Abstract
Seed-feeding Nysius insects (Hemiptera: Lygaeidae) have a symbiotic association with distinct intracellular bacteria, “Candidatus Schneideria nysicola” (Gammaproteobacteria). Although many other hemipteran insect groups generally rely on bacterial symbionts that synthesize all ten essential amino acids lacking in their plant sap diets, the nutritional role of Schneideria in Nysius hosts that specialize on a more nutritionally complete seed-based diet has remained unknown. To determine the nutritional and functional capabilities of Schneideria, we sequenced the complete Schneideria genomes from three distantly related endemic Hawaiian Nysius seed bug species. The complete Schneideria genomes are highly conserved and perfectly syntenic among Hawaiian Nysius host species. Each circular chromosome is ∼0.57 Mb in size and encodes 537 protein-coding genes. They further exhibit a strong A + T nucleotide substitution bias with an average G + C nucleotide content of 29%. The predicted nutritional contribution of Schneideria includes four B vitamins and five of the ten essential amino acids that likely match its hosts’ seed-based diet. Disrupted and degraded genes in Schneideria suggests that Hawaiian lineages are undergoing continued gene losses observed in the smaller genomes of the other more ancient hemipteran symbionts.
Keywords: symbiosis, Heteroptera, intracellular bacteria, nutrition, amino acids, genome size
Introduction
“Candidatus Schneideria nysicola” (Gammaproteobacteria; hereafter Schneideria) is a vertically transmitted, intracellular bacterial symbiont restricted to the seed bug genus, Nysius (Order: Hemiptera and Suborder: Heteroptera) (Matsuura, Kikuchi, Hosokawa, et al. 2012; Matsuura, Kikuchi, Meng, et al. 2012). Schneideria is permanently sequestered within insect host cells (bacteriocytes) in specialized organs (bacteriomes) located in the host abdomen (Matsuura, Kikuchi, Hosokawa, et al. 2012). Although Schneideria’s symbiotic role has remained unknown, nutrition supplementation is predicted because this is a common function of many obligate insect symbionts (Buchner 1965; Baumann 2005; Jing et al. 2020). Such nutrient synthesis is especially prevalent among symbionts associated with plant sap-feeding insects in the Auchenorrhyncha and Sternorrhyncha (Hemiptera: Suborders) (Sudakaran et al. 2017; Bennett 2021). In these groups, insect hosts generally maintain ancient, highly conserved relationships with symbionts that synthesize all ten essential amino acids (EAAs) (Shigenobu et al. 2000; Wu et al. 2006; McCutcheon and Moran 2007; Weglarz et al. 2018) that are lacking in their xylem and phloem diets and cannot be made by animals (Douglas 1993; Sandstrom and Moran 1999). In contrast to most auchenorrhynchan and sternorrhynchan species that feed on plant saps, species in the Nysius genus use a barbed proboscis to feed on plant seed tissues that have greater potential nutritional return (Schuh and Slater 1995). Nonetheless, Nysius species likely retain Schneideria to provide some nutrition.
Although plant sap-feeding insects are dependent on symbiont-derived nutrition, their symbionts are reciprocally dependent on them for most cellular functions (McCutcheon and Moran 2012). Because symbionts are permanently intracellular with no environmental phase, over time, they often lose up to 90% of their genes due to functional and nutritional streamlining and ongoing stochastic gene losses (Moran and Bennett 2014; McCutcheon et al. 2019). Schneideria is a relatively recently acquired symbiont compared with many other bacteriome-bound symbionts in hemipteran insects (Matsuura, Kikuchi, Meng, et al. 2012; Bennett 2021), and there are important unanswered questions about what nutritional and functional roles Schneideria may play in its Nysius host species.
To address these questions, we present the complete genome sequences of Schneideria from three endemic Hawaiian Nysius species. Endemic Hawaiian Nysius provide an ideal system to investigate symbiont genome function and evolution. They represent over 25% of the approximately 100 described species in this globally distributed genus. The Hawaiian species are known to specialize on specific host plants or on distinct plant parts and are restricted to narrow habitat and ecological ranges (e.g., high elevation mesic and/or rain forests; Zimmerman 1948). To understand the functional role and genomic evolution of Schneideria, we selected three host species that span the phylogenetic and morphological diversity of the Hawaiian Nysius radiation (see supplementary fig. 1, Supplementary Material online).
