Abstract
The regulation of intracellular ion concentrations is a fundamental property of living cells. Although many ion transporters have been identified, the systems that modulate their activity remain largely unknown. We have characterized two partially redundant genes from Saccharomyces cerevisiae, HAL4/SAT4 and HAL5, that encode homologous protein kinases implicated in the regulation of cation uptake. Overexpression of these genes increases the tolerance of yeast cells to sodium and lithium, whereas gene disruptions result in greater cation sensitivity. These phenotypic effects of the mutations correlate with changes in cation uptake and are dependent on a functional Trk1-Trk2 potassium transport system. In addition, hal4 hal5 and trk1 trk2 mutants exhibit similar phenotypes: (i) they are deficient in potassium uptake; (ii) their growth is sensitive to a variety of toxic cations, including lithium, sodium, calcium, tetramethylammonium, hygromycin B, and low pH; and (iii) they exhibit increased uptake of methylammonium, an indicator of membrane potential. These results suggest that the Hal4 and Hal5 protein kinases activate the Trk1-Trk2 potassium transporter, increasing the influx of potassium and decreasing the membrane potential. The resulting loss in electrical driving force reduces the uptake of toxic cations and improves salt tolerance. Our data support a role for regulation of membrane potential in adaptation to salt stress that is mediated by the Hal4 and Hal5 kinases.
Monovalent and divalent cations such as protons and potassium, sodium, calcium, and magnesium ions play multiple essential roles inside eukaryotic cells. Nonetheless, each of these ions must be maintained within a restricted concentration range to avoid toxicity (59, 68). The maintenance of internal ion concentrations in the face of widely ranging extracellular conditions is mediated by complex homeostatic pathways. Potassium, the major cellular cation, is actively retained so that it is present at very high concentrations internally. Potassium concentrations are the principal determinant of such physiological parameters as cell volume, turgor, and cytoplasmic ionic strength (which determines the optimal hydration layer of macromolecules and membranes) (63, 78). The threshold for the toxicity of other monovalent cations such as sodium and lithium is much lower than that for potassium. Sodium and lithium ions must be prevented from accumulating in the cytosol to protect essential and sensitive enzymes, such as the phosphatases of the Hal2 family (9, 46, 47).
Although many transporters for uptake and efflux of cations have been identified at the molecular level (2, 38, 63), the signal transduction pathways that regulate their activity and determine the homeostasis of intracellular ions remain largely uncharacterized. A better understanding of ion transporters and their regulation would have considerable practical value. In medicine this knowledge has revealed that inherited defects in ion homeostasis contribute to the risk of high blood pressure (36), and in agriculture the understanding of the proteins involved in ion homeostasis may permit the genetic engineering of crop plants with increased salt tolerance (63).
Regulatory circuits for ion transport may have several components that respond to different stimuli and may operate through multiple signal transduction pathways. The signals that trigger adaptive responses include turgor changes sensed by membrane proteins and changes in the intracellular concentrations of cations such as potassium, sodium, and calcium sensed by cytosolic proteins (63). Although knowledge about signaling pathways for ion homeostasis in eukaryotic cells is very limited, signaling components already identified include stretch-activated channels (21, 34), two-component signal transducers sensitive to osmotic stress (77), mitogen-activated protein kinase cascades activated by osmotic stress (15, 40, 77), “general” protein kinases such as protein kinase A (40), the calcium-activated protein phosphatase calcineurin (3, 44, 49), and transcription factors responsive to signals arising from osmotic stress (34, 54) and to calcium-calcineurin (41, 43, 67). The potential inputs modulating ion transporters may well include those operating during the cell growth and division cycle (13, 53), during metabolic switches (1, 40, 54), and in general stress responses (64).
Many of the recent advances in identifying ion transporters and their regulators have come from molecular genetic studies of the tolerance of the yeast Saccharomyces cerevisiae to salt stress (63). In this organism, the electrogenic plasma membrane H+-ATPase, encoded by the essential PMA1 gene, generates an electrochemical proton gradient that drives the secondary transport of nutrients (61, 62). Glucose metabolism can directly increase the expression of the PMA1 gene by a mechanism mediated via the Tuf1/Rap1/Grf1 transcription factor (6) and the product of the APA1 gene (16). In addition, this proton pump is activated at the protein level by glucose metabolism, which increases both its ATPase activity (60) and the H+/ATP coupling ratio (71). This regulation is effected by inactivation of an inhibitory domain at the carboxyl terminus of the enzyme (52), perhaps mediated via phosphorylation of Ser-899 (11). This regulatory domain also participates in the activation of the ATPase upon decreases in intracellular pH (4). The upregulation of Pma1 activity and consequent increased electrochemical gradient results in enhanced transport of nutrients. Increased activity of the plasma membrane ATPase may lead to an influx of toxic cations that move from the extracellular milieu into the cell.
Extrusion of toxic ions such as sodium and lithium in S. cerevisiae is dependent mainly on the ENA1/PMR2A gene. ENA1 is a member of a family of tandemly repeated genes that encode homologous P-type ATPases (23, 74). Transcription of ENA1, the only repeat that is highly expressed, is induced by sodium stress (17). This induction is mediated principally by the Hog1 mitogen-activated protein kinase (40) and the calcium-calmodulin-calcineurin (40, 44) pathways. In addition, ENA1 expression is modulated by the Hal3-Ppz1 regulatory subunit-protein phosphatase pair (13, 48, 53), by two signaling pathways triggered by glucose metabolism (the protein kinase A pathway [40] and the Snf1 pathway [1]), and by the Ure2-Gln3 pathway triggered by nitrogen metabolism (75). The Ena1 ATPase is also regulated posttranslationally by calcium-calmodulin via a calcineurin-independent mechanism (74).
By contrast, mechanisms that regulate cation uptake in S. cerevisiae remain poorly defined and the transporters primarily responsible for entry of Li+ and Na+ into the cell have yet to be identified. One clue is offered by the observation that the presence of high concentrations of Ca2+ (24) or K+ (49) in the medium confer enhanced resistance to Li+ and Na+ stress. The effects of Ca2+ may be mediated, at least in part, by its activation of calcineurin (41, 67), as well as by exchange and sequestration (8). However, the beneficial effects of K+ may be conferred either by competition or by exchange with other cations rather than as a consequence of its activation of a signaling pathway. Thus, it seems likely that K+ transport participates in ion tolerance directly or indirectly (20, 24).
The high-affinity potassium uptake system is encoded by the redundant transporter genes TRK1 and TRK2 (14, 32, 56). The Trk1-Trk2 system is activated by both potassium starvation and sodium stress (55), but transcriptional and/or posttranscriptional regulatory mechanisms remain uncharacterized. The sole link to a signaling pathway so far defined is that calcineurin is required for the activation of Trk1 and Trk2 by sodium stress (44). Importantly, by relaxing the electrical gradient induced by Pma1, the Trk1-Trk2 system is a major determinant of electrical membrane potential in S. cerevisiae (39) and consequently of cation uptake.
