Abstract
Aromatic aminotransferase II, product of the ARO9 gene, catalyzes the first step of tryptophan, phenylalanine, and tyrosine catabolism in Saccharomyces cerevisiae. ARO9 expression is under the dual control of specific induction and nitrogen source regulation. We have here identified UASaro, a 36-bp upstream element necessary and sufficient to promote transcriptional induction of reporter gene expression in response to tryptophan, phenylalanine, or tyrosine. We then isolated mutants in which UASaro-mediated ARO9 transcription is partially or totally impaired. Mutations abolishing ARO9 induction affect a gene called ARO80 (YDR421w), coding for a Zn2Cys6 family transcription factor. A sequence highly similar to UASaro was found upstream from the YDR380w gene encoding a homolog of bacterial indolepyruvate decarboxylase. In yeast, this enzyme is postulated to catalyze the second step of tryptophan catabolism to tryptophol. We show that ARO9 and YDR380w (named ARO10) have similar patterns of transcriptional regulation and are both under the positive control of Aro80p. Nitrogen regulation of ARO9 expression seems not directly to involve the general factor Ure2p, Gln3p, Nil1p, Uga43p, or Gzf3p. ARO9 expression appears, rather, to be mainly regulated by inducer exclusion. Finally, we show that Gap1p, the general amino acid permease, and Wap1p (Ycl025p), a newly discovered inducible amino acid permease with broad specificity, are the main aromatic amino acid transporters for catabolic purposes.
The yeast Saccharomyces cerevisiae can use tryptophan, phenylalanine, or tyrosine as the only source of cellular nitrogen (11). The main products of their catabolism are tryptophol, phenylethanol, and tyrosol, respectively, constituents of the mixture of alcohols collectively known as fusel oil in fermentations (41, 42, 66, 69). Fusel oil formation from amino acids is believed to proceed through the so-called Ehrlich pathway involving three enzymatic steps. A first transamination produces the α-keto-acid analog of the amino acid, a decarboxylation step yields an aldehyde, and a reduction step converts the aldehyde to a primary alcohol (79). Tryptophan is thus converted to tryptophol via the metabolic intermediates indole-3-pyruvate and indole-3-aldehyde (41, 69); tyrosine and phenylalanine are converted to tyrosol and phenylethanol in a similar way (42, 66). Yet as recently stressed for leucine catabolism by Dickinson et al. (15), this general scheme of amino acid degradation stems mainly from studies of metabolic intermediates and end product formation. Too little is known about the specific permeases and enzymes involved in aromatic amino acid utilization, their genetic determinants, and their regulation. Aromatic aminotransferase II, the product of the recently cloned ARO9 gene (34), is the first characterized enzyme proposed to be involved in the catabolism of aromatic amino acids. Specifically, (i) ARO9 gene transcription is induced by the presence of tryptophan, phenylalanine, or tyrosine in the growth medium and remains at a very low level in their absence (34, 41, 77); (ii) aromatic aminotransferase II catalyzes the first step of the catabolism of these amino acids (41, 77); (iii) the enzyme is dispensable for growth on phenylalanine or tyrosine as the only source of nitrogen, but aro9 mutants grow poorly on tryptophan or kynurenine (77); and (iv) preliminary expression studies (34) show that, in the presence of inducer, ARO9 expression levels are more than 10 times higher on urea medium than on ammonia medium, indicating that ARO9 is subject to an ammonia effect (80). Thus, as originally proposed by Kradolfer et al. (41), the physiological function of aromatic aminotransferase II is most likely to participate in the catabolism of aromatic amino acids, mainly tryptophan.
Like many other nitrogen-catabolic genes, ARO9 appears to be regulated in at least two complementary ways: by an induction mechanism, with aromatic amino acids acting as inducers, and by an ammonia effect which modulates expression levels according to nitrogen source quality.