Results and Discussion
Schneideria Has a Relatively Large Genome for an Intracellular Symbiont
The three Schneideria genomes from Hawaiian Nysius species are highly conserved and identical despite the phylogenetic and ecological diversity of their hosts. The genomes are perfectly syntentic with each other, and their circular chromosomes are ∼0.57 Mb in length with an average coding density of 93%. For scale, the Schneideria genomes are smaller than the 4.71 Mb size of the free-living Sodalis HS genome, but nearly five times larger than the 0.11 Mb size of the tiniest known bacterial genome of Nasuia found in leafhoppers (Oakeson et al., 2014; Bennett and Moran 2013). They further exhibit strong A + T nucleotide substitution bias with an average G + C nucleotide content of 29%. Finally, each Schneideria genome encodes 537 protein-coding genes, including the same four hypothetical genes of unknown function and four identical pseudogenes, a single rRNA operon (5S/16S/23S), and 39 tRNAs (table 1).
Table 1.
Comparison of the Schneideria Genome from the Seed Bug Nysius blackburni, to the Genomes of Sodalis HS (Free-Living Bacteria) (Oakeson et al. 2014), Sodalis BA (Obligate Symbiont of the Seed Bug Henestaris halophilus) (Santos-Garcia et al. 2017), and Sulcia ALF and Nasuia ALF (Obligate Co-symbionts of the Aster Leafhopper Macrosteles quadrilineatus) (Bennett and Moran 2013)
| Nasuia ALF | Sulcia ALF | Schneideria | Sodalis BA | Sodalis HS | |
|---|---|---|---|---|---|
| Chromosome size (Mb) | 0.11 | 0.19 | 0.57 | 1.62 | 4.71 |
| CDS (#) | 137 | 190 | 537 | 713 | 3993 |
| Pseudogenes (#) | 3 | 2 | 4 | 166 | 61 |
| rRNAs (#) | 3 | 3 | 3 | 3 | 23 |
| tRNAs (#) | 29 | 30 | 39 | 42 | 76 |
| Coding content (%) | 92 | 94 | 93 | 37 | 81 |
| GC content (%) | 17 | 24 | 29 | 45 | 58 |
| Accession number | CP006059 | CP006060 | CP074374 | PRJEB12882 | CP006569 |
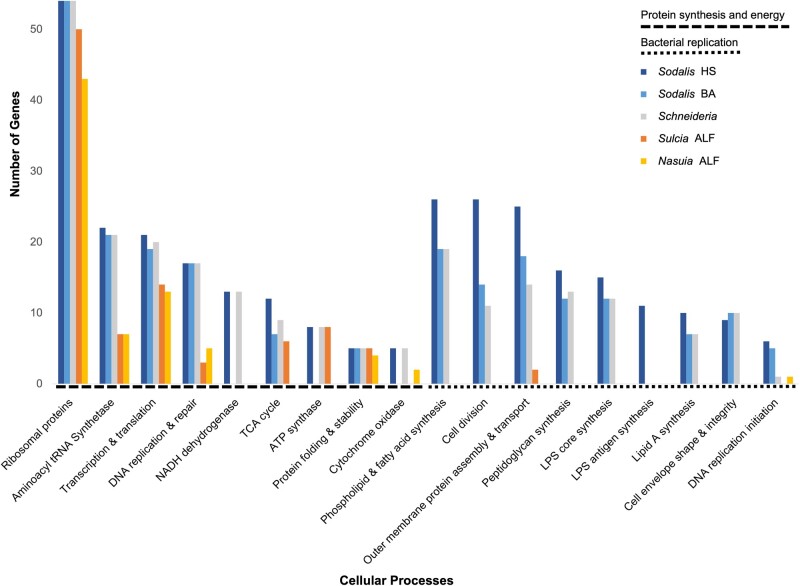
Schneideria’s genome is considerably larger than many other ancient symbionts found across hemipteran groups (e.g., “Ca. Carsonella rudii” in Sternorrhyncha [0.16 Mb] [Nakabachi et al. 2006], “Ca. Sulcia muelleri” in Auchenorrhyncha [∼0.15–0.27 Mb] [McCutcheon and Moran 2010], and “Ca. Evansia muelleri” in Coleorrhyncha [0.35 Mb] [Santos-Garcia et al. 2014], among many others; see also table 1). One of the main reasons Schneideria’s genome retains a relatively larger size, aside from vitamin and nutrition synthesis, is that it still encodes many genes involved in independent cell structures and cellular replication (e.g., cell division, phospholipid and fatty acid synthesis, outer membrane protein assembly and transport, peptidoglycan synthesis, etc.). These genes are highly conserved among the three Hawaiian lineages sequenced here. For example, each retains a relatively complete set of the fts cell division and mur peptidoglycan synthesis genes. Although these genes are primarily shared with the free-living Sodalis HS that has a much larger genome, the retention of these capabilities contrasts with many older hemipteran symbionts that typically lose all such functions (see fig. 1) (McCutcheon and Moran 2012). Schneideria’s expanded genome indeed appears to be functional. Previous transmission electron microscopy images show that it is still able to synthesize a cell wall and maintains a rod-shaped cell (Matsuura, Kikuchi, Hosokawa, et al. 2012; Matsuura, Kikuchi, Meng, et al. 2012). Symbionts that lose these capabilities typically have aberrant or shapeless cell morphologies (Buchner 1965).
Fig. 1.