The paucity of information on Trk1-Trk2 regulatory mechanisms in yeast leaves a significant gap in our knowledge of ion homeostasis in yeast. In the present work, we describe two novel protein kinases encoded by two paralogs, HAL4/SAT4 and HAL5, that appear to function by regulating the Trk1-Trk2 potassium transporter. These partially redundant protein kinases are important determinants of ion homeostasis and salt tolerance, perhaps because they modulate the electrical membrane potential and, indirectly, the uptake of other cations such as sodium.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Standard methods for yeast culture and manipulation were used (22). All agar plates contained 2% Bacto Agar (Difco). Synthetic minimal medium (SD) contained 2% glucose, 0.7% yeast nitrogen base without amino acids (Difco), and 50 mM succinic acid adjusted to pH 5.5 with Tris. This medium was supplemented with adenine (30 μg/ml), histidine (30 μg/ml), uracil (30 μg/ml), leucine (100 μg/ml), methionine (100 μg/ml), and tryptophan (80 μg/ml) as indicated. Rich medium (YPD) contained 1% yeast extract (Difco), 2% Bacto Peptone (Difco), and 2% glucose. NaCl, KCl, LiCl, sorbitol, or tetramethylammonium were added as indicated. CaCl2 and hygromycin B were added to the already autoclaved medium before it was poured. YPD plates at pH 3.5 were prepared by adjusting a twofold-concentrated YPD stock containing 50 mM succinic acid to the desired pH with Tris, autoclaving, and mixing with concentrated agar before pouring. Salt tolerance was determined by drop tests in solid medium as described previously (18). Briefly, yeast strains were grown to saturation in liquid SD with the required supplements and diluted with water (1/10, 1/100 and 1/1,000), and 3 μl was dropped on plates containing salts as indicated.
The S. cerevisiae strains used for this work are listed in Table 1. Strains were derived by standard genetic crosses or by transformation by the lithium acetate procedure (28).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| RS16 | MATa leu2-3,112 ura3-251,328,372 | 18 |
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 73 |
| SKY697 | W303-1A ena1-4::HIS3 | 13 |
| SKY655 | W303-1A hal4::LEU2 | |
| SKY656 | W303-1A hal5::HIS3 | |
| SKY637 | W303-1A hal4::LEU2 hal5::HIS3 | |
| SKY624 | W303-1A cmd1-3 cnb1::LEU2 | 13 |
| WΔ3 | W303-1A trk1::LEU2 trk2::HIS3 | 39 |
| JM74 | W303-1A [YEp351] | |
| JM75 | W303-1A [YEp351-HAL4] | |
| JM76 | W303-1A [YEp351-HAL5] | |
| PM90 | W303-1A ena1-4::HIS3 [YEp351] | |
| JM51 | W303-1A ena1-4::HIS3 [YEp351-HAL4] | |
| JM52 | W303-1A ena1-4::HIS3 [YEp351-HAL5] | |
| JM77 | W303-1A [YEp352] | |
| JM63 | W303-1A trk1::LEU2 trk2::HIS3 [YEp352] | |
| JM65 | W303-1A trk1::LEU2 trk2::HIS3 [YEp24-HAL4] | |
| JM66 | W303-1A trk1::LEU2 trk2::HIS3 [YEp24-HAL5] | |
| JM78 | W303-1A trk1::LEU2 trk2::HIS3 [YCp50-ENA1] | |
| JM79 | W303-1A hal4::LEU2 hal5::HIS3 [YCp50-HAL5] | |
| JM110 | W303-1A trk1::LEU2 trk2::HIS3 hal4::TRP1 hal5::KanMX |
Unless otherwise indicated, the strains are from this study.
Isolation of the HAL4 and HAL5 genes.
The screen for superresistance to NaCl and LiCl has been described previously (13, 18, 43). Briefly, a genomic library from S. cerevisiae in the yeast shuttle vector YEp24 (2μm origin, URA3 marker [7]) was transformed into strain RS16 and selected on SD plates containing leucine. Transformants were recovered and plated on the above-described medium containing 0.2 to 0.4 M LiCl or 1.2 M NaCl. A second screen was performed by transformation of the same gene library into the salt-sensitive W303-1A ena1-4::HIS3 derivative (SKY697). Transformants were isolated on SD plates containing adenine, leucine, and tryptophan and selected on the same medium containing methionine and 40 mM LiCl. In an alternative screen, the salt-sensitive strain W303-1A cmd1-3 cnb1::LEU2 (SKY624 [13]) was transformed with a yeast cDNA library in the pRS316-GAL1 promoter-cDNA vector (37) and selected for uracil prototrophy. Transformants were recovered and plated on rich medium containing 2% galactose and 0.6 M NaCl. In all cases, colonies exhibiting improved growth were selected and plasmids were isolated. Plasmids conferring superresistance to salt upon reintroduction into yeast were subjected to partial sequencing to obtain nucleotide sequences at the ends of the inserts. The complete sequence of each clone was inferred by comparison of these sequences with the Saccharomyces genome database (59a).
Cloning and disruption of HAL4 and HAL5.
HAL4 was isolated from genomic clone PM47 as a SacI-HpaI fragment (comprising 531 bp before the starting codon and 299 bp after the stop codon) and subcloned into the yeast shuttle vector YEp351 (2μm origin, LEU2 marker [26]) cut with SacI and SmaI. The HAL5 gene was amplified by PCR from genomic clone PM42 as the template and with primers P42-1 (517 bp upstream of the starting codon; 5′-GCGGATCCTGGTAGAGAAGAGCTGG) and P42-2 (201 bp downstream of the stop codon; 5′-GCGGATCCCCGGGACTTCTAATGAC); the BamHI sites are underlined (see Fig. 1). The PCR product was cut with BamHI and subcloned into YEp351. The BamHI fragment with HAL5 was also subcloned into plasmid YCp50 (centromeric, URA3 marker [22]).
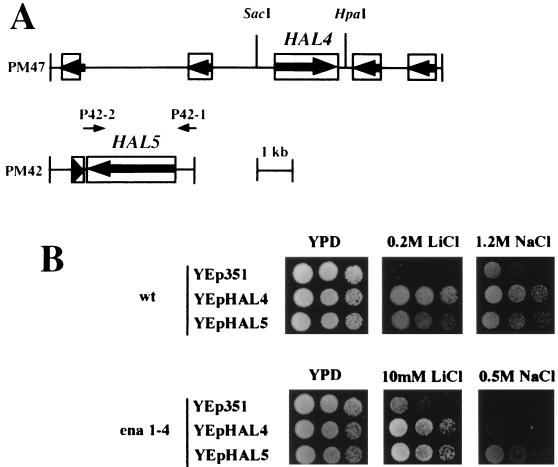
FIG. 1.
Isolation and halotolerance phenotypes of the HAL4 and HAL5 genes. (A) Genomic structure of the inserts of genomic clones PM47 and PM42 containing the HAL4 and HAL5 genes, respectively. ORFs are shown as boxes. Primers (not to scale) and restriction sites used for subcloning are indicated. (B) Overexpression of HAL4 and HAL5 confers Li+ and Na+ tolerance. The SacI-HpaI genomic fragment containing HAL4 and the PCR fragment amplified with the P42-2 and P42-1 primers containing HAL5 (see panel A) were subcloned into multicopy plasmid YEp351 (26) to give rise to overexpression plasmids YEp-HAL4 and YEp-HAL5, respectively. After transformation into strain W303-1A (wt) and its ena1-4::HIS3 derivative (ena1-4), salt tolerance was determined by drop tests in solid YPD containing the indicated concentrations of LiCl or NaCl. Cells transformed with empty YEp351 plasmid served as controls.
For disruption of HAL4 and HAL5, the XhoI-SacI fragments of the respective cDNAs were subcloned from the pRS316-GAL1 promoter vector (37) into pBluescript (Stratagene, La Jolla, Calif.). For HAL4, a 1,544-bp PstI-BssHII internal fragment was deleted and blunted with Klenow enzyme, and a BglII linker (New England Biolabs, Beverly, Mass.) was ligated into it. The BglII fragment of YEp13 containing the LEU2 gene (5) was ligated into the BglII site to make the knockout construct. An ApaI-SacI digest of this construct was used for transformation of W303 haploid cells of each mating type. For HAL5, a 1,433-bp EcoRI-NcoI internal fragment was deleted and blunted with Klenow, and a BglII linker was ligated into it. The BamHI fragment of pJH-H1 containing the HIS3 gene (29) was ligated into the BglII site to make the knockout construct. A HindIII-SacI digest was used for transformation of W303 haploid cells of each mating type. Gene disruptions were checked by Southern analysis. The double mutant MATa hal4 hal5 was made by crossing MATa hal4 and MATα hal5 mutants, sporulation, dissection, and genotyping of meiotic products.
