Abstract
The yeast UME3 (SRB11/SSN3) gene encodes a C-type cyclin that represses the transcription of the HSP70 family member SSA1. To relieve this repression, Ume3p is rapidly destroyed in cells exposed to elevated temperatures. This report demonstrates that Ume3p levels are also reduced in cultures subjected to ethanol shock, oxidative stress, or carbon starvation or during growth on nonfermentable carbons. Of the three elements (RXXL, PEST, and cyclin box) previously shown to be required for heat-induced Ume3p destruction, only the cyclin box regulates Ume3p degradation in response to these stressors. The one exception observed was growth on nonfermentable carbons, which requires the PEST region. These findings indicate that yeast cells contain multiple, independent pathways that mediate stress-induced Ume3p degradation. Ume3p destruction in response to oxidative stress, but not to ethanol treatment, requires DOA4 and UMP1, two factors required for 26S proteasome activity. This result for the first time implicates ubiquitin-mediated proteolysis in C-type cyclin regulation. Similarly, the presence of a membrane stabilizer (sorbitol) or the loss of phosphatidylinositol-specific phospholipase C (PLC1) protects Ume3p from oxidative-stress-induced degradation. Finally, a ume3 null allele suppresses the growth defect of plc1 mutants in response to either elevated temperature or the presence of hydrogen peroxide. These results indicate that the growth defects observed in plc1 mutants are due to the failure to downregulate Ume3p. Taken together, these findings support a model in which Plc1p mediates an oxidative-stress signal from the plasma membrane that triggers Ume3p destruction through a Doa4p-dependent mechanism.
The normal growth and development of all organisms requires the ability to sense and correctly respond to a myriad of environmental signals. In Saccharomyces cerevisiae, several systems are involved in extracellular signaling depending on the nature of the stimuli. For example, the RAS pathway activates cyclic AMP-dependent protein kinase A (PKA) in response to nutrient availability (50). RAS functions as a molecular switch for turning on and off PKA by alternating between the GDP (inactive) and the GTP (active) bound states. Maintenance of the GTP bound form is accomplished by either stimulating a GTP/GDP exchange factor or inhibiting the GTPase activating protein or GAP (reviewed in reference 3). However, strains unable to downregulate this pathway are more sensitive to the toxicity associated with heat shock or starvation (53). These results indicate that both the stimulation and inhibition of signaling pathways are important for the correct cellular response to changing environmental conditions.
Other signal transduction pathways perform more specialized roles in sensing changes in the local environment. The high -osmolarity glycerol (HOG) response pathway senses hypertonic conditions through a mitogen-activated protein (MAP) kinase module ending with the Hog1p MAP kinase (2). In response to hypotonic medium, protein kinase C (PKC1) is activated, which in turn stimulates the MAP kinase Mpk1p through a MAP kinase cascade separate from that utilized by the HOG pathway (11, 28). In addition to its requirement in hypotonic environments (29), the PKC1 pathway is also required for survival in response to heat shock (24). The phenotypes associated with pkc1 mutations are suppressed by the presence of an osmostabilizing agent (e.g., sorbitol) in the medium. These findings suggest that maintenance of membrane integrity under stress conditions is an essential function of Pkc1p.
In mammalian cells, Pkc is activated by the second messenger diacylglycerol, one product of the hydrolysis of phosphatidylinositol-4,5-bisphosphate by the phosphatidylinositol-specific phospholipase Cγ (Plcγ) (21). The other message generated by Plcγ, inositol 1,4,5-triphosphate or InP3, activates calmodulin-dependent kinases or phosphatases through the release of stored calcium (reviewed in reference 1). In the budding yeast cell, mutants lacking the Plcγ homolog PLC1 display several phenotypes, including growth defects at high temperature or in hypertonic medium (15, 37). These results also suggest a role for Plc1p in the maintenance of the plasma membrane. However, genetic experiments failed to identify a functional relationship between Plc1p and Pkc1p (15). In addition, a connection between Plc1p activation and calcium-dependent kinases or phosphatases (calcineurin) has not been directly demonstrated. Therefore, although a role for regulating membrane integrity is suggested by mutational studies, the actual function of Plc1p or a connection to a signaling pathway remains obscure.
The existence of multiple signal transduction pathways allows the cell to activate the appropriate gene expression program depending on the nature of the stimuli. In response to many types of stress, organisms ranging from bacteria to humans induce several highly conserved gene families collectively called heat shock proteins (Hsps; for a review, see reference 10). Heat shock gene induction occurs primarily through increased transcription that requires a highly conserved activator called heat shock factor (HSF; for a review, see reference 34). Although some detail is known concerning the induction of HSP genes, little information is available about the regulators involved in their repression. The yeast C-type cyclin Ume3p (Srb11p/Ssn3p) and the cyclin-dependent kinase (Cdk) that it regulates (Ume5p) were identified as negative regulators of several early meiotic genes (e.g., SPO13) (47). Subsequent studies revealed that this cyclin-Cdk complex is required for the repression of SSA1, an HSP70 family member (8). To relieve this repression, Ume3p is rapidly destroyed in cultures subjected to heat shock (8), suggesting that the downregulation of this cyclin is part of the normal cellular response to stress.
Three elements (RXXL, PEST, and cyclin box) are required for the rapid turnover rate of Ume3p in response to heat shock (8). The RXXL motif is similar to the destruction box required for the degradation of G2 cyclins via the ubiquitin system (18). Regions rich in proline, glutamic acid, serine, and threonine (PEST) residues are necessary for the rapid turnover of the budding yeast G1 cyclins (Cln1-3p) (41, 51), as well as other regulatory molecules (25). Similar to G2 cyclins, Cln destruction requires the ubiquitin pathway (12). However, heat shock-induced degradation of Ume3p was not dependent on several components of the ubiquitin pathway or the 26S proteosome itself (8), suggesting the presence of a destruction mechanism alternative to that observed for other cyclins. The third destruction element is located within the cyclin box, a region that confers Cdk binding specificity (38) but had not previously been shown to be involved in regulating protein turnover. Mutating each element individually increased the Ume3p half-life three- to fivefold in cultures subjected to heat shock, while the PEST-RXXL double mutant exhibited a greater than 15-fold increase in half-life (8). These findings suggest that more than one pathway is involved in controlling Ume3p levels. However, this study also demonstrated that the cyclin box mutation protected Ume3p from meiosis-induced degradation, indicating that the regulatory pathways sensing diverse stimuli (meiosis and heat shock) may share some components.
To further explore the regulation of Ume3p in response to stress, the present study examined Ume3p regulation in cultures subjected to several conditions known to induce cellular damage. These experiments revealed that Ume3p was destroyed in response to a subset of stress conditions (e.g., ethanol shock and oxidative stress). Furthermore, the cyclin box destruction element is required for the turnover of this cyclin under most of these stress conditions. This study also demonstrates that DOA4 and UMP1, two components of the 26S proteasome, are required for oxidative-stress-induced downregulation of Ume3p. In addition, Ume3p is protected from oxidative-stress-induced degradation in strains either lacking PLC1 or in cultures grown in sorbitol-containing medium. Finally, a ume3 null allele suppresses both the temperature-sensitive growth phenotype and hypersensitivity to oxidative stress observed in plc1 mutants. Taken together, these findings indicate that Ume3p is destroyed in response to a diverse set of exogenous stressors. Moreover, these results suggest that downregulating this cyclin is the major mechanism by which Plc1p protects the cell from stress.
MATERIALS AND METHODS
Media.