In S. cerevisiae, the synthesis of permeases and enzymes of many nitrogen-catabolic pathways is inducible. Examples include the pathways of arginine, proline, urea, and allantoin (for reviews, see references 47, 52, and 80) and 4-aminobutyrate (reference 76 and references therein) and serine/threonine (31) utilization. Induction is mediated in cis by a class of upstream activating sequences (UASs), specific to genes of a particular catabolic pathway. It has been demonstrated in several cases that these UASs are binding sites for specific trans-acting factors that can stimulate transcription of adjacent genes in response to inducer availability (31, 70). Many of these regulatory proteins, such as ArgRIIp (53), Uga3p (1), Put3p (48), and Cha4p (31), belong to the C6 zinc cluster family of fungal transcription factors, whose prototypes are Gal4p (50, 60) and Prp1p (51). As deduced from the analysis of the complete genome sequence, the family comprises no less than 56 members in S. cerevisiae, most of them of unknown function (64).
Inducer availability is by no means sufficient to ensure high-level transcription of the genes of a nitrogen catabolic pathway. S. cerevisiae can use some 30 different compounds as the sole source of cellular nitrogen (11). Some nitrogen sources such as glutamine and ammonia (good or preferred nitrogen sources) support optimal growth, while others, such as proline and urea (poor or secondary nitrogen sources), support slower growth. When a good nitrogen source is present, utilization of the poorer nitrogen sources is prevented. This general phenomenon, termed the ammonia effect (80) or nitrogen regulation (47), is the result of at least two distinct regulatory mechanisms: nitrogen catabolite repression (NCR), which affects the synthesis of enzymes and permeases responsible for the utilization of secondary nitrogen sources, and nitrogen catabolite inactivation (NCI), which down-regulates the activity of several permeases that import these substrates (24, 47, 80). NCR of susceptible genes is achieved through upstream 5′-GATA-3′ sequences variously characterized as UASNTR (58, 59), UASN (55), or UASGATA (4). Four distinct GATA factors can bind to these UASs and affect transcription: activators Gln3p and Nil1p/Gat1p and repressors Uga43p/Dal80p and Gzf3p/Nil2p/Deh1p (references 10, 62, and 71 and references therein). In addition, nitrogen repression of most NCR-sensitive genes is relieved by mutations in the URE2/GDHCR/USU locus (16, 22, 25). Ure2p is not a GATA factor, does not bind to DNA, and seems to act mainly by inactivating Gln3p, the main activator of nitrogen regulation (47). Hence, nitrogen source repression involves negative factors counteracting the function of positive factors when preferred nitrogen sources are available.
NCI, on the other hand, affects the activity of most permeases subject to NCR, including Gap1p and the proline permease Put4p (24). Addition of ammonium ions to cells growing on poor substrates such as proline or urea triggers rapid and complete inactivation (23) and degradation (30) of Gap1p. This process requires the Npi1p/Rsp5p ubiquitin ligase and the integrity of the C-terminal region of the permease (29, 30, 72). Ubiquitination is required for endocytic internalization of the permease and its subsequent degradation in the vacuole (72). In npi1 mutants, Gap1p ubiquitination is impaired and the permease remains located in the plasma membrane and active (23, 72).
Here we have sought to identify regulatory elements involved in the induction and nitrogen regulation of ARO9 expression. We have identified two elements required for specific induction of the ARO9 gene by aromatic amino acids: a UAS (UASaro) and the transcriptional activator Aro80p. We have further shown that the ARO10 (YDR380w) gene, encoding a putative indole-3-pyruvate decarboxylase, possesses an upstream sequence similar to UASaro and that this gene is also subject to Aro80p-dependent induction by tryptophan. Nitrogen control analyses indicate that the general factors of nitrogen transcriptional regulation do not directly affect ARO9 expression. Transcription of this gene appears, rather, to be regulated essentially through inducer exclusion. A preliminary report of this work has already appeared (32).
MATERIALS AND METHODS
Strains, media, and methods.