Distribution of genes in central cellular processes. Bars represent the number of genes related to protein synthesis and energy, and bacterial replication in Schneideria from the seed bug Nysius blackburni, Sodalis HS (free-living bacteria) (Oakeson et al. 2014), Sodalis BA (obligate symbiont of the seed bug Henestaris halophilus) (Santos-Garcia et al. 2017), and Sulcia ALF and Nasuia ALF (obligate co-symbionts of the Aster Leafhopper Macrosteles quadrilineatus) (Bennett and Moran 2013).
Schneideria Provides a Limited Nutritional Profile to Its Seed-Feeding Hosts
Schneideria retains the genetic capabilities to synthesize four B vitamins and five EAAs that animals generally require from an exogenous source (fig. 2).
Fig. 2.
Essential amino acid and vitamin contributions of Schneideria. Genes in black font are present, genes in gray font are absent, and dotted lines are pathway links. Nutrients in black font are likely produced by the symbiont. Abbreviations for nutrients are as follows: Pan, pantothenate; Bio, biotin; Fol, folate; Rib, riboflavin; Pyr, pyridoxine; Ile, isoleucine; Val, valine; Leu, leucine; Lys, lysine; Thr, threonine; Met, methionine; Arg, arginine; His, histidine; Trp, tryptophan; Phe, phenylalanine.
On a broad level, the nutritional contribution of Schneideria to its Nysius hosts is less complete than in many other insect symbioses where the host receives all ten EAAs from either a single symbiont (e.g., Buchnera aphidicola in the pea aphid Acyrthosiphonpisum; Shigenobu et al. 2000), or from the parallel contributions of two or more obligate co-symbionts as is common among the Auchenorrhyncha (e.g., “Ca. Sulcia muelleri” and “Ca. Baumannia cicadellinicola” in the glassy-winged sharpshooter Homalodisca vitripennis that provide eight and two EAAs, respectively; Wu et al. 2006; McCutcheon and Moran 2007).
Several questions still remain about the complete capabilities of Schneideria to furnish several B vitamins and EAAs, as the pathways retained are incomplete (fig. 2). For example, the pantothenate B vitamin pathway appears to lack a key gene involved in an intermediate catabolic step (panE) and the phenylalanine EAA pathway is missing the final aminotransferase gene (aspC; see fig. 2). It is not uncommon for insect symbiont genomes to lose such essential nutrition synthesis genes involved in key initiation, intermediate, and even final catabolic steps (reviewed by Hansen and Moran [2014]). Although it is unclear how these pathways are ultimately completed in each case, recent studies have indicated that the host insect is likely able to contribute eukaryotic genes to the symbiosis that can bridge missing steps. A prominent example is in the synthesis of the EAA phenylalanine. Like Schneideria, many other bacterial symbionts of insects have lost the aspC gene, which is essential to the final catabolic step in phenylalanine synthesis. Gene expression data from bacteriocytes indicate that insect hosts dramatically upregulate a widely conserved insect gene capable of filling in for the missing aspC gene (Hansen and Moran 2011; Sloan et al., 2014). Similar patterns of host support and compensation of B vitamin synthesis pathways have been observed in other insect symbioses such as those found in mealybugs and related insects (Husnik and McCutcheon 2016). Remarkably, some of the genes involved in these cases have been horizontally acquired by insects from other infecting bacteria (reviewed by Mao et al. [2018]). In the Nysius, it seems likely that this general evolutionary process operates to similarly shore-up gene losses in the Schneideria genome.