Cultures were grown on either rich YPDA medium (20 g of yeast extract, 10 g of peptone, and 20 g of dextrose per liter supplemented with 2 ml of 0.5% adenine sulfate per liter) or synthetic dextrose (SD; 1.7 g of yeast nitrogen base, 5 g of ammonium sulfate, and 20 g of dextrose per liter supplemented with amino acids and uracil as required). SD medium with 2% ethanol, glycerol, or acetate substituted for dextrose was used as the nonfermentable carbon source medium. For carbon or nitrogen depletion studies, the dextrose or ammonium sulfate was omitted from the SD medium, respectively. Phosphate-limiting medium was produced by adding 20 ml of 1 M MgSO4 and 20 ml of concentrated NH4OH per liter of YPDA, stirring the solution at 25°C for 30 min and then filtering the medium twice through Whatman number 1 paper to remove the Mg(NH4)PO4 precipitate. SD medium containing 10% sorbitol (wt/vol) was used in the membrane-stabilizing experiments. Hydrogen peroxide plates were prepared as described previously (26).
Plasmids and strains.
The yeast strains used in this study are listed in Table 1. To monitor SSA1 mRNA expression, an ssa1-lacZ reporter gene (pZDO25) was used (45). The RAS2 (B1569) and RAS2val19 (B1695) expression constructs were a kind gift from C. Davis and J. Broach (Princeton University). The RAS mutants containing the G19D, E99K, D126N, or E130K substitutions (53) were a gift from J. Cannon (University of Missouri—Columbia). The UME3 open reading frame was tagged with the c-myc epitope (14) and placed under the control of the ADH1 promoter on a single-copy vector to form pKC337 (8). This construct is able to complement a ume3 mutation, indicating that Ume3p is still functional. The Ume3p mutant derivatives pKC220 (L28A), pKC359 (PESTΔ), and pKC202 (A110V) disrupting the RXXL, PEST, and cyclin box destruction signals, respectively, were described previously (8). The PLC1 deletion strains were constructed by using PCR-based one-step transplacement (30) with oligonucleotides complementary to regions just upstream of the initiation codon (5′-AAACGTACAACGGTAAGGTCATTCACGCAGTGTATATGA-3′) and downstream of the termination codon (5′-CGCGTATTTATGAATATGTGTATTTGGCCGGAAAAAGAT-3′). Deletion alleles were confirmed by using PCR methodologies of yeast genomic DNA with primers internal to the selective marker (his5+) and in the PLC1 promoter region.
TABLE 1.
Strain genotypes used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| RSY10 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 48 |
| RSY391 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ume3::LEU2 | 8 |
| 1500-1A | MATa ade1 cln1-del cln2-del cln3-del leu2::GAL1::CLN1 his2 trp1 ura3 | F. Cross |
| W303hog1Δ | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hog1::LEU2 | M. Gustin |
| MHY623 | MATa his3Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 doa4::LEU2 | 5 |
| RSY531 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 plc1::his5+ | This study |
| RSY532 | MATa ade2 ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ume3::LEU2 plc1::his5+ | This study |
| JD59 | MATa his3Δ-200 leu2-3,112 lys2-801 trp1-Δ63 ura3-53 ump1::HIS3 | 40 |
| JTY2304 | MATa ade2-101och his3Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 plc1Δ::HIS3 | J. Thorner |
Ume3p stability assays.
To examine Ume3p levels in response to various stresses, cultures harboring pKC337 (and pZDO425 when indicated) were grown to mid-log phase (5 × 106 cells/ml) in SD medium and subjected to the treatments described below. Cells were grown (500 ml per assay time point) and treated at 30°C except for the experiments described in Fig. 1, which were performed at 23°C. For each assay, the experimental conditions used followed previously published protocols. Since the stress response is induced with different kinetics depending of the type of stimulus, the treatments were first analyzed in a time course experiment to identify the optimal time after administration of the treatment to examine Ume3p levels. After each treatment described below, the cells were harvested by centrifugation followed by flash freezing in a dry ice-ethanol bath. For heat shock analysis, cells were washed in water, pelleted in a Falcon tube, vortexed briefly to disperse the cells, and kept at 37°C for 12 min. (42, 45). Ethanol shock was performed by adding ethanol (6%, final concentration) and then incubating the culture with shaking for the times indicated in the text (39, 42). For oxidative stress, either 0.4 mM hydrogen peroxide (H2O2, final concentration) or 1 mM menadione (bisulfate salt; Sigma) (7, 22) was added directly to the culture. For both treatments, the cells were incubated with shaking for the indicated times. UV irradiation was examined by exposing a shaking, thin-cell culture to a 100-J/m2 dose of irradiation as described previously (36). After treatment, the cells were incubated in the dark to prevent photoreactivation. Samples were taken before and after UV treatments every 15 min for 1 h. Cells were subjected to hypertonic stress by resuspending a washed culture in growth medium containing 0.4 M NaCl, followed by incubation for 240 min (2, 52). Samples were removed for analysis every 30 min. Nutrient deprivation (13) was accomplished by resuspending washed cells in medium lacking phosphates, carbon, or nitrogen. The cultures were harvested for analysis after a 4-h incubation at 30°C with shaking. To examine Ume3p levels in cultures growing on nonfermentable carbons, dextrose-grown cultures were washed and then resuspended in minimal medium containing 2% of the indicated carbon source and incubated for 4 h. Cell survival was monitored at each time point by serially diluting an aliquot in water followed by spotting on SD plates.
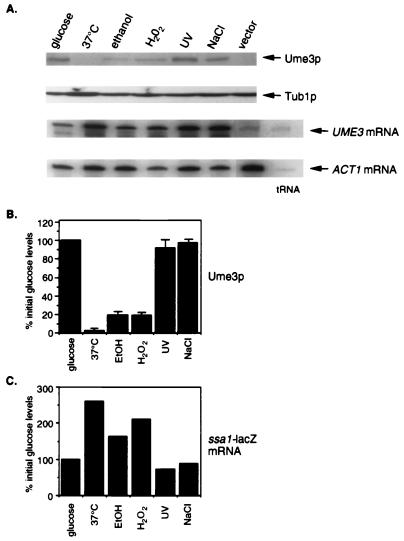
FIG. 1.
Ume3p regulation by environmental stress. (A) Mid-log-phase RSY10 cultures (glucose) were transferred to the conditions indicated (see Materials and Methods for details). UME3 protein (Ume3p) and transcript levels were determined by Western blot and S1 nuclease protection studies, respectively. ACT1 and Tub1p levels serve as loading controls for mRNA and protein, respectively. The vector lane controls for the nonspecific cross-reactivity of the myc monoclonal antibody. The tRNA lane indicates bands corresponding to nonspecific duplex formation of the S1 nuclease probe itself. (B) Quantitation of the results from panel A were determined from phosphorimager analysis after standardization against Tub1p levels. The values presented are the averages from at least two independent experiments with Ume3p levels in glucose medium being set at 100%. (C) ssa1-lacZ levels in response to stress. Total RNA preparations from the cultures described in panel A were blotted and probed with 32P-labeled lacZ. Quantitation of the signals was done by phosphorimager analysis, and results are presented relative to the internal ACT1 mRNA controls. ssa1-lacZ levels in glucose prior to stress were set at 100%.
Protein synthesis and cell cycle inhibition experiments.
Cells harboring pKC337 were grown to 6 × 106 cells/ml in SD medium. The culture was split in half; one half was treated with cycloheximide (100 μg/ml) for 30 min, and the other half was left untreated as the control (24). Both cultures were then subjected to either ethanol (6%) or H2O2 (0.4 mM) shock as described above for the time points indicated. Protein extracts were prepared and analyzed as indicated below. To analyze the regulation of Ume3p during cell cycle arrest, strain 1500-1A, in which all three G1 cyclins (CLN1, CLN2, and CLN3)are deleted but which contains CLN1 under the control of the galactose-inducible promoter (gal1), was used. 1500-1A harboring pKC337 was grown to mid-log phase in galactose medium, and the gal-CLN1 gene was repressed by the addition of glucose (2%, final concentration). Samples were taken when glucose was added and at 1-h increments for 4 h. Ume3p was detected as described above.
Northern blot analysis.