The S. cerevisiae strains used are listed in Table 1. All are isogenic with wild-type strain Σ1278b (MATα) (6). The laboratory’s standard minimal medium has been described previously (36); the nitrogen source was either 10 mM ammonium sulfate or another nitrogenous compound at 1 g/liter; the carbon source was 3% glucose. Inducing media were supplemented with 0.5 g of tryptophan, phenylalanine, or tyrosine per liter. When needed, 50 μg of uracil or leucine per ml was added to the medium. Complex medium 868 has been described previously (17). Standard yeast genetic techniques were applied (68). Yeast cells were transformed by the lithium acetate method (21, 35). The Escherichia coli strain used in this work was JM109. All E. coli and DNA manipulations were performed according to standard procedures (5, 63).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference(s) |
|---|---|---|
| Σ1278b | MATα | 6 |
| 23346c | MATa ura3 | Laboratory’s collection |
| 26854a | MATa ure2 ura3 | 25 |
| 26992c | MATα ure2 uga43Δ ura3 | 4 |
| 27038a | MATa npi1 ura3 | 23, 29 |
| 30078c | MATα uga43Δ ura3 | 12 |
| 30312d | MATα aro9Δ ura3 | 34 |
| 30332d | MATa ura3 | Laboratory’s collection |
| 30505b | MATα gln3Δ ura3 | 4 |
| 30622a | MATα wap1-1 gap1-92 ura3 | This study |
| 30622b | MATa gap1-92 ura3 | This study |
| 30622c | MATa ura3 | This study |
| 30622d | MATα wap1-1 ura3 | This study |
| 30629c | MATa gap1Δ::Kanrura3 | 33 |
| 30633c | MATa gap1Δ::Kanrwap1Δ::Kanrura3 | This study |
| 30637a | MATa wap1Δ::Kanrura3 | This study |
| 30701a | MATa aro80Δ::Kanrura3 | This study |
| 30768d | MATα npi1 gap1Δ::Kanrura3 | Laboratory’s collection |
| 30831b | MATα ure2 npi1 ura3 | Laboratory’s collection |
| 32054d | MATa aro8Δ-1 aro80-2 ura3 | This study |
| 32102c | MATα wap1-1 gap1-92 ura3 | This study |
| 32164c | MATa gln3Δ nil1Δ::Kanrura3 | Laboratory’s collection |
| 32173c | MATα ure2 aro80Δ::Kanrura3 | This study |
| 33089d | MATα gap1pgr ura3 | 23 |
| 50000b | MATa uga43Δ gzf3Δ::LEU2 ura3 | 71 |
| 50036c | MATα ure2 gzf3Δ::LEU2 ura3 | 71a |
| 50054a | MATα ure2 uga43Δ gzf3Δ::LEU2 ura3 | 71a |
| IHI602 | MATa aro80-1 ura3 | This study |
| IHI606 | MATa aro80-2 ura3 | This study |
| IHI611 | MATa gap1-92 wap1-1 ura3 | This study |
| IHI614 | MATa aro80-3 ura3 | This study |
| IHI708 | MATα aro80Δ::Kanrura3 | This study |
| IHI726 | MATα wap1Δ::Kanrgap1-92 ura3 | This study |
| SBS10 | MATa gzf3Δ::LEU2 ura3 | 71 |
| SBS21 | MATα nil1Δ::Kanrura3 | 71 |
Formation of 5′ and internal deletions in the ARO9 upstream region.
Construction of the YCpARO9-lacZ plasmid has been described previously (34). In the plasmid, the upstream ARO9 DNA fragment is flanked at the 5′ end by a portion of the pFL38 polylinker. To isolate the 5′ nested deletions of the ARO9-lacZ reporter gene, the plasmid was cleaved at the polylinker BamHI and SphI restriction sites, treated with exonuclease III and S1 nuclease, and religated, as instructed for the double-stranded nested deletion kit (Pharmacia LKB).
Plasmid pII472 is a derivative of YCpARO9-lacZ from which the 36 bp of the UASaro element have been specifically deleted. The deletion, spanning positions −168 through −133 relative to the ATG of the open reading frame (Fig. 1), was introduced by using the Altered Sites in vitro mutagenesis system (Promega). A 4.3-kb EcoRI-EcoRI fragment, comprising 1.3 kb of the 5′-flanking region of ARO9 and 3 kb of the 5′ coding region of the lacZ gene, was extracted from plasmid YCpARO9-lacZ and inserted into the EcoRI site of the pALTER-1 phagemid. Single-stranded DNA was produced, and the 36-bp sequence was deleted by site-directed mutagenesis with the appropriate oligonucleotide. The accuracy of the deletion was checked by sequencing, and then the mutagenized EcoRI-EcoRI fragment was reinserted into the EcoRI-EcoRI site of plasmid YCpAJ153 (3).