For disruption of HAL4 and HAL5 in the WΔ3 strain (trk1::LEU2 trk2::HIS3), we disrupted HAL5 by the PCR-kanMX method (72). The primers used for amplification of the disruption cassette were DHAL51 (5′-CAGCAAGAATAA TAACTAGCAACGTATCTTCCCCGTCTATCAGCTGAAGCTTCGTAC GC) and DHAL52 (5′-CATCCAGGAGCTTTGTAGTAATTGTTCAATGGTGATTCTCGCATAGGCCACTAGTGGATCTG). For disruption of HAL4, the BglII-BamHI fragment of pJH-W1 containing the TRP1 gene (29) was ligated into the BglII site of the pBluescript construct described above and digested with XhoI and SacI before transformation.
Measurement of intracellular cation concentrations.
Cells were grown in YPD to an absorbance at 660 nm of 0.6 to 0.7, centrifuged for 5 min at 1,900 × g, resuspended at the same concentration in YPD containing the indicated concentration of salt, and incubated at 30°C. Aliquots of 10 ml were taken at several time points, centrifuged in plastic tubes for 5 min at 2,000 rpm and 4°C, and washed twice with 10 ml of ice-cold washing solution (20 mM MgCl2 and iso-osmotic sorbitol) by resuspension and subsequent centrifugation. The cell pellets were resuspended with 1 ml of cold washing solution, centrifuged again, and taken up in 0.5 ml of 20 mM MgCl2. Ions were extracted by heating the cells for 15 min to 95°C. After centrifugation, aliquots of the supernatant were analyzed with an atomic absorption spectrometer (Varian) in flame emission mode. For sodium efflux experiments, the cells were loaded for 2.5 h with the indicated concentrations of NaCl as described above, centrifuged, washed once, and resuspended at the same concentration in YPD without salt. Aliquots of 10 ml were processed as indicated above.
Measurement of methylammonium and rubidium uptake.
Cells were grown in SD containing 0.2 M KCl and the required supplements to an absorbance at 660 nm of 0.5 to 0.6, washed twice to remove potassium, and resuspended at the same concentration in a potassium starvation medium containing 2% glucose, 50 mM succinic acid (adjusted to pH 5.5 with Tris), and 1% casein hydrolysate (Merck). After a 4-h incubation, the cells were centrifuged, washed, resuspended with water, and stored in ice. For uptake measurements, the cells were diluted to a final absorbance at 660 nm of about 10 in a reaction medium containing 4% glucose and 50 mM succinic acid adjusted to pH 5.5 with Tris. After a 5-min preincubation, either [14C]methylammonium (0.2 mM and 2.5 μCi/ml final concentration) or 86RbCl (0.2 mM and 0.22 μCi/ml final concentration) were added from concentrated stock solutions. At the indicated times, the transport reaction was stopped by diluting samples of 100 μl with 10 ml of ice-cold 20 mM MgCl2 and cells were collected by vacuum filtration through a 0.45-μm-pore-size nitrocellulose filter (Millipore HAWP) and washed three times in the filter with 20 mM ice-cold MgCl2. Moist filters were transferred to scintillation cocktail, and radioactivity was monitored using a Pharmacia Wallac 1410 liquid scintillation counter.
RESULTS
Screens for superresistance to salt stress reveal a protein kinase that enhances tolerance by a novel mechanism.
We screened a library of yeast genes that are expressed from a 2μm plasmid to identify genes that confer salt tolerance upon overexpression (18). The halotolerance genes previously characterized by this high-gene-dosage strategy enhance salt resistance by two mechanisms. The elevated dose increases expression of the ENA1 cation extrusion pump (13, 43, 57), or the genes correspond to salt toxicity targets such as HAL2/MET22, which encodes a nucleotidase that dephosphorylates 3′-phosphoadenosine 5′-phosphate and 3′-phosphoadenosine 5′-phosphosulfate, intermediates in the sulfate assimilation pathway (19, 46, 47).
A high-copy-number genomic library was transformed into wild-type strain RS16, and over 200,000 transformants were selected in both SD and YPD supplemented with either 0.2 to 0.4 M LiCl or 1.2 M NaCl (18, 43). A total of 78 plasmids which, by retransformation, could confer salt tolerance were isolated. About half of them contained genomic clones which carried the previously described halotolerance genes HAL1 (18), HAL2 (19), HAL3 (13), and ENA1 (23). In addition, several plasmids containing two new genes were identified. Restriction analysis and recloning of the inserts revealed that one of these genes was YJL165c (45). In accordance with the other genes recovered in these screens, we have named this open reading frame (ORF) HAL5 for its role in halotolerance. HAL5 encodes a protein kinase that belongs to a subfamily unique to S. cerevisiae (27). This kinase subfamily includes three kinases (Npr1, Ptk1/Stk1, and Ptk2/Stk2) known to regulate plasma membrane transporters. Npr1 regulates ammonia-sensitive amino acid permeases (70) and Ptk1/Stk1 and Ptk2/Stk2 regulate polyamine transport (30, 31, 50).
The second gene is another member of this protein kinase-encoding family, YCR101c; it encodes a putative protein kinase, SAT4, involved in salt tolerance (53, 65). This gene was identified as an NPR1 homolog during the systematic sequencing of the yeast genome, and its disruption was previously reported to confer slight sodium sensitivity (65). In accordance with the other HAL genes, we suggest renaming the gene from SAT4 to HAL4, to refer to the screen for halotolerance. The original genomic inserts including HAL4 and HAL5 are depicted in Fig. 1A.
To avoid genes operating through the ENA1 or HAL2/MET22 systems, we performed the high-copy-number screen with a recipient in which the ENA1-4 tandem array is deleted (ena1-4). Salt-tolerant transformants isolated in this background should not be regulators of ENA1. By selecting for salt tolerance in the presence of methionine, the major salt toxicity target encoded by HAL2/MET22 was bypassed (19). A selection from 100,000 transformants yielded 96 recombinant plasmids that could confer lithium tolerance upon retransformation into the ena1-4 strain. As expected, a high proportion (65%) of these high-copy-number suppressor plasmids cross-hybridized with ENA1 and were not investigated further. However, 30 of the recovered plasmids were HAL5, which we had isolated earlier. The fact that only HAL5 but not HAL4 was isolated in the screen with the ena1-4 mutant and methionine-supplemented medium may be explained by the observation that the ena1 mutant is more sensitive to salt than is the wild type and that HAL5 confers greater salt tolerance than does HAL4 (Fig. 1B). The fact that only HAL4 and HAL5 arose in these selections suggests that they are the major proteins conferring salt tolerance under these conditions.
The Hal5/Npr1 subfamily of yeast protein kinases includes a gene product more closely related to Hal5p and Hal4p than to other members of the subfamily (27). The ORF (YKL168c) encoding this product was never isolated in our screens. We cloned this ORF with its own promoter in a multicopy plasmid, but this construct was unable to confer salt tolerance in our assay.
Salt tolerance conferred by overexpression of HAL4/SAT4 or HAL5 is independent of ENA1 and calcium signaling.