Total RNA preparations and S1 protection assays were performed essentially as previously described (47). UME3 (8) and ACT1 (49) S1 nuclease protection assay probes have been described. SSA1-lacZ Northern blot analyses were performed as described earlier (32) with 10 μg of total RNA. The 4.0-kbp BamHI lacZ fragment from p(spo13)28 (4) was used to probe Northern blots to monitor ssa1-lacZ expression. The lacZ signals were quantitated relative to internal ACT1 levels.
Western blot analysis.
To visualize either myc-tagged Ume3p or the mutant derivatives, extracts were prepared and analyzed by Western blot as previously described (8). Unless otherwise indicated, Ume3p was immunoprecipitated from 250 μg of soluble extracts prepared from the samples indicated in the text. Ume3p was detected with 125I-labeled antibodies (Dupont) directed against mouse monoclonal myc antibody. Quantitation of signals derived from these studies was accomplished by using a phosphorimager (Fuji, Inc.) and is reported in arbitrary PSL units. Tubulin (Tub1p) was detected as previously described (8) with antibody obtained from F. Solomon (Massachusetts Institute of Technology).
RESULTS
Ume3p is destroyed in response to a subset of environmental stresses.
Previous studies from our laboratory found that the C-type cyclin Ume3p and its cyclin-dependent kinase Ume5p are required for the full repression of the HSP70 family member SSA1 (8). To relieve this repression, Ume3p is destroyed in response to heat shock. To further investigate the role that Ume3p and Ume5p play in controlling the stress response in yeast cells, the levels of this cyclin were measured in cultures exposed to a variety of adverse environmental conditions. Wild-type strain RSY10 harboring a plasmid (pKC337) containing the myc-tagged derivative of UME3 was grown to mid-log phase and then exposed to several stress-inducing treatments (see Materials and Methods for details). Similar to our previous report, transfer of the culture from 23 to 37°C caused a rapid reduction in Ume3p levels compared to that in the untreated control (glucose lane, Fig. 1A [quantitated in Fig. 1B]). This downregulation of Ume3p occurred posttranscriptionally since UME3 mRNA levels were not significantly altered. As expected, exposure to an elevated temperature also induced ssa1-lacZ mRNA levels as determined by quantitative Northern blot analysis (Fig. 1C). ssa1-lacZ transcript levels were monitored in order to distinguish SSA1 mRNA from that of the constitutive and 97% identical SSA2 gene (45).
For the remaining stresses examined, the time points presented represent the maximum effect on Ume3p levels for a given stress as determined by time course experiments. These additional studies were necessary since the cell responds to each stress with different kinetics (10). Subjecting RSY10 to ethanol shock also caused a reduction in Ume3p levels, but this was not as severe as that observed with heat shock. Consistent with the partial reduction in Ume3p levels, a reduced but significant increase in ssa1-lacZ mRNA was detected. Cultures exposed to oxidative stress in the form of H2O2 also exhibited reduced Ume3p levels, with a corresponding increase in ssa1-lacZ mRNA. However, irradiation with UV light (100 J/m2) or exposure to hypertonic medium (0.4 M NaCl) had no effect on Ume3p or ssa1-lacZ mRNA levels. These results indicate that Ume3p is destroyed in response to several, but not all, of the conditions that induce cellular stress. Moreover, consistent with its role in SSA1 repression, the downregulation of this cyclin corresponds to an increase in ssa1-lacZ expression.
Ume3p is regulated by exogenous carbon.
In addition to agents that induce cellular damage, the stress response is also activated when cells are deprived of essential nutrients (reviewed in reference 31). To examine whether Ume3p is regulated by starvation, the levels of this cyclin were monitored in cultures incubated in medium lacking glucose, phosphate, or nitrogen (see Materials and Methods for details). In cultures deprived of a carbon source, Ume3p levels were reduced to less than 10% of those observed in the glucose control (Fig. 2A). As observed in cells exposed to stress, the reduction of Ume3p occurs at the posttranscriptional level since UME3 mRNA levels were not affected. However, cultures starved for nitrogen or phosphates did not exhibit any reduction in Ume3p concentrations. Therefore, as in the studies described above, Ume3p is downregulated in response to a subset of starvation conditions.
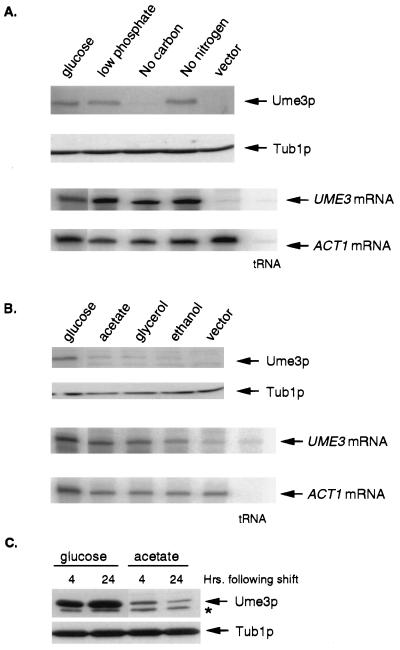
FIG. 2.
Analysis of Ume3p regulation in response to changing nutritional conditions. (A) Starvation response. Mid-log-phase RSY10 glucose cultures were harvested and resuspended in either complete prewarmed medium (glucose) or medium depleted of the indicated nutrient and incubated for 4 h (see Materials and Methods for details). Total protein and RNA samples were prepared and analyzed as described in Fig. 1. Tub1p and ACT1 mRNA levels were used as protein and RNA loading controls, respectively. (B) Response to nonfermentable carbon source. Mid-log-phase glucose cultures were harvested, washed, and transferred to medium containing the indicated carbon source. These cultures were incubated for 4 h prior to harvesting. The vector lane controls for the nonspecific cross-reactivity of the myc monoclonal antibody. The tRNA lane indicates bands derived from self-annealed probe protected from S1 nuclease. (C) Ume3p levels in continuous acetate cultures. Mid-log-phase glucose cultures were harvested and transferred to either fresh glucose- or acetate-based medium as indicated. Samples were taken at 4 and 24 h after transfer, and Ume3p levels were determined. The asterisk denotes a nonspecific cross-hybridizing band.
Previous observations in our laboratory suggested that Ume3p levels were reduced in cultures grown on acetate versus those grown on glucose medium (8). To more carefully investigate this observation, mid-log-phase cultures of RSY10 were harvested, washed, and resuspended in defined growth medium containing one of three different nonfermentable carbon sources (acetate, ethanol, or glycerol [see Materials and Methods for details]). After a 4 h incubation, the cultures were harvested and the Ume3p levels were determined. These cultures exhibited an approximate 70% reduction in Ume3p levels compared to the levels of this cyclin in cells transferred to fresh glucose medium (Fig. 2B). As observed above, the reduction in Ume3p levels occurs at the protein level since UME3 mRNA expression was not significantly altered. These results indicate that Ume3p is downregulated during aerobic growth. Two pieces of data indicate that the reduction in Ume3p levels is not due to the transient cell cycle arrest associated with the switch from glucose to a nonfermentable carbon source. First, the reduction in Ume3p levels is observed even after multiple (>15) generations in acetate medium (Fig. 2C). Second, a 4-h cell cycle arrest due to depletion of G1 cyclin activity does not alter Ume3p levels (data not shown). These results indicate that the downregulation of this cyclin is an adaptive response to oxidative respiration and is not a consequence of a temporary stoppage in cell division.
Destruction of Ume3p is mediated through separate destruction signals.