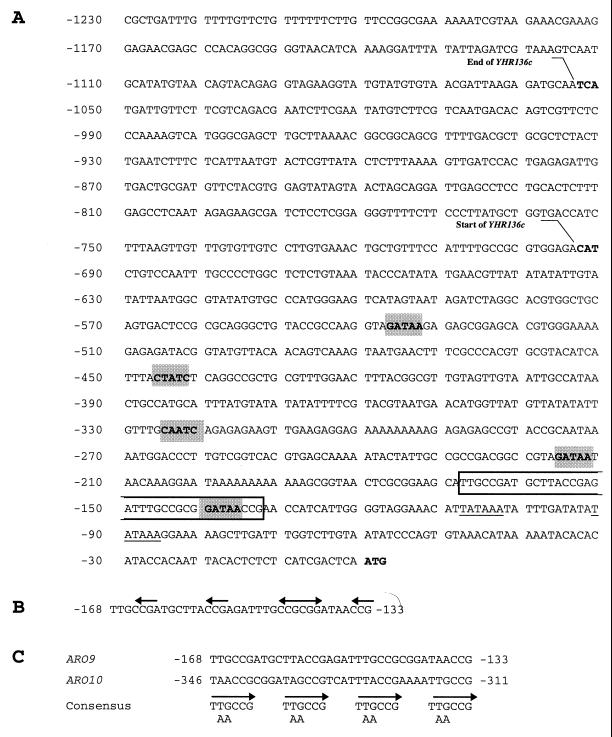
FIG. 1.
(A) Sequence of the 5′-flanking region of the ARO9 gene. The sequence includes the SPL2 (YHR136c) gene, recently shown to encode a novel inhibitor of the Pho80p/Pho85p cyclin-dependent protein kinase (20). Putative TATAAA sequences are underlined, GAT(A/T)(A/G) (75) sequences are shaded, and the UASaro sequence is boxed. (B) Sequence of the UASaro indispensable for ARO9 gene induction. The arrows indicate the CCG triplets. (C) Comparison of UASaro and a similar sequence located upstream from the ARO10 gene. The arrows indicate the direct repeats.
Plasmids carrying heterologous lacZ fusions.
Plasmid PAL1-15 carries a heterologous UASaro-CYC1-lacZ fusion gene in which the TATA box and transcriptional start site are provided by the CYC1 gene and the UAS is the UASaro element of the ARO9 gene. Plasmid PAL1-15 was constructed by introducing UASaro at the XhoI restriction site position of the CYC1 promoter in plasmid pLG670-Z (28). This was done in several steps with the Altered Sites in vitro mutagenesis system (Promega). First, a BamHI-SalI fragment of the CYC1 promoter was isolated and inserted into the pALTER-1 phagemid restricted with BamHI and SalI. UASaro was then introduced into the CYC1 promoter by directed mutagenesis with an oligonucleotide comprising the 36-bp element flanked by targeting sequences of CYC1. The correct insertion of UASaro into pALTER-1 was checked by sequencing, and finally the BamHI-SalI fragment containing the CYC1-UASaro-CYC1 construct was isolated from pALTER-1 and inserted into plasmid pLG670-Z (28), yielding plasmid PAL1-15. Using appropriate oligonucleotides, we followed the same procedure to obtain the PAL series of heterologous fusions carrying variously altered forms of the 36-bp sequence (PAL60, PAL50, PAL2-13, and PAL2-10). PAL1-31 and PAL1-46 were fortuitously obtained in the course of PAL1-15 construction. All the constructs were sequenced on both strands.
Isolation of mutants affected in the ARO9 gene induction.
Cells of the ura3 strain 30332d bearing episomal plasmid PAL1-15 were mutagenized with ethane methylsulfonate and plated on proline minimal medium supplemented with 500 μg of tryptophan per ml and 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml, the medium being buffered at pH 7.0 (28). About 32,500 colonies were screened, white or pale blue colonies were restreaked, and 17 isolates were chosen for enzymatic tests.
Isolation, subcloning, and deletion of the WAP1 (YCL025c) gene.