Schneideria’s complete nutrient provisioning profile (i.e., fully or partially retained EAA and B vitamin nutrition synthesis pathways) likely reflects its hosts’ diet. Nysius seed bugs apparently do not require all ten EAAs from Schneideria. As seed feeders, hosts presumably have a more complete diet than other hemipterans that feed strictly on nutritionally depauperate xylem and phloem plant saps (Douglas 1993; Schuh and Slater 1995; Sandstrom and Moran 1999). For example, Nysiusterrestris are found on Chenopodium oahuense in the Amaranthaceae family that also includes quinoa, which is known to have enriched levels of protein and associated EAAs (Lutz and Bascunan-Godoy 2017). Similarly, Nysius blackburni and Nysiusrubescens are found on an endemic Hawaiian relative of the blueberry, Vaccinium reticulatum in the Ericaceae family. Like the seeds of Chenopodium, blueberries (i.e., the seeds) are also known to be enriched for EAAs compared with phloem and xylem (McCusker et al. 2014).
Schneideria Exhibits Recent Gene Losses Typical for Intracellular Nutritional Symbionts
Previous phylogenetic work indicates that the Nysius-Schneideria symbiosis is likely younger than many other more ancient obligate insect-bacteria symbioses found among the Auchenorrhyncha and Sternorrhyncha (Matsuura, Kikuchi, Meng, et al. 2012). The potential recency of the Schneideria symbiosis among Nysius species may explain its larger genome size (table 1) and relative cellular autonomy compared with other small obligate symbionts (fig. 2). Genome shrinkage typical of obligate symbionts in insects passes through several stages, including the initial bulk loss of unessential or redundant genes, which then transitions to ongoing stochastic losses primarily driven by genetic drift (Moran 1996; McCutcheon and Moran 2010, 2012; Bennett and Moran 2015; Wernegreen 2017; McCutcheon et al. 2019). Schneideria appears to already be well past these initial stages, as it has lost many genes that are present in its free-living relative, Sodalis HS (Oakeson et al. 2014). Many of these same genes have also been parallelly lost in Sodalis BA, which is the symbiont of another heteropteran seed bug species, Henestaris halophilus (Santos-Garcia et al. 2017). Although Schneideria and Sodalis BA have fewer genes than Sodalis HS, both symbionts have substantially more genes (especially those involved in bacterial replication) than are present in the smaller and more ancient symbionts (fig. 2; McCutcheon and Moran 2012; Bennett and Moran 2013).
Schneideria appears to be further experiencing ongoing loss of genes involved in its expanded capabilities in cell envelope synthesis and related functions (fig. 2). Specifically, genes predicted to be recently inactivated, but still identifiable as fragments, in all three Schneideria strains include tmk required for cell growth (Chaperon 2006) and three genes involved in cell membrane biosynthesis (hldE, pgpA, and kdsD). The hldE gene catalyzes reactions that help produce the LPS inner core region (Kneidinger et al. 2002), the pgpA gene helps catalyze a phospholipid in cell membranes (Icho and Raetz 1983), and the kdsD gene encodes arabinose‐5‐phosphate isomerase which plays a critical role in the biosynthesis of the lipopolysaccharide (LPS) that forms the outer membrane layer of gram-negative bacterial cells (Raetz and Whitfield 2002).