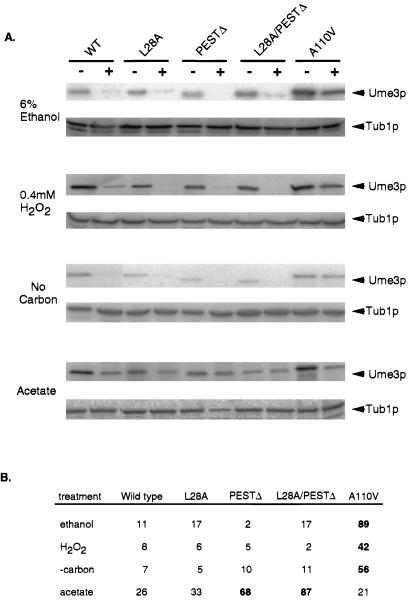
A previous study identified three domains (RXXL, PEST, and cyclin box) that are involved in the destruction of Ume3p in response to heat shock (8). We next investigated the role of these elements in controlling Ume3p levels in response to ethanol shock, oxidative stress, carbon starvation, or growth in acetate medium. Cultures containing the indicated UME3 allele were grown to mid-log phase in glucose medium, split, and either treated or not with the indicated stress as described above. To control against differences in overall stability between the different mutant cyclins, these experiments were evaluated by calculating the treated versus untreated sample percentages for each experiment. The values presented are averages from at least two separate experiments (standard deviation, <17%). As was observed previously, wild-type Ume3p levels were reduced to approximately 10% or less of those of the untreated controls in response to ethanol shock, oxidative stress, or carbon starvation (Fig. 3A [quantitated in Fig. 3B]). Reduction of Ume3p levels in acetate cultures (26% of untreated control) was less severe than for the other stressors. The L28A substitution mutant, which interrupts the RXXL motif, did not alter the degradation pattern of Ume3p in response to any of the conditions tested. Deleting the PEST region (PESTΔ) stabilized Ume3p approximately threefold in acetate medium (68% of control compared to 26% for wild type), but this mutation had no effect on cyclin levels in response to the other treatments. We previously demonstrated that mutating both the RXXL and the PEST domains had a synergistic effect on Ume3p stability in response to heat shock (8). Under the conditions examined in this study, the double PESTΔ/L28A mutant exhibited a modest, but not statistically significant, increase in Ume3p levels in acetate cultures. No increase in Ume3p stability was observed with ethanol shock, oxidative stress, or carbon starvation with the double mutant. In contrast, the single amino acid substitution in the cyclin box (A110V) stabilized Ume3p six- to eightfold under all of the conditions tested except growth on acetate. Again, no differences were observed in either the mRNA or tubulin levels (data not shown), indicating that the changes in Ume3p levels occurred at the posttranslational level. These results indicate that different destruction signals are utilized depending on the nature of the stress stimuli. Moreover, these findings suggest that the regulatory pathways mediating Ume3p degradation in response to heat shock and other stress stimuli are not identical.
FIG. 3.
cis-Acting destruction signals required for stress-induced degradation of Ume3p. (A) The levels of wild-type Ume3p (WT) or mutant derivatives (see text for details) were determined in cultures either subjected to various conditions as indicated (+) or left untreated (−). L28A, RXXL mutation; PESTΔ, PEST region deletion; A110V, cyclin box mutation. (See reference 8 for details on the mutation constructions.) Ume3p levels (arrows) were determined as described previously. Treatment conditions were as follows: ethanol, sample taken 10 min after addition of 6% ethanol to culture; H2O2, sample taken 120 min after addition of 0.4 mM hydrogen peroxide; no carbon, sample taken 4 h after transfer to minimal medium lacking a carbon source; acetate, sample taken 4 h after transfer to acetate-containing minimal medium. Tub1p levels served as protein loading controls. (B) Relative Ume3p levels after treatment. The treated/ untreated sample percentages from at least two separate experiments are listed. The standard deviations were ≤17% for all averages. Values significantly exceeding the wild-type levels (indicating Ume3p stabilization) are shown in boldface type.
Stress-induced destruction of Ume3p does not require new protein synthesis.
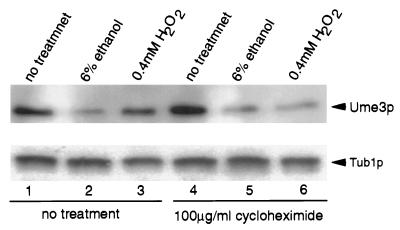
The differential utilization of cis-acting destruction elements described above suggests that Ume3p is regulated differently with respect to heat, ethanol, or oxidative stress. In addition, the turnover rates for Ume3p in response to the different stresses vary considerably. For example, Ume3p levels are reduced to their lowest point less than 3 min after heat shock but not until after 10 min with ethanol shock or after 120 min for H2O2 treatment. The decreased decay kinetics may reflect the requirement for new protein synthesis (e.g., heat shock proteins) for Ume3p destruction in response to ethanol or oxidative stress that is not necessary with heat shock. To address this possibility, a log-phase RSY10 culture harboring pKC337 was split, with half of the culture treated with cycloheximide to inhibit protein synthesis (see Materials and Methods for details). These cycloheximide-treated and untreated cultures were then subjected to either ethanol shock or oxidative stress as before, and Ume3p levels were monitored. As observed previously, Ume3p levels were reduced in stressed cultures allowed to continue protein synthesis compared to the nonstressed control (Fig. 4, lanes 1 to 3). Preincubation of the culture with cycloheximide did not significantly alter the degradation profiles (lanes 4 to 6). Therefore, we conclude that new protein synthesis is not required for the stress-induced regulation of Ume3p. Moreover, significant differences in Ume3p levels were observed in the cycloheximide-treated cultures exposed to ethanol shock and the control cultures (compare lanes 4 and 5). This finding indicates that reductions in Ume3p levels in response to ethanol shock are due to enhanced turnover and not to stress-induced cessation of translation.
FIG. 4.
New protein synthesis is not required for Ume3p destruction in response to stress. A log-phase culture (RSY10) harboring pKC337 was split, and one-half was treated with cycloheximide for 30 min. Both untreated (lanes 1 to 3) and treated (lanes 4 to 6) cultures were split again and either harvested directly (lanes 1 and 4) or subjected to ethanol (lanes 2 and 5) or oxidative (lanes 3 and 6) stress as described in the Fig. 3 legend. Protein extracts were prepared, and Ume3p levels (arrow) were determined as described in Materials and Methods. Tub1p levels were used as loading controls.
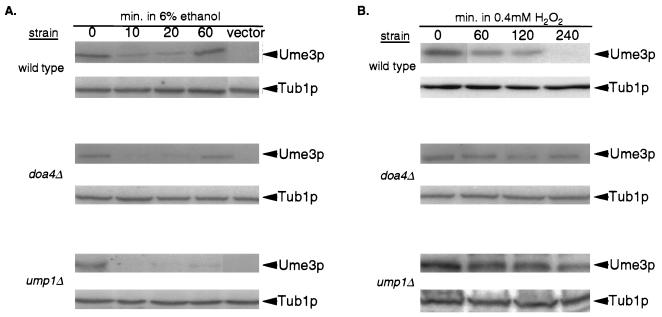
Ume3p degradation in response to oxidative stress requires a component of the 26S proteasome.
Cyclins that regulate cell cycle progression are selectively destroyed via ubiquitin-mediated proteolysis by the 26S proteasome (reviewed in reference 12). To test whether Ume3p degradation in response to ethanol or H2O2 was dependent on this system, pKC337 was introduced into a doa4 mutant or a wild-type control. Doa4p, a component of the 26S proteasome, performs the necessary deubiquitination of substrates prior to their destruction (35). Both wild-type and doa4 mutant cultures harboring the UME3 expression plasmid pKC337 were subjected to either ethanol shock or oxidative stress. In wild-type cultures subjected to ethanol shock, Ume3p levels were quickly reduced within 10 min but recovered to nearly pretreatment levels by 60 min (Fig. 5A). The return to preshock levels is due to the adaptation of the cell to this level of ethanol treatment. A similar response is observed for Ume3p levels in cultures exposed to mild heat shock (8). In the doa4 mutant, a similar expression pattern was observed, indicating that Doa4p is not required for Ume3p degradation in response to ethanol shock. However, Ume3p levels were stabilized in doa4 mutants subjected to H2O2 treatment compared to the wild-type control (Fig. 5B). As before, no significant effect was observed in either UME3 mRNA (data not shown), indicating that these findings were not due to increased transcription. Since Doa4p has been implicated in activities independent of the proteasome (17), we examined the effect of a second proteasome mutant (ump1) on Ume3p degradation. Ump1p is required for normal maturation of the proteasome (40). As with the doa4 strains, the loss of Ump1p activity has no effect on Ume3p turnover in response to ethanol shock (Fig. 5A), while again the cyclin was stabilized in response to H2O2 treatment (Fig. 5B). Quantitation of these signals revealed that the half-life of Ume3p increased threefold from 40 to approximately 120 min (r = 0.99) in the ump1 mutant. These results indicate that Ume3p degradation in response to oxidative stress is mediated through the ubiquitin-26S proteasome pathway. These findings provide the first evidence that a C-type cyclin is controlled by this system. These findings further separate the stress response pathways sensing ethanol- or H2O2-generated damage.