The halotolerance effects of the individual protein kinase genes are indicated in Fig. 1B. Overexpression of both genes in multicopy plasmids conferred sodium and lithium tolerance to wild-type Ena1-4+ strains such as RS16 and W303, as well as to W303 ena1-4 null mutants. No tolerance effects were observed in osmotic stress media prepared with 1.2 M KCl or 1.8 M sorbitol (data not shown). Therefore, these genes are not connected to osmotic tolerance but more probably are associated with sodium or lithium homeostasis. As with HAL5, the tolerance conferred by overexpression of HAL4 is also observed in methionine-supplemented media, where the Hal2 toxicity target is not essential to viability (data not shown). Therefore, HAL4 and HAL5 are new mediators of salt tolerance; they mediate tolerance by a mechanism that is independent of ENA1 and HAL2 and that may involve the regulation of membrane transporters for cation uptake.
HAL4 and HAL5 also suppress the salt sensitivity of the W303 cmd1-3 cnb1 double mutant, which is deficient in both calmodulin and calcineurin activity (13). Calcium regulates the transcription of the ENA1 gene through activation of both calcineurin and calmodulin, and it also regulates the activity of the Ena1 ATPase through activation of calmodulin (74). The W303 cmd1-3 cnb1 strain containing either HAL4 or HAL5 expressed from the GAL1 promoter (37) was plated on high-salt plates. These strains showed galactose-dependent salt tolerance.
hal4 and hal5 mutants are sensitive to sodium and lithium.
Since we were concerned that the salt resistance phenotypes we observed might be an indirect consequence of overexpressing the genes, we constructed strains lacking the HAL4 or HAL5 function. Loss of function rather than overexpression may offer a more reliable reporter of the possible participation of the Hal4 and Hal5 protein kinases in the control of ion homeostasis and salt tolerance under physiological conditions.
Strains containing a null mutation in either HAL4 or HAL5 displayed marked sodium and lithium sensitivity but grew normally in standard YPD. Importantly, the mutants showed no growth defect when subjected to osmotic stress by inoculation into medium containing 1.2 M KCl (Fig. 2) or 1.8 M sorbitol (results not shown), pointing to ion homeostasis rather than osmotic tolerance as the likely regulatory target of these protein kinases. Salt sensitivity was dramatically enhanced in the hal4 hal5 double mutant, suggesting that both proteins are required for maximum salt tolerance. In view of their sequence identity, it was reasonable to assume that HAL4 and HAL5 may be partially redundant, i.e., capable of performing the same function. In support of this hypothesis, a construction in which HAL5 was cloned into the YCp50 centromeric vector was able to suppress the salt-sensitive phenotype of both the hal5 and the hal4 single mutants completely. However, only partial suppression of the salt sensitivity of the hal4 hal5 double mutant was conferred by this clone (data not shown).
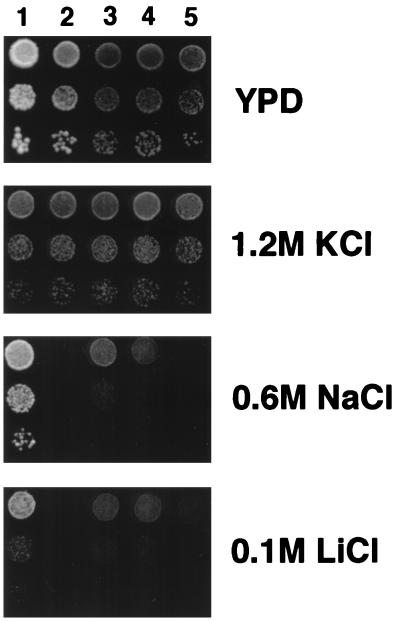
FIG. 2.
Yeast hal4 and hal5 mutants are sensitive to lithium and sodium. Strain W3031-A (lane 1) and its derivatives SKY697 (ena1-4::HIS3) (lane 2), SKY655 (hal4::LEU2) (lane 3), SKY656 (hal5::HIS3) (lane 4), and SKY637 (hal4::LEU2 hal5::HIS3) (lane 5) were tested for salt tolerance in YPD plates containing KCl, LiCl, or NaCl as indicated.
These results indicate that Hal4 and Hal5 are important determinants of ion homeostasis and salt tolerance in S. cerevisiae and that these homologous protein kinases are partially redundant. One model is that the Hal4 and Hal5 kinases cooperate in a single pathway modulating cation transport. Accordingly, we refer in the following to the Hal4 and Hal5 protein kinases as a single activity (Hal4-Hal5) involved in halotolerance.
The Hal4-Hal5 protein kinases regulate cation uptake instead of efflux.
Measurements of intracellular ion concentrations in yeast cells subjected to salt stress indicate that overexpression of HAL4 or HAL5 reduces lithium and sodium accumulation and increases intracellular potassium accumulation (Table 2). These alterations in ion homeostasis may explain the salt-tolerant growth of these strains and could reflect failure to either limit cation uptake or increase efflux. The major inducible efflux pathway in yeast is mediated by upregulation of ENA1 expression.
TABLE 2.
Effect of overexpression of HAL4 and HAL5 on the intracellular cation concentrations of cells incubated with LiCl and NaCla
| Strain | Plasmid | No salt, [K+]ib | 0.1 M LiClb
|
1 M NaClb
|
||
|---|---|---|---|---|---|---|
| [Li+]i | [K+]i | [Na+]i | [K+]i | |||
| wt | YEp351 | 330 (±30) | 69 (±3) | 383 (±6) | 283 (±10) | 92 (±5) |
| YEpHAL4 | 330 (±20) | 46 (±4) | 399 (±3) | 225 (±9) | 151 (±8) | |
| YEpHAL5 | 351 (±12) | 53 (±4) | 410 (±5) | 250 (±50) | 165 (±7) | |
| ena1-4 | YEp351 | 125 (±12) | 331 (±10) | 199 (±13) | ||
| YEpHAL4 | 113 (±6) | 300 (±30) | 260 (±20) | |||
| YEpHAL5 | 120 (±20) | 310 (±40) | 289 (±11) | |||
Strain W303-1A (wt), and its ena1-4 derivative (SKY697) were transformed with either empty plasmid YEp351 or YEp351 carrying HAL4 (YEpHAL4) or HAL5 (YEpHAL5). Cells were grown in YPD to exponential phase and treated for 3 h with the indicated concentrations of either LiCl or NaCl.
The intracellular concentrations (millimolar units) of potassium ([K+]i), lithium ([Li+]i), and sodium ([Na+]i) were determined as described in Materials and Methods. Results are the averages of three determinations (± standard deviations).
To test the possibility that the Hal4-Hal5 protein kinases regulate Ena1, we assayed β-galactosidase activity in cells transformed with an ENA1-lacZ fusion (13). We observed no increase in β-galactosidase activity in wild-type yeast compared to that in the hal4 hal5 mutant strain. Furthermore, mutants carrying the ena1-4 allele in combination with the hal4 and/or hal5 mutation displayed much greater salt sensitivity than did an ena1-4 mutant alone. These results suggest that the Hal4-Hal5 kinases can modulate ion transport independently of the expression and function of Ena1.
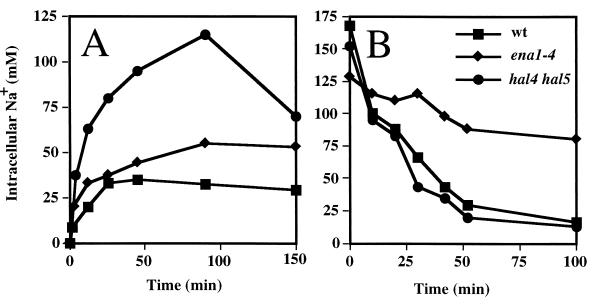
Direct comparison between hal4 hal5 mutants and ena1 mutants (Fig. 3) suggests that the Hal4-Hal5 protein kinases regulate sodium uptake rather than efflux. Net sodium accumulation was increased in both the ena1-4 mutant and in the hal4 hal5 mutant compared to uptake in wild-type yeast (Fig. 3A). On the other hand, sodium efflux after cell loading was altered only in the ena1-4 mutant (Fig. 3B). Therefore, the small increase in net sodium accumulation observed in the ena1-4 mutant might be explained by reduced cation efflux during the loading interval rather than by increased influx. In light of the above results, the significant increase in sodium uptake exhibited by the hal4 hal5 mutant is probably due to failure to appropriately regulate cation influx.