The processes of cell growth and membrane biosynthesis are disrupted and likely being lost in all three Schneideria strains, which suggests that Schneideria is evolving to have increased dependency on its host for cell membrane structures and integrity. For instance, in Escherichia coli, a loss of kdsD does not result in the disruption of LPS synthesis because gutQ is a paralogous gene that can perform the same function as kdsD (Meredith and Woodard 2005); however, gutQ is not present in the Schneideria genomes. The loss of LPS biosynthesis ability is common among insect symbionts (e.g., Koga and Moran 2014; Oakeson et al. 2014), and although Schneideria appears to still make most of its own cell envelope, it is clearly losing some genes involved in that process. Multiple losses of genes involved in related functions indicate that the Schneideria genome is expected to continue decreasing in size over evolutionary time as it sheds cellular functions and increases its dependence on its host (Baumann 2005; McCutcheon and Moran 2007; Moran and Bennett 2014; Bennett et al. 2015).
Materials and Methods
Insect Material and DNA Extraction
Target Nysius hosts were selected for comparative symbiont genomic sequencing based on their phylogenetic relationships (supplementary fig. 1, Supplementary Material online). This sampling allows us to assess the conserved functional role and genomic stability of Schneideria. Host phylogenies were estimated with two mitochondrial loci (Cytochrome Oxidase I and II) using maximum likelihood in RAxML v8.2.11 with a GTR + CAT model and 100 bootstrap replicates. Nysius blackburni, N. rubescens, and N. terrestris were chosen because they span the group’s phylogenetic diversity permitting genomic comparisons across the Hawaiian Nysius radiation (Zimmerman 1948). Nysiusblackburni, N. rubescens, and N. terrestris were collected in their natural habitats near Hilo, Hawai‘i, placed directly into 95% ethanol, and stored at −20 °C. Nysiusblackburni were collected from Vaccinium reticulatum (Ericaceae) understory litter, N. rubescens were collected from V. reticulatum branches and leaves, and N. terrestris were collected from Chenopodium oahuense (Amaranthaceae) branches and leaves. Bacteriomes were dissected and pooled for genomic sequencing from 20 N. blackburni, 13 N. rubescens, and 10 N. terrestris. Total genomic DNA was extracted with a PureLink Genomic DNA Kit (Invitrogen) and total DNA concentrations were determined with a Qubit v3 Fluorometer (Invitrogen).
Genome Sequencing, Assembly, and Annotation
The three Schneideria strains were sequenced with Illumina Miseq using a 2 × 300-bp paired-end protocol at the University of California Berkeley, QB3 Vincent J. Coates Genomics Sequencing Laboratory. The Illumina raw reads were trimmed with Trimmomatic v0.32 (Bolger et al. 2014) (settings: ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). The quality of the trimmed reads was assessed with FastQC (Andrews 2010). All reads were then de novo assembled in SPAdes v3.6.2 (Bankevich et al. 2012) with a k-mer length of 127 (settings: -k 127 -m 60 --careful --only-assembler -1). Approximately 13,229,453 (N. blackburni), 10,823,547 (N. rubescens), and 7,553,158 (N. terrestris) quality-filtered reads were obtained from the Illumina MiSeq run for each Schneideria strain. Coverage mapping of the genome assemblies resulted in unbroken scaffolds with high sequencing depths between 180× and 1,100×. No other bacterial genomes were recovered that meet general expectations for symbiont status.
Schneideria genomes and scaffolds were extracted based on size, coverage, GC content, and gene predictions from NCBI-BLASTP (Altschul et al 1990). After genomes were extracted and their circularity confirmed, we assessed assembly quality by mapping trimmed reads back to the bacterial contigs in Geneious v11.1.5 using Bowtie2 (Kearse et al. 2012; Langmead and Salzberg 2012). Through this process we identified several coverage breaks in the initial assemblies. These regions were compared against mapping reads and edited to reflect the dominant read-supported sequence. To confirm that the ambiguities were resolved, filtered reads were then re-mapped back to edited contigs to verify that there were no persistent coverage breaks.