FIG. 5.
Doa4p and Ump1p are required for Ume3p degradation in response to oxidative stress. Log-phase wild-type (RSY10), doa4 mutant (MHY623) and ump1 mutant (JD59) cultures harboring pKC337 were subjected to ethanol shock (A) or oxidative stress (B) as described previously. Samples were taken prior to stress treatment (0 min) and at subsequent times as indicated. Protein extracts from these samples were prepared, and Ume3p levels were determined by Western blot analysis. The vector lane controls for nonspecific binding of the myc monoclonal antibody. Tub1p levels served as protein loading controls.
Ume3p destruction is not regulated through the RAS pathway.
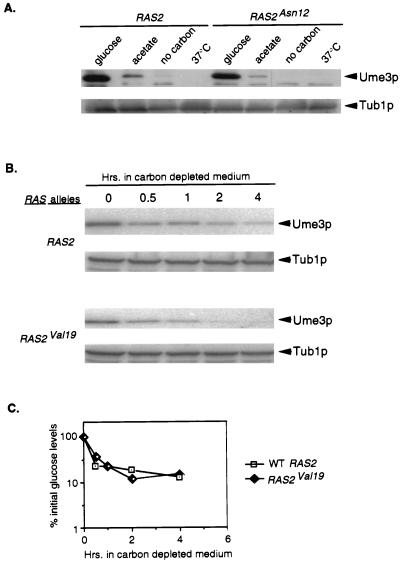
The results described above suggest the existence of both independent and overlapping regulatory systems that govern the destruction of Ume3p in response to a variety of stresses. One candidate global regulator is the RAS pathway, which regulates the activity of cyclic AMP-dependent PKA. Studies have shown that Ras2p is downregulated (GDP form) in cultures respiring, during carbon starvation, and in response to heat shock (reviewed in reference 3). This study and a previous report (8) have demonstrated that all three of these conditions lead to Ume3p degradation. To test the possibility that the downregulation of the RAS pathway is required for Ume3p degradation, an activated allele of RAS2 (RAS2Asn126) was transformed into a wild-type strain harboring the UME3 plasmid pKC337. The RAS2Asn126 mutant protein is guanine nucleotide binding deficient, thus allowing strong, constitutive activation of PKA (53). This allele functioned as expected in our strain background since cells expressing this mutant failed to arrest in the G1 phase and rapidly lost viability in response to nutrient deprivation (data not shown). In parallel, the wild-type RAS2 was introduced into the same strain and was used as a control. These cultures were grown to mid-log phase, harvested, and then either transferred to medium containing acetate as the carbon source or transferred to medium lacking carbon entirely or else subjected to heat shock. These experiments revealed that Ume3p levels were still reduced to a similar extent in response to these stresses regardless of the RAS allele present (Fig. 6A). These findings indicate that the downregulation of RAS2 is not important for Ume3p degradation in response to these stressors.
FIG. 6.
Effect of RAS activity on Ume3p regulation in response to stress. (A) Cultures expressing Ume3p and either wild-type RAS2 or the constitutively active RAS2Asn126 allele were either harvested directly (glucose), switched to acetate medium or medium lacking any carbon source, or subjected to a 37°C heat shock (see Fig. 2 legend). Protein extracts were prepared, and Ume3p levels were determined by Western blot analysis (see Materials and Methods for details). Tub1p levels served as protein loading controls. (B) Cultures harboring either the wild-type (RAS2) or the activated (RAS2val19) allele of RAS2 were grown to mid-log-phase in glucose medium, harvested, and then resuspended in carbon-depleted medium. Samples were taken at the indicated times, and Ume3p levels were determined. (C) Decay kinetics of Ume3p. The Ume3p signals derived from the experiments in panel B were quantitated by phosphorimaging and plotted with 100% representing the preshift (0-h) values.
Since the RAS2Asn126 mutation represents a specific defect in RAS regulation, we examined the effect of other constitutively active RAS2 alleles. Specifically, RAS mutants containing either the G19V (GTPase negative), E99K (GAP interaction defective), or E130K (increased GDP dissociation rate) substitutions (53) were tested for an effect on Ume3p turnover during carbon starvation. To monitor Ume3p levels more closely, a complete time course experiment was performed. As with the previous findings, no difference in the decay kinetics for Ume3p was observed for the RAS2Val19 mutation (Fig. 6B [quantitated in Fig. 6C]) or in the other activated alleles (data not shown). Taken together, these findings indicate that Ume3p degradation by carbon starvation is not dependent on the downregulation of the RAS pathway. These findings argue that Ume3p is regulated by pathways outside of RAS control.
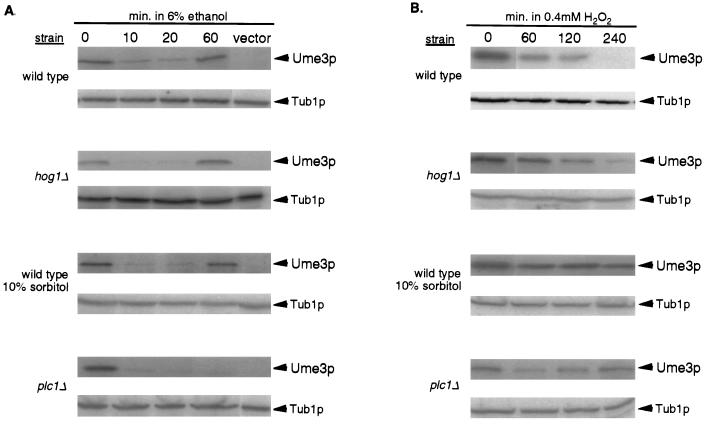
Ume3p is protected from oxidative-stress-induced degradation in the presence of the membrane-stabilizing agent sorbitol.
Ethanol or H2O2 treatment damages a number of cellular components, including membranes. Both the HOG and PKC1 pathways play an important role in maintaining plasma membrane integrity in response to stress (for a review, see reference 20). For example, the HOG pathways protect the cell from hypertonic media, while PKC1 is important for survival under hypotonic conditions (11). A recent study revealed that PKC1 activation by heat shock is inhibited by the addition of the osmostabilizing agent sorbitol to the medium (24). Further experiments led those authors to propose that sorbitol prevented inward stretching of the plasma membrane, an event believed to activate PKC1. To investigate whether Ume3p degradation in response to oxidative stress or ethanol shock was dependent on changes in membrane integrity, the levels of this cyclin were monitored in cultures subjected to these stressors but in the presence of 10% sorbitol. The addition of sorbitol had no effect on Ume3p degradation in ethanol-shocked wild-type cultures (Fig. 7A). However, cultures growing in sorbitol failed to destroy Ume3p in response to H2O2 treatment (Fig. 7B). Quantitation of these results indicated that the stabilization of Ume3p was nearly complete over the length of this experiment. These findings suggest that changes in membrane integrity in response to H2O2 exposure may trigger Ume3p degradation. Since sorbitol is able to compensate for many types of membrane defects, the exact nature of the H2O2 damage is yet to be determined. However, these findings support our conclusions from the doa4 mutant study that ethanol and H2O2 stresses are recognized differently by the cell and that this difference may be due to the nature of the damage generated by these agents.
FIG. 7.