FIG. 3.
Effect of ENA1-4 and HAL4-HAL5 on Na+ uptake and efflux. (A) Sodium uptake. Cells from wild-type yeast (W303-1A) the ena1-4 mutant (SKY697), and the hal4 hal5 mutant (SKY637) were grown overnight in YPD, and 0.8 M NaCl was added at time zero. Samples were taken at the indicated times for determination of the intracellular sodium concentration. (B) Sodium efflux. Cells from the yeast strains described for panel A were loaded with either 0.8 M (wild type) or 0.4 M (ena1-4 and hal4 hal5 mutants) NaCl for 2.5 h, washed, and resuspended in NaCl-free medium at time zero. Samples were taken at the indicated times for determination of the intracellular sodium concentration. Results are the averages of three determinations, and the standard deviations were less of 15%. The experiment was repeated twice with similar results.
The Hal4-Hal5 protein kinases regulate the Trk1-Trk2 potassium transport system.
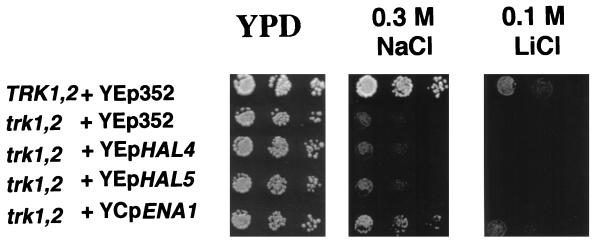
We tested whether the effects of the Hal4-Hal5 kinases were mediated by regulation of Trk1-Trk2-dependent potassium and/or sodium transport. The Trk1-Trk2 system is involved in high-affinity potassium uptake (14, 63). We found that overexpression of either HAL4 or HAL5 from a high-copy-number plasmid did not confer salt tolerance to a trk1 trk2 mutant strain (Fig. 4) whereas it does to a TRK+ strain. By contrast, even the modest overexpression of the ENA1 gene induced by a single additional copy carried on a centromere-based vector significantly increases the sodium and lithium tolerance of the trk1 trk2 mutant (Fig. 4). These results suggest that the effects of the Hal4-Hal5 protein kinases on ion homeostasis and salt tolerance are mediated via the Trk1-Trk2 potassium uptake system.
FIG. 4.
Overexpression of HAL4 and HAL5 is unable to confer salt tolerance in a trk1 trk2 mutant. Strain WΔ3 (trk1 trk2 double mutant) was transformed with control plasmid YEp352 (2μm origin, URA3 marker) (26), with YEp24 (2μm origin, URA3 marker) carrying HAL4 (YEpHAL4) or HAL5 (YEpHAL5), and with YCp50 carrying ENA1 (YCpENA1) (17). Salt tolerance was measured by drop test in YPD containing NaCl or LiCl as indicated. As a control for wild-type tolerance, we used strain W303-1A (TRK1 TRK2) transformed with empty plasmid YEp352.
Since the hal4 hal5 mutant exhibited increased sodium uptake, a plausible mechanism for the effects of Hal4-Hal5 on Trk1-Trk2 and salt tolerance is that under conditions of salt stress, these protein kinases phosphorylate the potassium transporters to inhibit their activity, thereby preventing the inappropriate influx of sodium and lithium. Indeed, our present studies show that the presence of potassium in the medium markedly decreases the apparent toxicity of sodium and lithium ions to wild-type cells. Perhaps the uptake of potassium via Trk1-Trk2 prevents the influx of sodium and lithium ions by competition.
The above model, however, was not compatible with the observation that trk1 mutants exhibit increased lithium uptake (57). Thus, an alternative model, i.e., that the Hal4-Hal5 protein kinases may be activators of Trk1-Trk2 and that the uptake of sodium and lithium during salt stress may be inversely proportional to Trk1-Trk2 activity, was considered. If Hal4-Hal5 kinases are required for Trk1-Trk2 function, then hal4 hal5 mutants might be deficient in Trk1-Trk2-dependent potassium accumulation. In fact, the hal4 hal5 double mutant showed a growth defect in SD which could be compensated by supplementation of the medium with 0.2 M KCl (Fig. 5). This potassium requirement was not as striking as for the trk1 trk2 mutant (Fig. 5), but it supports the idea that the Hal4-Hal5 protein kinases are required for proper functioning of the Trk1-Trk2 potassium uptake system. This hypothesis was tested directly by measuring 86Rb+ uptake in potassium-starved cells. Rubidium has been used as a tracer of potassium transport in S. cerevisiae, and its Trk1-Trk2-dependent uptake is activated by potassium starvation (55, 56). As previously observed (32), the trk1 trk2 mutant was markedly deficient in Rb+ uptake in comparison to wild-type controls (Fig. 6). Importantly, rubidium uptake in Trk1+-Trk2+ cells was dependent on the gene dosage of the Hal4-Hal5 protein kinases: overexpression of HAL5 increased Rb+ uptake whereas the hal4 hal5 double mutation greatly decreased it (Fig. 6). These results suggest a pathway in which Trk1-Trk2 is activated by the Hal4-Hal5 protein kinases to increase potassium import, resulting in a decrease in sodium and lithium uptake as a secondary effect.
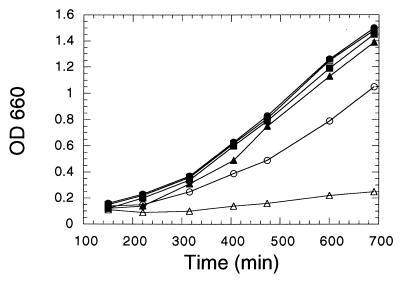
FIG. 5.
Growth of hal4 hal5 mutants is limited by potassium. Cells from wild-type yeast (W303-1A; squares), hal4 hal5 mutant cells (SKY637; circles) and the trk1 trk2 mutant (WΔ3; triangles) were grown overnight in SD containing 0.2 M KCl and the required supplements, washed, and resuspended at time zero in the same medium without (open symbols) or with (solid symbols) 0.2 M KCl. Growth was recorded by measurement of the absorbance at 660 nm (OD 660) at the indicated times.
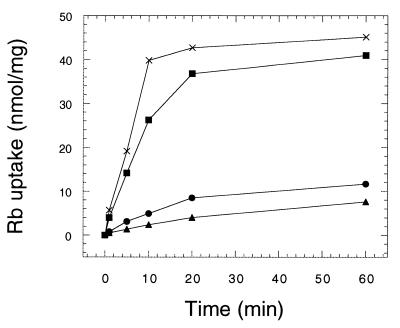
FIG. 6.
Rubidium uptake, an indicator of Trk1-Trk2 activity, is modulated by the gene dosage of HAL4 and HAL5. Cells from wild-type yeast (W303-1A; squares), wild-type yeast transformed with YEp351-HAL5 (JM76; crosses), the hal4 hal5 mutant (SKY637; circles), and the trk1 trk2 mutant (WΔ3; triangles) were grown to exponential phase in SD containing 0.2 M KCl and the required supplements, washed, and incubated for 4 h in potassium starvation medium. After further washing, the uptake of 0.2 mM 86Rb was determined at the indicated times as described in Materials and Methods.
Membrane hyperpolarization and sensitivity to toxic cations.