The initial annotation of the Schneideria genome was performed with open reading frame (ORF) predictions using Glimmer v3 in Geneious v11.1.5 (Delcher et al. 2007). ORFs were initially identified with RAST (Aziz et al. 2008) and verified with individual NCBI-BLASTP searches (Altschul et al 1990). Finally, gene names, functions, and pathways were further verified with the EcoCyc database (Keseler et al. 2017). We then compared the genomic features of Schneideria between the three Schneideria strains (N. blackburni, N. rubescens, and N. terrestris), against the bacteriome-associated symbiont “Candidatus (Ca.) Sodalis baculum” (hereafter referred to as Sodalis BA) in the seed bug Henestaris halophilus (Hemiptera: Lygaeidae), and against the free-living Sodalis praecaptivus HS (hereafter referred to as Sodalis HS). We also compared the genomic content of Schneideria against two of the smallest and oldest known bacteriome-associated symbionts in the Hemiptera: “Ca. Sulcia muelleri” and “Ca. Nasuia deltocephalinicola” (hereafter referred to as Sulcia ALF and Nasuia ALF, respectively) in the sap-feeding Aster Leafhopper (ALF) Macrosteles quadrilineatus (Hemiptera: Cicadellidae).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank D. Rubinoff, M. Shintaku, M. Wright, and the University of Hawai‘i system for supporting this project, and M. Sheffield and A. Stever for assisting with Nysius collections. The authors also thank anonymous reviewers for their constructive input. Funding and logistical support for this research was provided by the Office of Maunakea Management, University of Hawai‘i at Hilo, HI, USA (3801415 to H.S.). Permits were issued by Hawai‘i Volcanoes National Park (United States National Park Service) (HAVO-2018-SCI-0044), the Hawai‘i Department of Forestry and Wildlife, the Hawai‘i Department of Land and Natural Resources, and the Natural Area Reserve (I1039; I1207; I2437).
Data Availability
The data for the chromosome sequences for the three strains of “Candidatus Schneideria nysicola” reported in this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/nucleotide/ (last accessed August 1, 2021) and can be accessed with the following accession numbers: CP074374, CP074435, and CP074100. The data for the Cytochrome Oxidase I and II genes reported in the supplemental materials are also available in the GenBank Nucleotide Database and can be accessed with the following accession numbers: Nysius blackburni (MZ362403; MZ362404); Nysius abnormis (MZ367506; MZ367507; MZ367516; MZ367517); Nysius coenosulus (MZ367504; MZ367505; MZ367514; MZ367515); Nysius communis (MZ367500; MZ367501; MZ367510; MZ367511); Nysius palor (MZ367502; MZ367503; MZ367512; MZ367513); Nysius plebeius (MN599979); Nysius rubescens (MZ362405; MZ362406); Nysius terrestris (MZ362407; MZ362408); and Nysius wekiuicola (MZ367508; MZ367509; MZ367518; MZ367519).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S.2010. FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed May 9, 2019.
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P.2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 59:155–189. [DOI] [PubMed] [Google Scholar]
- Bennett GM.2021. Evolving integrated multipartite symbioses between plant-sap feeding insects (Hemiptera) and their endosymbionts. In: Bosch TCG, Hadfield MG, editors. Cellular dialogues in the holobiont. Chapter 11. Milton (ON: ): CRC Press. p. 173–200. [Google Scholar]
- Bennett GM, McCutcheon JP, McDonald BR, Moran NA.. 2015. Lineage-specific patterns of genome deterioration in obligate symbionts of sharpshooter leaf-hoppers. Genome Biol Evol. 8(1):296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5(9):1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran N.. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 112(33):10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P.1965. Endosymbiosis of animals with plant microorganisms. New York: Wiley Interscience. [Google Scholar]
- Chaperon DN.2006. Construction and complementation of in frame deletions of the essential Escherichia coli thymidylate kinase gene. Appl Environ Microbiol. 72(2):1288–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL.. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23(6):673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE.1993. The nutritional quality of phloem sap utilized by natural aphid populations. Ecol Entomol. 18(1):31–38. [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 108(7):2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2014. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 23(6):1473–1496. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP.. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA. 113(37):E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T, Raetz CRH.. 1983. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol. 153(2):722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing TZ, Qi FH, Wang ZY.. 2020. Most dominant roles of insect gut bacteria: digestion, detoxification, or essential nutrient provision? Microbiome 8(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, et al. 2017. EcoCyc: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45(D1):D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneidinger B, et al. 2002. Biosynthesis pathway of ADP-l-glycero-beta-d-manno-heptose in Escherichia coli. J Bacteriol. 184(2):363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA.. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8(6):1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz M, Bascunan-Godoy L.. 2017. The revival of quinoa: a crop for health. In: Waisundara V, Shiomi N, editors. Superfood and functional food-an overview of their processing and utilization. Chapter 3. Rijeka, Croatia: Intech. p. 37–54. [Google Scholar]
