the analysis of functionally important residues in Ku80 is the identification of mutations in Ku80-defective mutants. Previously, such analysis has not been informative, since the changes identified resulted in large deletions or truncations. Here we report the identification of a point mutational change in the xrs-2 mutant. Additionally, we report that the mouse GenBank sequence (U68181) has an error at position 410 and that a proline at this position is completely conserved between species. The proline-to-leucine change at this site in xrs-2 impairs the ability of the Ku80 protein to interact with Ku70, possibly by affecting protein conformation, as discussed above. Our results also show that this mutational change does not act as a dominant-negative mutation since the defect in xrs-2 cells can be corrected by wild-type Ku80 cDNA.
In conclusion, we show here that the C-terminal region of Ku80 significantly enhances the binding of DNA-PKcs to DNA and DNA-PK activity. Significantly, xrs-6 cells expressing Ku80 constructs lacking the C-terminal 178 amino acids acquire the phenotype of SCID cells even though they express wild-type DNA-PKcs protein. These results suggest that an interaction between DNA-PKcs and Ku80 is required for DSB repair and coding joint formation and that this interaction requires the C-terminal region of Ku80.
ACKNOWLEDGMENTS
We thank S. P. Jackson and W. H. Reeves for providing antibodies. We thank S. P. Jackson, D. Gell, H. Beamish, and A. R. Lehman for advice and critical reading of the manuscript.
This work was supported by a grant from the Kay Kendall Leukaemia Fund. Additional work in the P.A.J. laboratory is funded by European Union grant F13PCT920007, by the Human Frontiers Science Programme, and by a grant from the UKCCCR Radiation Research Programme. G.E.T. is a special fellow of the Leukemia Society of America and is supported by grant CA76409 from the National Cancer Institute. S.T.R. was supported by a Marshall Scholarship.
REFERENCES
- 1.Biedermann K A, Sun J, Giaccia A J, Tosto L M, Brown J M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 3.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 4.Cao Q P, Pitt S, Leszyk J, Baril E F. DNA-dependent ATPase from HeLa cells is related to human Ku autoantigen. Biochemistry. 1994;33:8548–8557. doi: 10.1021/bi00194a021. [DOI] [PubMed] [Google Scholar]
- 5.Cary R B, Chen F, Shen Z, Chen D J. A central region of Ku80 mediates interaction with Ku70 in vivo. Nucleic Acids Res. 1998;26:974–979. doi: 10.1093/nar/26.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Peterson S R, Story M D, Chen D J. Disruption of DNA-PK in Ku80 mutant xrs-6 and the implication in DNA double-strand break repair. Mutat Res. 1996;362:9–19. doi: 10.1016/0921-8777(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 7.Dvir A, Peterson S R, Knuth M W, Lu H, Dynan W S. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dynan W S, Hoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errami A, Smider V, Rathmeli W K, He D M, Hendrickson E A, Zdzienicka M Z, Chu G. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol Cell Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. DNA-dependent protein-kinase activity is absent in xrs-6 cells—implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulop G M, Phillips R A. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;374:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 12.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 13.Getts R C, Stamato T D. Absence of a Ku-like DNA end binding-activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 14.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement of DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 15.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Z, Johnston C, Reeves W H, Carter T, Wyche J H, Hendrickson E A. Characterization of a Ku86 variant protein that results in altered DNA binding and diminished DNA-dependent protein kinase activity. J Biol Chem. 1996;271:14098–14104. doi: 10.1074/jbc.271.24.14098. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson E A, Qin X-Q, Bump E A, Schatz D G, Oettinger M, Weaver D T. A link between double-strand break-related repair and V(D)J recombination: the scid mutation. Proc Natl Acad Sci USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 19.Jeggo P A. DNA-PK: at the cross-roads of biochemistry and genetics. Mutat Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 20.Jeggo P A, Holliday R. Azacytidine induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol Cell Biol. 1986;6:2944–2949. doi: 10.1128/mcb.6.8.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeggo P A, Kemp L M. X-ray sensitive mutants of Chinese hamster ovary cell line. Isolation and cross-sensitivity to other DNA damaging agents. Mutat Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 22.Jeggo P A, Smith-Ravin J. Decreased stable transfection frequences of six X-ray-sensitive CHO strains, all members of the xrs complementation group. Mutat Res. 1989;218:75–86. doi: 10.1016/0921-8777(89)90013-x. [DOI] [PubMed] [Google Scholar]