The uptake of potassium could restrict the uptake of sodium and lithium if the resulting net influx of positive charge significantly dissipates the plasma membrane electrical potential in yeast (62). In this model, the plasma membrane potential is inversely correlated with Trk1-Trk2 activity; activation of Trk1-Trk2 by overexpression of the Hal4-Hal5 protein kinases would result in depolarization of the plasma membrane. The absence of Hal4-Hal5 would lead to inactivation of Trk1-Trk2 and subsequent hyperpolarization, which might increase the rate of “leakage” of toxic cations into the cell.
Support for this mechanism of Hal4-Hal5 modulation of ion homeostasis was obtained by measuring [14C]methylammonium uptake as an indicator of plasma membrane electrical potential (69). Remarkably, overexpression of HAL5 decreased methylammonium accumulation. By contrast, uptake was greatly increased in the hal4 hal5 mutant (Fig. 7). As expected, the trk1 trk2 mutation also resulted in increased methylammonium uptake (Fig. 7). This indicator of hyperpolarization for the trk1 trk2 mutant is consistent with independent results obtained with fluorescent probes of membrane potential (39).
FIG. 7.
Methylammonium uptake, an indicator of membrane potential, is modulated by the gene dosage of HAL4 and HAL5. Strains, symbols, culture treatments, and transport procedures were as described in the legend to Fig. 6, except that 0.2 mM [14C]methylammonium was used as the transported substrate instead of 86Rb.
The constitutively hyperpolarized state of the hal4 hal5 mutant provides an attractive mechanism to explain both the increased uptake of sodium (Fig. 3) and the decreased sodium tolerance (Fig. 2). This mechanism may also explain the sensitivity of the hal4 hal5 mutant to other toxic cations such as lithium, calcium, hygromycin B, tetramethylammonium, and protons (Fig. 8). Interestingly, the trk1 trk2 mutant exhibited the same range of sensitivity, reinforcing the hypothesis that the Hal4-Hal5 protein kinases are activators of Trk1-Trk2. By comparison, the ena1-4 mutant was sensitive only to sodium and lithium, not to calcium, hygromycin B, tetramethylammonium, or low pH (Fig. 8). No increased sensitivity of hal4 hal5 or trk1 trk2 mutants was observed with KCl (a nontoxic salt) or sorbitol, pointing out that no loss in tolerance of osmotic stresses was involved.
FIG. 8.
Similar sensitivities to toxic cations of hal4 hal5 and trk1 trk2 mutants. Strain W303-1A (lane 1) and its derivatives the ena1-4 mutant (SKY697) (lane 2), the hal4 hal5 mutant (SKY637) (lane 3), and the trk1 trk2 mutant (WΔ3) (lane 4) were spotted on YPD plates containing the indicated concentrations of toxic cations or acidic buffer, and growth was monitored after 2 days. Hyg B, hygromycin B; TMA, tetramethylammonium.
DISCUSSION
The search for effectors of pathways mediating the tolerance of the budding yeast S. cerevisiae to high concentrations of sodium or lithium ions has provided novel insights into eukaryotic ion homeostatic mechanisms (63). In the present work, we have characterized two novel paralogous genes, HAL4 and HAL5, which encode partially redundant protein kinases required for ion homeostasis. We found that overexpression of either HAL4 or HAL5 confers enhanced tolerance to sodium and lithium. This tolerance is due at least in part to a reduction in the uptake of toxic cations into the cell. Reciprocally, cells lacking the HAL4 and/or HAL5 gene are more sensitive to these toxic cations than are wild-type cells. Hal4 and Hal5 are distinct from the majority of the other proteins found previously to participate in sodium and lithium tolerance, in that their ability to mediate halotolerance does not require the ENA1 sodium- and lithium-exporting ATPase. Instead, genetic and physiological evidence indicates that the Hal4-Hal5 protein kinases modulate the major potassium transporter of yeast cells, Trk1-Trk2 (14, 32, 56), and, indirectly, the uptake of toxic cations such as sodium.
The most compelling evidence for this novel mechanism of ion homeostasis is that overexpression of Hal4-Hal5 can confer salt tolerance only to cells capable of expressing the Trk1-Trk2 potassium transporters. In addition, the fact that mutants lacking HAL4 and HAL5 fail to grow on potassium-poor media and have decreased rubidium uptake suggest that Hal4-Hal5 kinases are positive effectors required for the normal function of the Trk1-Trk2 potassium uptake system. A likely mechanism by which activation of Trk1-Trk2 might confer enhanced salt tolerance is that the enhanced Trk1-Trk2 activity increases potassium uptake, resulting in a collapse of the membrane potential. The reduction in the membrane potential may slow the electrically driven influx of sodium and lithium across the plasma membrane.
This novel mechanism of salt tolerance, revealed by a study of the Hal4-Hal5 protein kinases, has highlighted the importance of Trk1-Trk2 in setting the electrical membrane potential of yeast cells. Our data suggest that a key determinant of ion homeostasis in S. cerevisiae is the activity of the Trk1-Trk2 system and its inverse relationship with membrane potential. The dependence of membrane potential on potassium transport has recently been proposed based on studies of the trk1 trk2 mutant by using a fluorescent reporter of membrane potential (39). Our results with methylammonium uptake as an indicator of membrane potential (69) provide independent evidence for this relationship. We interpret the effect of Trk1-Trk2 activity on membrane potential as reflecting the fact that Trk1-Trk2 is a major “consumer” of membrane potential in S. cerevisiae. The yeast membrane potential is determined by the relative activities of the electrogenic proton-pumping Pma1 ATPase and of the secondary transporters driven by the electrochemical proton gradient (62). Relaxation of the membrane potential due to Trk1-Trk2 may involve not only potassium transport but also proton transport (39).
The Trk1-Trk2 transporter system is not likely to be the pathway by which most other cations, including toxic cations such as lithium, sodium, calcium, tetramethylammonium, protons, and hygromycin B, enter the cell. Instead, “low-affinity” uptake pathways allow potassium and other cations to enter the cell under the influence of a negative-inside membrane potential. This uptake may be due to additive contributions of transporters selective for glucose (33, 35, 39) and other carbon sources, amino acids (76), choline, ammonium, and divalent cations (39), each of which is also likely to transport small cations nonspecifically. Where this has been measured, these transporters exhibit a nonspecific (and marginal) cation transport activity that can be increased by either overexpression or specific mutations within the transmembrane helices (35, 39).
We can only speculate about the signal transduction pathway connecting the Hal4-Hal5 protein kinases with the Trk1-Trk2 potassium transporter. At the physiological level, Trk1-Trk2 is known to be regulated by both potassium starvation and high sodium stress (55), but regulatory mechanisms, whether transcriptional or posttranscriptional, remain poorly characterized. Calcineurin is involved in the high-sodium activation of Trk1-Trk2 (44), but the Hal4-Hal5 protein kinases can mediate their effects in calcineurin mutants and probably define a second and independent pathway. Hal4-Hal5 protein kinases appear to be required for the potassium starvation response of Trk1-Trk2, a phenomenon independent of calcineurin (44) that, as with potassium transport in Escherichia coli (10), could be mediated by turgor decrease. It would be interesting to investigate if known turgor sensors of S. cerevisiae such as Sln1 or Sho1 (77) function upstream of Hal4-Hal5 to regulate Trk1-Trk2. Northern analysis indicates that the HAL4 and HAL5 genes are expressed at low, constitutive levels that do not increase during salt stress (data not shown), and therefore the Hal4-Hal5 protein kinases, if responsive to ion stress or other stimuli, are probably subject to regulation at the protein level.