Addition of osmostabilizing agents or loss of PLC1 activity stabilize Ume3p in response to oxidative stress. (A) Ethanol-shock-induced Ume3p degradation was monitored in wild-type cultures (RSY10) grown in SD medium (wild type) in the presence of 10% sorbitol or in hog1 (W303 hog1Δ) or plc1 (JTY2304) mutants as indicated. Samples were taken prior to (0 min) and at various times after treatment as indicated. Protein extracts were prepared, and Ume3p levels were determined by Western blot analysis. The vector lane controls for nonspecific cross-hybridization of the myc monoclonal antibody used to detect the epitope-tagged Ume3p derivative. Tub1p levels served as protein loading controls. (B) Oxidative stress. The experiments described in panel A were repeated, except that 0.4 mM H2O2 was added to the cultures to generate oxidative stress. Samples were taken at the indicated times.
Phospholipase C (PLC1) is required for oxidative-stress-induced degradation of Ume3p.
One model consistent with the results described above is that oxidative-stress-induced Ume3p degradation is triggered by changes in membrane integrity. These changes may result in increased membrane fluidity or stretching as described above. In addition, membrane damage can also affect mechanosensitive transporters (19), resulting in the increased influx of small molecules such as calcium (24). To further investigate the nature of the signal transduction system regulating Ume3p turnover, two candidate pathways that are activated by these stimuli were examined. First, Ume3p levels were monitored in a hog1 mutant strain that is defective in sensing hypertonic conditions (2). In response to either ethanol shock or oxidative stress, Ume3p was destroyed with similar kinetics in the hog1 mutant compared to wild type (Fig. 7). These findings indicate that the HOG pathway is not involved in transmitting either stress signal to the Ume3p destruction system. Next, the regulation of Ume3p was examined in mutants lacking PLC1. Plc1p activity requires calcium, and mutants lacking this gene, similar to hog1 strains, are sensitive to hyperosmotic medium (15). Similar to hog1 strains, Ume3p was downregulated normally in the plc1 mutant exposed to ethanol shock (Fig. 7A). However, Ume3p was protected from degradation in the plc1 mutant in response to oxidative stress (Fig. 7B). The degree of stabilization was nearly complete and was similar to the results obtained in the presence of sorbitol or in a doa4 mutant. These results indicate that the PLC1 is a component of the Ume3p degradation pathway in response to this type of oxidative stress (see Discussion). This finding is the first example of a candidate upstream regulatory protein involved in Ume3p degradation. Moreover, these findings, combined with the sorbitol experiments, suggest that H2O2-induced degradation of Ume3p is triggered by changes in membrane integrity, perhaps through a Plc1p-dependent pathway.
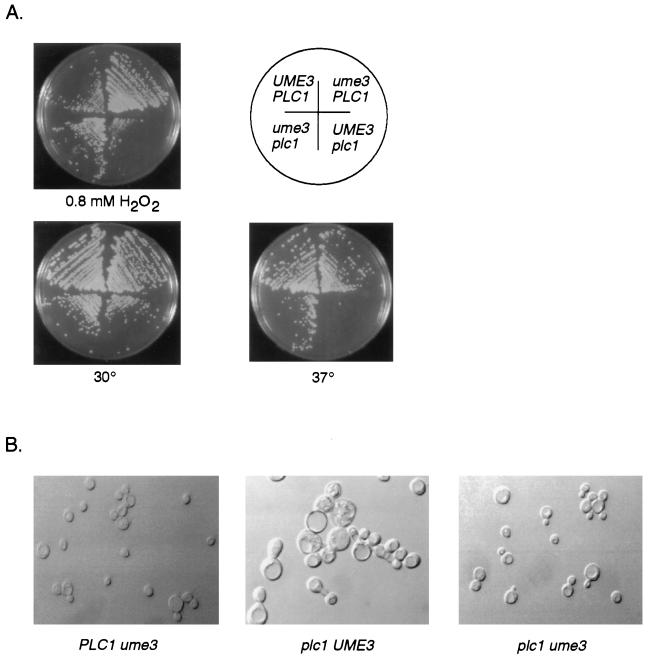
Loss of Ume3p activity suppresses growth defects in plc1 mutants.
The results described above indicate that Plc1p is required for Ume3p degradation in response to oxidative stress. To investigate the physiological significance of this regulation, we first tested the requirement of PLC1 for viability in strains exposed to stress-inducing levels of H2O2. A strain deleted for PLC1 (RSY531; see Materials and Methods for details) and the wild-type control were streaked onto rich plates containing 0.8 mM H2O2 and incubated at 30°C. The plc1 mutant was viable under these conditions but grew significantly slower than the control (Fig. 8A), indicating a role for PLC1 in the cellular response to oxidative stress. Next, we examined whether this growth defect was due to the inability of the plc1 mutant to downregulate Ume3p. To address this question, the UME3 gene was deleted in the plc1 mutant and again plated on medium containing H2O2. These experiments revealed that the double mutant (RSY532) was able to grow at rates similar to either the wild-type strain or a ume3 single mutant. These results formally suggest that the growth defect in plc1 mutants exposed to H2O2 is due to the inability to destroy Ume3p.
FIG. 8.
Loss of Ume3p activity suppresses the hypersensitivity of plc1 mutants to hyperthermia and oxidative stress. (A) Suppression of growth defects in plc1 mutants. Isogenic strains with the indicated genotypes were streaked onto rich medium and plated at 30°C or 37°C or were grown at 30°C in the presence of 0.8 mM H2O2. The cells were incubated for 2 days and photographed. (B) Suppression of cell morphology defects in plc1 mutants. Cells obtained from the 37°C plate shown in panel A were examined with Numarski optics at a 400× final magnification. The genotypes are listed below each image.
Loss of Plc1p activity results in a variety of other phenotypes, including temperature-sensitive growth and aberrant large-cell morphology (15, 37). To investigate whether aberrant stabilization of Ume3p also contributes to these phenotypes, epistasis studies were again performed. The wild type, both single mutants, and the ume3 plc1 double mutant were streaked onto rich plates and incubated at 30 or 37°C. After a 2-day incubation, the wild type and the ume3 single mutant grew on both plates (Fig. 8A). As expected, the plc1 mutant was not viable at 37°C. Interestingly, the ume3 plc1 double mutant was able to grow at 37°C at a rate similar to that of the ume3 single mutant. These results indicate that, similar to oxidative stress, failure to destroy Ume3p in response to elevated temperatures may account for the growth defect in plc1 strains. Moreover, cells taken from the 37°C plates were examined for the cell morphology abnormality associated with plc1 mutations. As previously reported (15), loss of Plc1p activity resulted in large cells with unusual bud development (Fig. 8B). As with the temperature-sensitive growth defect, the ume3 null allele again suppressed this plc1 phenotype. Although more-complicated models can be envisioned, the most straightforward interpretation of these findings is that a central task of Plc1p in response to stress is to trigger the degradation of Ume3p.
DISCUSSION
The yeast C-type cyclin Ume3p is required for the full repression of genes involved in the stress response (SSA1) or meiosis (SPO13). This study demonstrates that Ume3p is destroyed in response to ethanol shock, oxidative stress, or carbon deprivation and that its levels are significantly reduced during growth in a nonfermentable carbon-based medium. However, subjecting cultures to nitrogen or phosphate starvation, hypertonic stress, or UV irradiation did not affect the levels of this cyclin. These results indicate that Ume3p is selectively destroyed by a subset of stress conditions. The degradation of Ume3p corresponded with an increase in SSA1 transcript accumulation supporting previous genetic data indicating a negative regulatory role for this cyclin. Moreover, of the three elements required for heat-shock-induced destruction, only the cyclin box is required for Ume3p degradation in response to these stresses. The lone exception is the reduction of Ume3p levels during growth on nonfermentable carbon sources which utilizes the PEST region. In addition, we report that Ume3p is protected from oxidative-stress-induced degradation in strains growing in osmostabilized medium, suggesting that changes in membrane integrity may signal the destruction of this cyclin. Finally, mutants lacking either phospholipase C (PLC1) or two factors required for 26S proteasome activity (Doa4p and Ump1p) fail to destroy Ume3p in response to H2O2 treatment. These findings for the first time identify a potential signaling pathway that triggers Ume3p degradation, as well as the destruction machinery itself. Moreover, deletion of UME3 relieves the requirement for PLC1 in cells grown either at high temperature or in the presence of hydrogen peroxide. Taken together, these findings support a model in which oxidative stress generates a Plc1p-dependent signal at the plasma membrane that leads to the destruction of Ume3p via ubiquitin-mediated proteolysis.