- 23.Jeggo P A, Taccioli G E, Jackson S P. Menage à trois: double strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 24.Jin S, Weaver D T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–368. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Lieber M R, Hesse J E, Lewis S, Bosma G C, Rosenberg N, Mizuuchi K, Bosma M J, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 27.Lin J D, Muhlmann-Diaz M C, Stackhouse M A, Robinson J F, Taccioli G E, Chen D J, Bedford J S. An ionizing radiation sensitive mutant cell line: irs-20. IV. Genetic complementation, V(D)J recombination, and the SCID phenotype. Radiat Res. 1997;147:166–171. [PubMed] [Google Scholar]
- 28.Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, Griffith A J, Hardin J A. Isolation and characterization of cDNA encoding the 80-kDa subunit protein of the human autoantigen Ku (p70/p80) recognized by autoantibodies from patients with scleroderma-polymyositis overlap syndrome. Proc Natl Acad Sci USA. 1990;87:1777–1781. doi: 10.1073/pnas.87.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuta R, Taccioli G E, Alt F W. The V(D)J recombination defect in the xrs-6 cell line results from a point mutation in the Ku80 gene. Int Immunol. 1996;8:1467–1471. doi: 10.1093/intimm/8.9.1467. [DOI] [PubMed] [Google Scholar]
- 30.Muller C, Salles B. Regulation of DNA-dependent protein kinase activity in leukemic cells. Oncogene. 1997;15:2343–2348. doi: 10.1038/sj.onc.1201402. [DOI] [PubMed] [Google Scholar]
- 31.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 32.Osipovich O, Durum S K, Muegge K. Defining the minimal domain of Ku80 for interaction with Ku70. J Biol Chem. 1997;272:27259–27265. doi: 10.1074/jbc.272.43.27259. [DOI] [PubMed] [Google Scholar]
- 33.Paillard S, Strauss F. Site-specific proteolytic cleavage of Ku protein bound to DNA. Proteins Struct Funct Genet. 1993;15:330–337. doi: 10.1002/prot.340150310. [DOI] [PubMed] [Google Scholar]
- 34.Pergola F, Zdzienicka M Z, Lieber M R. V(D)J recombination in mammalian cell mutants defective in DNA double strand break repair. Mol Cell Biol. 1993;13:3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priestley A, Beamish H J, Gell D, Amatucci A G, Muhlmann-Diaz M C, Singleton B K, Smith G C M, Blunt T, Schalkwyk L C, Bedford J S, Jackson S P, Jeggo P A, Taccioli G E. Molecular and biochemical characterisation of DNA dependent protein kinase defective rodent mutant, irs-20. Nucleic Acids Res. 1998;26:1965–1973. doi: 10.1093/nar/26.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves W H, Sthoeger Z M. Molecular cloning of cDNA encoding the p70 (Ku) lupus autoantigen. J Biol Chem. 1989;264:5047–5052. [PubMed] [Google Scholar]
- 38.Scholer H, Haslinger A, Heguy A, Holtgreve H, Karin M. In vivo competition between a metallothionein regulatory element and the SV40 enhancer. Science. 1986;232:76–80. doi: 10.1126/science.3006253. [DOI] [PubMed] [Google Scholar]
- 39.Singleton B K, Priestley A, Gell D, Blunt T, Jackson S P, Lehmann A R, Jeggo P A. Molecular and biochemical characterisation of mutants defective in Ku80. Mol Cell Biol. 1997;17:1264–1273. doi: 10.1128/mcb.17.3.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Singleton, B. K., P. A. Jeggo, D. Gell, and S. P. Jackson. Unpublished data.
- 40.Taccioli G E, Cheng H-L, Varghese A J, Whitmore G, Alt F W. A DNA repair defect in Chinese hamster ovary cells affects V(D)J recombination similarly to the murine scid mutation. J Biol Chem. 1994;269:7439–7442. [PubMed] [Google Scholar]
- 41.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene. Role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 42.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 43.Taccioli G E, Rathbun G, Shinkai Y, Oltz E M, Cheng H, Whitmore G, Stamato T, Jeggo P, Alt F W. Activities involved in V(D)J recombination. Curr Top Microbiol Immunol. 1992;182:108–114. doi: 10.1007/978-3-642-77633-5_13. [DOI] [PubMed] [Google Scholar]
- 44.Tuteja N, Tuteja R, Ochem A, Taneja P, Huang R K, Marusic L, Chen J Q, Zhang J W, Wang S, Pongor S, Falaschi A. Human DNA helicase-II—a novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Dong X, Myung K, Hendrickson E A, Reeves W. Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J Biol Chem. 1998;273:842–848. doi: 10.1074/jbc.273.2.842. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Lieber M R. Protein-protein and protein-DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol Cell Biol. 1996;16:5186–5193. doi: 10.1128/mcb.16.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaneva M, Kowalewski T, Leiber M R. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaneva M, Wen J, Ayala A, Cook R. cDNA-derived amino acid sequence of the 86-kDa subunit of the Ku antigen. J Biol Chem. 1989;264:13407–13411. [PubMed] [Google Scholar]
- 49.Zhu C M, Bogue M A, Lim D S, Hasty P, Roth D B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]