It is relevant that Hal4-Hal5 kinases belong to a subfamily of yeast protein kinases whose other members are also dedicated to regulation of membrane transporters (27). The best studied, Npr1, regulates the Gap1 general amino acid permease (70). Active Gap1 permease is phosphorylated by Npr1. Feeding yeast cells with ammonia or glutamine causes rapid dephosphorylation and inactivation of Gap1. Subsequently, a slow disappearance of the Gap1 protein (66), probably mediated by the Npi1 ubiquitin-protein ligase (25), is observed. The direct phosphorylation of Trk1 by Hal4-Hal5 is under investigation, although its detection is complicated by the low abundance of Trk1 protein (14). Two other protein kinases of the Hal4-Hal5 family, Ptk1/Stk1 and Ptk2/Stk2, regulate polyamine transport mediated by unidentified transporters (30, 31, 50). Interestingly, stk2 mutants are not only defective in polyamine uptake but also tolerant to toxic concentrations of sodium and lithium (30). It is tempting to speculate that the phenotypes of the Stk1 and Stk2 protein kinases are also mediated by changes in membrane potential. However, the activities of Stk1 and Stk2 and those of Hal4-Hal5 would have opposite effects on this parameter. A pathway for regulation of Trk1-Trk2 by the Stk1 and Stk2 protein kinases is under investigation.
A plausible adaptive response in yeast cells confronted with high concentrations of toxic cations in the environment would be to relax their electrical membrane potential. This could be effected either by activating the Trk1-Trk2 potassium transporter or by inhibiting the Pma1 proton pump. Indeed, high sodium stress activates Trk1-Trk2 (55), and the calcium-activated protein phosphatase calcineurin participates in this regulation (44), along with its better-characterized role in elevating the expression of the ENA1 sodium and lithium extrusion pump (40, 44). Modulation of the membrane potential via Trk1-Trk2 activity might explain the role of calcineurin in tolerance to toxic cations not transported by Ena1, such as manganese and hygromycin B (75). The participation of the Hal4-Hal5 protein kinases in the response of Trk1-Trk2 to high sodium stress is also under investigation.
Consistent with these ideas, pma1 mutants with reduced electrogenic activity (51) confer resistance to Li+, Na+, Mn2+, and hygromycin B (42, 75), as would be expected from a reduction in membrane potential (negative inside) and concomitant decrease in the uptake of toxic cations. Interestingly, overexpression of yeast casein kinase I (encoded by the YCK1 and YCK2 genes) improves sodium tolerance (58), and this protein kinase has been reported to downregulate Pma1 activity (12). The signals modulating casein kinase I activity remain unknown. All the evidence presented here points to Hal4-Hal5 as an activator of Trk1-Trk2 rather than an inhibitor of Pma1. When we compared enzymatic activities of membranes extracted from wild-type cells and hal4 hal5 cells, we observed no change in Pma1 activity (data not shown). In addition, the hal4 hal5 trk1 trk2 quadruple mutant was not more sensitive to potassium limitation (growth in SD with no potassium supplementation) and lithium toxicity (10 mM LiCl in YPD) than was the trk1 trk2 mutant (data not shown). Thus, we conclude that these studies have identified a novel mechanism promoting tolerance to toxic cations via enhanced uptake of potassium.
The mechanism of salt tolerance promoted by the Hal4-Hal5 protein kinases impinges on a fundamental process of ion homeostasis: the coordinate regulation of potassium transport and electrical membrane potential. Given the similarity between plants and fungi in terms of basic ion transport mechanisms (61), it is a reasonable speculation that similar protein kinases may be present in plant cells as part of their ion homeostasis and ion-signaling regulatory circuits.
ACKNOWLEDGMENTS
This work was supported by grants from the Spanish CICYT (Madrid, BIO96-1196) and European Union (Brussels, BIO4-CT96-0775) to R.S. and by NIH grant GM40266 to G.R.F. J.M.M. is a fellow of the Conselleria de Educacio i Ciencia (Valencia), and G.R. was a fellow of the Spanish Ministerio de Educacion y Ciencia (Madrid). S.J.K. was supported in part by a fellowship from the Helen Hay Whitney Foundation.
We thank A. Rodriguez-Navarro (Madrid) for the WΔ3 strain and Avelino Corma (Instituto de Tecnologia Quimica, Valencia, Spain) for making available his atomic absorption spectrophotometer.
J.M.M. and M.P.L. contributed equally to this work.
REFERENCES
- 1.Alepuz P M, Cunningham K W, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- 2.André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 3.Aperia A, Ibarra F, Svensson L-B, Klee C, Greengard P. Calcineurin mediates α-adrenergic stimulation of Na+,K+-ATPase activity in renal tubule cells. Proc Natl Acad Sci USA. 1992;89:7394–7397. doi: 10.1073/pnas.89.16.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benito B, Portillo F, Lagunas R. In vivo activation of the yeast plasma membrane ATPase during nitrogen starvation. Identification of the regulatory domain that controls activation. FEBS Lett. 1992;300:271–274. doi: 10.1016/0014-5793(92)80861-a. [DOI] [PubMed] [Google Scholar]
- 5.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 6.Capieaux E, Vignais M-L, Sentenac A, Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem. 1989;264:7437–7446. [PubMed] [Google Scholar]
- 7.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham K W, Fink G R. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol Rev. 1986;39:73–78. [Google Scholar]
- 11.Eraso P, Portillo F. Molecular mechanism of regulation of yeast plasma membrane H+-ATPase by glucose. Interactions between domains and identification of new regulatory sites. J Biol Chem. 1994;269:10393–10399. [PubMed] [Google Scholar]
- 12.Estrada E, Agostinis P, Vandenheede J R, Goris J, Merlevede W, Francois J, Goffeau A, Ghislain M. Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I. J Biol Chem. 1996;271:32064–32072. doi: 10.1074/jbc.271.50.32064. [DOI] [PubMed] [Google Scholar]
- 13.Ferrando A, Kron S J, Rios G, Fink G R, Serrano R. Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol Cell Biol. 1995;15:5470–5481. doi: 10.1128/mcb.15.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaber R F, Styles C A, Fink G R. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2848–2859. doi: 10.1128/mcb.8.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galcheva-Gargova Z, Derijard B, Wu I-H, Davis R J. An osmosensing signal transduction pathway in mammalian cells. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Arranz M, Maldonado A M, Mazon M J, Portillo F. Transcriptional control of yeast plasma membrane H+-ATPase by glucose. Cloning and characterization of a new gene involved in its regulation. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 17.Garciadeblas B, Rubio F, Quintero F J, Bañuelos M A, Haro R, Rodriguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 18.Gaxiola R, de Larrinoa I F, Villalba J M, Serrano R. A novel and conserved salt-induced protein is an important determinant of salt tolerance in yeast. EMBO J. 1992;11:3157–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gläser H-U, Thomas D, Gaxiola R, Montrichard F, Surdin-Kerjan Y, Serrano R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993;12:3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez M J, Luyten K, Ramos J. The capacity to transport potassium influences sodium tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1996;135:157–160. doi: 10.1111/j.1574-6968.1996.tb07982.x. [DOI] [PubMed] [Google Scholar]
- 21.Gustin M C, Zhou X-L, Martinac B, Kung C. A mechanosensitive ion channel in the yeast plasma membrane. Science. 1988;242:762–765. doi: 10.1126/science.2460920. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 23.Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 24.Haro R, Bañuelos M A, Quintero F J, Rubio F, Rodriguez-Navarro A. Genetic basis of sodium exclusion and sodium tolerance in yeast. A model for plants. Physiol Plant. 1993;89:868–874. [Google Scholar]