We have previously demonstrated that three domains (RXXL, PEST, and cyclin box) mediate the heat shock-induced destruction of Ume3p (8). This study revealed that these domains can be divided into separate categories, depending on the nature of the stresses to which they respond. First, the cyclin box is required for Ume3p destruction in response to heat shock, ethanol shock, oxidative stress, and carbon starvation. The PEST region responds to heat shock and growth on nonfermentable carbons, while the RXXL motif is only involved in heat-induced Ume3p turnover. This strategy of utilizing multiple destruction signals has also been observed in the transcriptional repressor α2 (6). These findings allow us to make two conclusions. First, the separate utilization of the PEST, RXXL, and cyclin box domains indicates that multiple independent pathways trigger Ume3p degradation. Second, the previous finding that all of these domains are involved in heat-shock-induced degradation suggests that hyperthermia activates more than one pathway simultaneously. These findings indicate that, like higher organisms, yeasts possess multiple avenues by which external stresses are recognized, even though these pathways lead to similar changes in gene expression. Moreover, the destruction signal utilization described here and previously (8) suggests that these stresses can be categorized into at least three separate sets. One is heat shock, which targets Ume3p degradation through all three elements. The second includes ethanol shock, oxidative stress, and carbon starvation, since they only recognize the cyclin box. The last set includes growth on nonfermentable carbon sources, which utilizes the PEST region.
The results from this study indicate that sequences within the cyclin box domain are required for Ume3p degradation in a majority of the stresses tested. What role is the cyclin box playing in regulating Ume3p stability? Since the cyclin box specifies cyclin-Cdk association, a prerequisite for kinase activation, one possibility is that mutations within the cyclin box alter Ume3p-Ume5p interaction which in turn negatively impacts Ume3p-Ume5p kinase activity. It has been shown that activation of the Cdc28p Cdk by the CLN2 or CLN3 G1 cyclins is required for the rapid turnover of these cyclins (55, 27). However, several pieces of data suggest that the cyclin box mutation described here, A110V, does not significantly alter Ume3p-Ume5p activity. First, this mutant cyclin is able to complement the loss of SSA1 or SPO13 repression in a ume3 mutant host (8). In addition, the ability of the A110V mutant cyclin (Ume3pA110V) to coimmunoprecipitate Ume5p is similar to that of wild type (9). Moreover, immunoprecipitated Ume3pA110V-Ume5p kinase is able to phosphorylate the carboxyl-terminal repeat of RNA polymerase II in vitro with an efficiency similar to that of the wild-type cyclin (9). Finally, Ume3p degradation in response to heat shock occurs in the absence of Ume5p entirely (8). Currently, we cannot discount the possibility that another Cdk is bound by Ume3p in the absence of Ume5p that supplies some missing kinase function. However, the most straightforward interpretation of these results is that Ume5p activity is not necessary for the rapid turnover of Ume3p. Therefore, it is possible that the cyclin box region is playing another role in addition to binding and activating a Cdk. The analysis of the human cyclin B nuclear export identified an element adjacent to the cyclin box that binds directly the CRM1 export protein (56). Perhaps more relevant were studies involving the so-called P-box, which resides within the cyclin box of vertebrate B-type cyclins. In the absence of cdc2, this domain directs cyclin B binding to, and the stimulation of, the cdc25 phosphatase (16, 57). These data are consistent with the cyclin box, or elements contained within its boundaries, possessing activities in addition to Cdk activation. Therefore, it is possible that the Ume3p “cyclin box” destruction element may possess an activity distinct from the binding of Ume5p. Given its role in Ume3p turnover, this element may bind a transport protein that shuttles Ume3p to the proteasome or is recognized by modification enzymes (e.g., protein kinases). Understanding the function of this domain may provide insight into how stress stimuli trigger Ume3p destruction.
Previous studies support the presence of two separate systems regulating the oxidative stress response in yeast cells (for a review, see reference 43). For example, treatment of yeast cells with sublethal concentrations of either H2O2 or menadione (a superoxide-generating agent) protects the cell from lethal doses of the same oxidant. However, hydrogen peroxide treatment does not protect cells from a subsequent challenge with high concentrations of menadione (22). In addition, Yap1p, a transcription factor that induces the expression of several stress response genes, is required for adaptive protection by H2O2 pretreatment but not for menadione-induced acquired tolerance (46). Finally, hydrogen peroxide treatment induces the expression of SSA1, while menadione does not (23). This latter observation is consistent with our finding that H2O2, but not menadione treatment (data not shown), reduces Ume3p levels. These findings support a model in which Ume3p is regulated by the branch of the oxidative-stress response pathway that recognizes hydrogen peroxide-induced damage but not that of menadione.
In addition to H2O2 and other exogenous free-radical generators, reactive oxygen species (ROS) are also produced as natural by-products of oxidative respiration (reviewed in reference 33). Therefore, the observed downregulation of Ume3p in cultures growing on nonfermentable carbons (e.g., acetate) may be due to the production of ROS. Interestingly, our analysis demonstrates that H2O2 treatment triggers Ume3p destruction through the cyclin box destruction signal, while growth on acetate medium utilized the PEST domain. If ROS produced from oxidative respiration enhances Ume3p turnover, then the cell may be recognizing the two stresses differently. The possible existence of separate pathways is also consistent with different effects that H2O2 and oxidative respiration have on Ume3p levels. Respiration elicits a signal that reduces Ume3p levels to approximately 20 to 30% of that of glucose-grown cells. However, H2O2 treatment is more severe, reducing Ume3p levels to less than 10% of the pretreatment levels (see Fig. 3). These observations are consistent with the extent of oxidative damage being indirectly proportional to the resulting Ume3p levels. ROS generated by respiration are handled by the basal antioxidant mechanisms. Increased damage, as would be induced by the H2O2 treatments described in this study, induces the oxidative-stress response. There are many ROS known (e.g., superoxide anion, hydroxyl radical, and hydrogen peroxide) that can damage a variety of cellular components, including lipids, proteins, and nucleic acids. Therefore, it may not be surprising that the cell may possess three systems (menadione, H2O2, and respiration) that monitor and respond to a variety of levels and types of damage caused by ROS. Such a strategy may allow the cell to fine-tune its stress response to a given stimulus.
This study demonstrates for the first time that components of the 26S proteasome (Doa4p and Ump1p) are important for Ume3p destruction in response to oxidative stress. Therefore, as with cyclins that control mitotic cell cycle progression, Ume3p levels also appear to be regulated by ubiquitin-mediated proteolysis. Interestingly, this and a previous study (8) found that ethanol stress or heat shock still induces normal Ume3p decay in doa4 or ump1 mutants. This result may indicate that there are several different “types” or specialized proteasomes in yeast cells and that Doa4p or Ump1p is involved in destroying Ume3p only within a specific subset of conditions. Indeed, there is evidence that there are proteasome complexes that may perform more restricted tasks (44). Another possibility is that there are multiple pathways that Ume3p can take that will lead to degradation. For example, cells subjected to heat shock utilize multiple cis-acting elements, some of which may lead to the destruction of Ume3p independent of Doa4p. According to this model, the nature or severity of the damage may dictate the avenue (or avenues) that becomes competent to direct Ume3p degradation. It has been shown that lethal concentrations of H2O2 damage DNA, whereas lower amounts cause more subtle effects on lipids and proteins (54). Due to the relatively low cellular concentrations of this cyclin, any one of these destruction pathways may be sufficient to reduce Ume3p levels below the limits of detection. In the case of H2O2 stress, our protocol may trigger only one of the available Ume3p destruction pathways that contain PLC1 and DOA4. More-severe treatments that damage a wider spectrum of cellular components may trigger additional pathways that are able to destroy Ume3p independently of PLC1 or DOA4. This possibility is supported by the findings that doa4 mutants are unable to protect Ume3p in cultures subjected to 50-fold-higher concentrations of H2O2 (data not shown). This observation is consistent with a model in which, as more damage to different cellular components (e.g., lipid, protein, and nucleic acids) occurs, additional degradation pathways are activated.