- Mao M, Yang X, Bennett GM.. 2018. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in an leafhopper host. Proc Natl Acad Sci USA. 50:E11691–E11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Kikuchi Y, Hosokawa T, et al. 2012. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. 6(2):397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T.. 2012. Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl Environ Microbiol. 78(12):4149–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker S, Buff PR, Yu Z, Fascetti AJ.. 2014. Amino acid content of selected plant, algae and insect species: a search for alternative protein sources for use in pet foods. J Nutr Sci. 3:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Boyd BM, Dale C.. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol. 29(11):R485–R495. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 104(49):19392–19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Meredith TC, Woodard RW.. 2005. Identification of GutQ from Escherichia coli as a d-arabinose 5-phosphate isomerase. J Bacteriol. 187(20):6936–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA.1996. Accelerated evolution and Muller’s ratchet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM.. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68:195–215. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314(5797):267. [DOI] [PubMed] [Google Scholar]
- Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 6(1):76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C.. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem. 71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Garcia D, et al. 2014. Small but powerful, the primary endosymbiont of moss bugs, Candidatus Evansia muelleri, holds a reduced genome with large biosynthetic capabilities. Genome Biol Evol. 6(7):1875–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom J, Moran NA.. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 91(1):203–210. [Google Scholar]
- Santos-Garcia D, Silva FJ, Morin S, Dettner K, Kuechler SM.. 2017. The all-rounder sodalis: A new bacteriome-associated endosymbiont of the lygaeoid bug Henestaris halophilus (heteroptera: Henestarinae) and a critical examination of its evolution. Genome Biol Evol. 9(10):2893–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh RT, Slater JA.. 1995. True bugs of the world (Hemiptera: Heteroptera) classification and natural history. Ithaca (NY): Cornell University Press. [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H.. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407(6800):81–86. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31(4):857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Kost C, Kaltenpoth M.. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25(5):375–390. [DOI] [PubMed] [Google Scholar]
- Weglarz KM, Havill NG, Burke GR, von Dohlen CD.. 2018. Partnering with a pest: genomes of hemlock woolly adelgid symbionts reveal atypical nutritional provisioning patterns in dual-obligate bacteria. Genome Biol Evol. 10(6):1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ.2017. In it for the long haul: evolutionary consequences of persistent endosymbiosis. Curr Opin Genet Dev. 47:83–90. [DOI] [PubMed] [Google Scholar]
- Wu D, et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4(6):e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E.1948. Heteroptera. Insects of Hawaii. A manual of the insects of the Hawaiian Islands, including an enumeration of the species and notes on their origin, distribution, hosts, parasites, etc. Vol. 3. Honolulu (HI: ): University of Hawaii Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for the chromosome sequences for the three strains of “Candidatus Schneideria nysicola” reported in this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/nucleotide/ (last accessed August 1, 2021) and can be accessed with the following accession numbers: CP074374, CP074435, and CP074100. The data for the Cytochrome Oxidase I and II genes reported in the supplemental materials are also available in the GenBank Nucleotide Database and can be accessed with the following accession numbers: Nysius blackburni (MZ362403; MZ362404); Nysius abnormis (MZ367506; MZ367507; MZ367516; MZ367517); Nysius coenosulus (MZ367504; MZ367505; MZ367514; MZ367515); Nysius communis (MZ367500; MZ367501; MZ367510; MZ367511); Nysius palor (MZ367502; MZ367503; MZ367512; MZ367513); Nysius plebeius (MN599979); Nysius rubescens (MZ362405; MZ362406); Nysius terrestris (MZ362407; MZ362408); and Nysius wekiuicola (MZ367508; MZ367509; MZ367518; MZ367519).