- 25.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 26.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 27.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:14–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 28.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 30.Kaouass M, Audette M, Ramotar D, Verma S, de Montigny D, Gamache I, Torossian K, Poulin R. The STK2 gene, which encodes a putative Ser/Thr protein kinase, is required for high-affinity spermidine transport in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2994–3004. doi: 10.1128/mcb.17.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakimura Y, Maruyama T, Nozaki T, Wada Y, Ohsumi Y, Igarashi K. Cloning of the gene encoding a putative serine/threonine protein kinase which enhances spermine uptake in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;216:985–992. doi: 10.1006/bbrc.1995.2717. [DOI] [PubMed] [Google Scholar]
- 32.Ko C H, Gaber R F. TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4266–4273. doi: 10.1128/mcb.11.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko C H, Gaber R F. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:638–648. doi: 10.1128/mcb.13.1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon H M, Handler J S. Cell volume regulated transporters of compatible osmolytes. Curr Opin Cell Biol. 1995;7:465–471. doi: 10.1016/0955-0674(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 35.Liang H, Ko C H, Herman T, Gaber R F. Trinucleotide insertions, deletions, and point mutations in glucose transporters confer K+ uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:926–935. doi: 10.1128/mcb.18.2.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lifton R P. Molecular genetics of human blood pressure variation. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular cell biology. New York, N.Y: W. H. Freeman & Co.; 1995. pp. 633–668. [Google Scholar]
- 39.Madrid R, Gomez M J, Ramos J, Rodriguez-Navarro A. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 40.Marquez J A, Serrano R. Multiple transduction pathways regulate the sodium-extrussion gene PMR2/ENA1 during salt stress. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- 41.Matheos D P, Kinsbury T J, Ahsan U S, Cunningham K W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCusker J H, Perlin D, Haber J E. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendizabal I, Ríos G, Mulet J M, Serrano R, de Larrinoa I F. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998;425:323–328. doi: 10.1016/s0014-5793(98)00249-x. [DOI] [PubMed] [Google Scholar]
- 44.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 45.Mewes H W, Albermann K, Bähr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maieri A, Oliver S G, Pfeiffer F, Zollner A. Overview of the yeast genome. Nature. 1997;387(Suppl.):7–61. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 46.Murguía J R, Bellés J M, Serrano R. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 47.Murguía J R, Bellés J M, Serrano R. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem. 1996;271:29029–29033. doi: 10.1074/jbc.271.46.29029. [DOI] [PubMed] [Google Scholar]
- 48.Nadal E, Clotet J, Posas F, Serrano R, Gomez N, Ariño J. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc Natl Acad Sci USA. 1998;95:7357–7362. doi: 10.1073/pnas.95.13.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK-506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nozaki T, Nishimura K, Michael A J, Maruyama T, Kakinuma Y, Igarashi K. A second gene encoding a putative serine/threonine protein kinase which enhances spermine uptake in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1996;228:452–458. doi: 10.1006/bbrc.1996.1681. [DOI] [PubMed] [Google Scholar]
- 51.Perlin D, Brown C, Haber J E. Membrane potential defect in hygromycin B-resistant mutants of Saccharomyces cerevisiae. J Biol Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- 52.Portillo F, de Larrinoa I F, Serrano R. Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett. 1989;247:381–385. doi: 10.1016/0014-5793(89)81375-4. [DOI] [PubMed] [Google Scholar]
- 53.Posas F, Camps M, Ariño J. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J Biol Chem. 1995;270:13036–13041. doi: 10.1074/jbc.270.22.13036. [DOI] [PubMed] [Google Scholar]
- 54.Proft M, Serrano R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos J, Haro R, Rodriguez-Navarro A. Regulation of potassium fluxes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1029:211–217. doi: 10.1016/0005-2736(90)90156-i. [DOI] [PubMed] [Google Scholar]
- 56.Ramos J, Alijo R, Haro R, Rodriguez-Navarro A. TRK2 is not a low-affinity potassium transporter in Saccharomyces cerevisiae. J Bacteriol. 1994;176:249–252. doi: 10.1128/jb.176.1.249-252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rios G, Ferrando A, Serrano R. Mechanisms of salt tolerance conferred by overexpression of the HAL1 gene in Saccharomyces cerevisiae. Yeast. 1997;13:515–528. doi: 10.1002/(sici)1097-0061(199705)13:6<515::aid-yea102>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 58.Robinson L C, Hubbard E J A, Graves P R, DePaoli-Roach A A, Roach P J, Kung C, Haas D W, Hagedorn C H, Goebl M, Culbertson M R, Carlson M. Yeast casein kinase I homologues: an essential gene pair. Proc Natl Acad Sci USA. 1992;89:28–32. doi: 10.1073/pnas.89.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothstein A. Membrane function and physiological activity of microorganisms. In: Hoffman J F, editor. The cellular functions of membrane transport. Englewood Cliffs, N.J: Prentice-Hall; 1964. pp. 23–39. [Google Scholar]
- 59a.Saccharomyces Genome Database. http://genome-www.stanford.edu/Saccharomyces/.
- 60.Serrano R. In vivo glucose activation of yeast plasma membrane ATPase. FEBS Lett. 1983;156:11–14. doi: 10.1016/0014-5793(83)80237-3. [DOI] [PubMed] [Google Scholar]
- 61.Serrano R. Structure and function of plasma membrane ATPase. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:61–94. [Google Scholar]
- 62.Serrano R. Transport across yeast vacuolar and plasma membranes. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Genomic dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 523–585. [Google Scholar]
- 63.Serrano R. Salt tolerance in plants and microorganisms: toxicity targets and defense mechanisms. Int Rev Cytol. 1996;165:1–52. doi: 10.1016/s0074-7696(08)62219-6. [DOI] [PubMed] [Google Scholar]
- 64.Siderius M, Mager W H. General stress response: in search of a common denominator. In: Hohmann S, Mager W H, editors. Yeast stress responses. Berlin, Germany: Springer-Verlag KG; 1997. pp. 213–230. [Google Scholar]
- 65.Skala J, Purnelle B, Crouzet M, Aigle M, Goffeau A. The open reading frame YCR101 located on chromosome III from Saccharomyces cerevisiae is a putative protein kinase. Yeast. 1991;7:651–655. doi: 10.1002/yea.320070614. [DOI] [PubMed] [Google Scholar]
- 66.Stanbrough M, Magasanik B. Transcriptional and posttranscriptional regulation of the general amino-acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stathopoulos A, Cyert M S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stein W D. Channels, carriers, and pumps. An introduction to membrane transport. New York, N.Y: Academic Press, Inc.; 1990. [Google Scholar]
- 69.Vallejo C, Serrano R. Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast. 1989;5:307–319. doi: 10.1002/yea.320050411. [DOI] [PubMed] [Google Scholar]
- 70.Vandenbol, M., J.-C. Jauniaux, and M. Grenson. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encode a protein kinase homologue. Mol. Gen. Genet. 222:393–399. [DOI] [PubMed]
- 71.Venema K, Palmgren M G. Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J Biol Chem. 1995;270:19659–19667. doi: 10.1074/jbc.270.33.19659. [DOI] [PubMed] [Google Scholar]
- 72.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 73.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyperrecombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 74.Wieland J, Nitsche A M, Strayle J, Steiner H, Rudolph H K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Withee J L, Sen R, Cyert M S. Ion tolerance of Saccharomyces cerevisiae lacking the Ca2+/CaM-dependent phosphatase (calcineurin) is improved by mutations in URE2 or PMA1. Genetics. 1998;149:865–878. doi: 10.1093/genetics/149.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright M B, Ramos J, Gomez M J, Moulder K, Scherrer M, Munson G, Gaber R F. Potassium transport by amino acid permeases in Saccharomyces cerevisiae. J Biol Chem. 1997;272:13647–13652. doi: 10.1074/jbc.272.21.13647. [DOI] [PubMed] [Google Scholar]
- 77.Wurgler-Murphy S M, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 78.Wyn Jones R G, Pollard A. Proteins, enzymes and inorganic ions. In: Laüchli A, Pirson A, editors. Encyclopedia of plant physiology, new series. 15B. Berlin, Germany: Springer; 1983. pp. 528–562. [Google Scholar]