This study demonstrates that a ume3 null allele can suppress the hypersensitivity of plc1 mutants to elevated temperatures and oxidative stress. These findings suggest that the downregulation of Ume3p is an important function of Plc1p in cells exposed to these types of stresses. Given the role of Ume3p in repressing the HSP70 family member SSA1 (8), one model consistent with these findings is that the failure to destroy Ume3p maintains repression on SSA1, thus lowering the ability of the cell to contend with adverse environmental conditions. In light of other studies, however, this model appears to be too simplistic. For example, deletion of any one of the HSP70 genes does not significantly affect cell viability in response to stress (10). Only when multiple HSP70 genes are mutated is an effect observed. However, Ume3p does not regulate either the inducible (SSA3) or the constitutive (SSA2) HSP70 genes (unpublished results). These findings suggest that the requirement for Ume3p destruction is not due solely to the regulation of the HSP70 family of heat shock proteins. Therefore, the Ume3p-Ume5p kinase is most likely involved in the regulation of additional loci, perhaps outside the heat shock protein families, that are involved in the stress response. Given the pleiotropic effects of heat shock or oxidative stress on cell homeostasis, the list of potential genes regulated by this cyclin-Cdk may be extensive.
ACKNOWLEDGMENTS
We thank M. Gustin and J. Flick for helpful discussions and J. Thorner for bringing the plc1 mutant phenotypes to our attention. We thank J. Broach and J. Cannon for activated RAS alleles, J. Thorner for plc1 mutant strains, F. Cross for the triple cln deletion strain, R. J. Dohmen for the ump1 mutant strain, and V. Guacci for tubulin antibodies. We also thank E. Golemis and J. Chernoff for critical reading of the manuscript.
K.F.C. was supported by NIH grant CA-09035-23. This work was supported by NSF grant MCB-9513479 to R.S. and an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Berridge M J. Inositol triphosphate and calcium signaling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 2.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 3.Broach J R. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham L E, Wang H-T, Elder R T, McCarroll R M, Slater M R, Esposito R E. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugated enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 7.Collinson L P, Dawes I W. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- 8.Cooper K F, Mallory M J, Smith J S, Strich R. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, K. F., and R. Strich. Functional analysis of the yeast C-type cyclin Ume3p and the RNA polymerase II holoenzyme interaction. Gene Expr., in press. [PMC free article] [PubMed]
- 10.Craig E A. The heat-shock response of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 501–538. [Google Scholar]
- 11.Davenport K R, Sohaskey M, Kamada Y, Levin D E, Gustin M C. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies R J, Chau V, Kirschner Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Destruelle M, Holzer H, Klionsky D. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol Cell Biol. 1994;14:2740–2754. doi: 10.1128/mcb.14.4.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flick J S, Thorner J. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5861–5876. doi: 10.1128/mcb.13.9.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galaktionov K, Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclin. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- 17.Galan J, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 19.Gustin M C, Zhou X-L, Martinac B, Kung C. A mechanosensitive ion channel in the yeast plasma membrane. Science. 1988;242:762–765. doi: 10.1126/science.2460920. [DOI] [PubMed] [Google Scholar]
- 20.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 21.Hokin L E. Receptors and phosphoinsitol-generated second messengers. Annu Rev Biochem. 1985;54:205–235. doi: 10.1146/annurev.bi.54.070185.001225. [DOI] [PubMed] [Google Scholar]
- 22.Jamieson D J. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamieson D J, Rivers S L, Stephen D W S. Analysis of Saccharomyces cerevisiae proteins induced by peroxide and superoxide stress. Microbiology. 1994;140:3277–3283. doi: 10.1099/13500872-140-12-3277. [DOI] [PubMed] [Google Scholar]
- 24.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 25.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krems B, Charizanis C, Entian K D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- 27.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1600. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 28.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin D E, Fields F O, Kunisawa R, Bishop J M, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 30.Longtine M S, McKenzie A R, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules of versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Mager W H, De Kruijff A J J. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 34.Morimoto R I, Sarge K D, Abravaya K. Transcriptional regulation of heat shock genes. J Biol Chem. 1992;267:21987–21990. [PubMed] [Google Scholar]
- 35.Papa F, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;25:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 36.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 37.Payne W E, Fitzgerald-Hayes M. A mutation in PLC1, a candidate phosphoinositide-specific phospholipase C gene from Saccharomyces cerevisiae, causes aberrant mitotic chromosome segregation. Mol Cell Biol. 1993;13:4351–4364. doi: 10.1128/mcb.13.7.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pines J. Cyclins and cyclin-dependent kinases: take your partners. Trends Biochem Sci. 1993;18:195–197. doi: 10.1016/0968-0004(93)90185-p. [DOI] [PubMed] [Google Scholar]
- 39.Piper P W, Talreja K, Panaretou B, Moradas-Ferreira P, Byrne K, Praekelt U M, Meacock P, Recnacq M, Boucerie H. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology. 1994;140:3031–3038. doi: 10.1099/13500872-140-11-3031. [DOI] [PubMed] [Google Scholar]
- 40.Ramos P C, Hockendorff J, Johnson E S, Varshavsky A, Dohmen R J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 41.Salama S R, Hendricks K B, Thorner J. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol Cell Biol. 1994;14:7953–7966. doi: 10.1128/mcb.14.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoro N, Thiele D J. Yeast stress responses. In: Hohmann S, Mager W H, editors. Molecular biology intelligence unit. R. G. Austin, Tex: Landes Co.; 1997. pp. 171–203. [Google Scholar]
- 44.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 45.Slater M R, Craig E A. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1906–1916. doi: 10.1128/mcb.7.5.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephen D, Rivers S, Jamieson D. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 47.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strich R, Woontner M, Scott J F. Mutations in ARS1 increase the rate of simple loss of plasmids in Saccharomyces cerevisiae. Yeast. 1986;2:169–178. doi: 10.1002/yea.320020305. [DOI] [PubMed] [Google Scholar]
- 49.Surosky R T, Esposito R E. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3948–3958. doi: 10.1128/mcb.12.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatchell K, Robinson L C, Breitenbach M. RAS2 of Saccharomyces cerevisiae is required for glyconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci USA. 1985;82:3785–3789. doi: 10.1073/pnas.82.11.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson B A, Khalil M, Tamonoi R, Cannon J F. New activated RAS2 mutations identified in Saccharomyces cerevisiae. Oncogene. 1993;8:3441–3445. [PubMed] [Google Scholar]
- 54.Woodford D V, Parish H H, Moradas-Ferreira P. Hydrogen peroxide induces DNA damage in Saccharomyces cerevisiae. Yeast. 1995;11:149. [Google Scholar]
- 55.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J-M, Chin K-V, Hait W N. Involvement of phospholipase c in heat-shock-induced phosphorylation of P-glycoprotein in multidrug resistant human breast cancer cells. Biochem Biophys Res Commun. 1995;210:21–30. doi: 10.1006/bbrc.1995.1622. [DOI] [PubMed] [Google Scholar]
- 57.Zheng X F, Ruderman J V. Functional analysis of the P box, a domain in cyclin B required for the activation of Cdc25. cell. 1993;75:155–164. [PubMed] [Google Scholar]