S. cerevisiae 32102c, derived from IHI611, carries mutations gap1-92 and ura3, plus a mutation affecting an unknown locus (see Results). The strain is unable to grow on minimal medium containing either tryptophan or phenylalanine at a 500-μg/ml concentration as the only source of nitrogen. The mutant was transformed with a genomic library from wild-type strain Σ1278b (49), and transformants growing on tryptophan or phenylalanine were selected. A second cycle of transformation was carried out with purified plasmids, and the extremities of the inserts were sequenced. From transformants selected on tryptophan, only genomic fragments carrying the GAP1 gene (38) were obtained (four different complementing clones). From transformants selected on phenylalanine, 11 different genomic fragments were analyzed. Of these, one carried the GAP1 gene. The 10 other genomic inserts were partially overlapping fragments from a region of chromosome III (56). YCL025c was the only open reading frame common to all of them. YCL025c was isolated as a 3-kb BsmI-BsmI subfragment and cloned into vector pFL38 (8) at the SmaI restriction site. The resulting pII406 plasmid, when introduced into cells of strain 32102c, restored their ability to grow on phenylalanine medium.
Chromosomal deletion of the YCL025c gene was performed by homology-directed gene replacement with PCR-based techniques described by Wach et al. (78) as described in reference 33. Correct targeting of the kanMX2 cassette into the YCL025c gene was checked by PCR. Further work (see Results and reference 33) showed that YCL025c encodes an inducible amino acid permease with broad substrate specificity. YCL025c was consequently named WAP1, for wide-specificity amino acid permease.
Isolation, subcloning, and deletion of the ARO80 (YDR421w) gene.
S. cerevisiae 32054d (aro80-2 aro8Δ-1 ura3), auxotrophic for phenylalanine and tyrosine, was transformed with a wild-type Σ1278b genomic library (49), and prototrophic transformants were selected. Among them, two different growth phenotypes were observed on aromatic amino acid substrates because both aro80 ARO8 and ARO80 aro8 single mutant strains are prototrophs. Only transformants displaying the Aro8− phenotype (77) were further analyzed, yielding complementing plasmids pII242 and pII251. Sequencing revealed that their inserts overlapped over almost their entire length and came from a region of chromosome IV extending from bp 1305529 to bp 1316570 (37). This chromosomal region comprises two internal open reading frames: YDR420w, corresponding to the HKR1 gene, and YDR421w (see Results). The function of the latter is unknown, but the deduced polypeptide contains a Zn2Cys6 binuclear cluster domain and other features characteristic of Gal4p family transcriptional regulators (64). YDR421w was cloned as a 5.6-kb BamHI-BamHI fragment extracted from plasmid pII242 and inserted into the BamHI restriction site of centromeric plasmid pFL38 (8). The resulting pII270 plasmid complemented the auxotrophy of strain 32054d (aro80-2 aro8Δ-1 ura3) and the growth defects of the aro80 mutants on medium containing an aromatic amino acid as the sole source of nitrogen.
Chromosomal deletion of the ARO80 gene was also performed by homology-directed gene replacement (78). The primers used for the amplification were 5′-TGTCTGCTAAGAAAAGGCCTTCGGGAAACGCAGCATTTGAGCGG CCGCCAGCTGAAGCTTCGTACGC-3′ and 5′-CGAATAGTGCGGTTG TCTTGGTTGATGACGTAATTCTTTGGCGGCCGCATAGGCCACTAGT GGATCTG-3′. Correct targeting of the cassette into the ARO80 gene was checked by PCR.
Tryptophan uptake.
The time course of l-[side chain-3-14C]tryptophan (specific radioactivity, 56 mCi/mmol; from DuPont NEN) uptake was determined as previously described for 4-aminobutyric acid accumulation (27), with radiolabeled amino acids at a 20 μM final concentration.
Enzyme assays.
Tryptophan-phenylpyruvate transamination activity was assayed as described elsewhere (77). β-Galactosidase was assayed according to the method of Miller (54); the unit used was nanomoles of o-nitrophenol formed per minute per milligram of protein. Protein concentrations were determined according to the method of Lowry et al. (43) with bovine serum albumin as the standard.
RNA preparation and gene expression analysis by RT-PCR.
Cells were grown to a density of approximately 107 cells/ml on minimal medium with the nitrogen source specified in the legends to the figures. Total RNA was isolated as instructed for the RNeasy Mini kit by the supplier (Qiagen); the final RNA concentration was approximately 1 μg/μl. The purified RNA was treated with RNase-free DNase I (Boehringer Mannheim) to eliminate any trace of DNA. Reverse transcription-PCR (RT-PCR) was performed with approximately 10 ng of RNA per reaction with the Titan One RT-PCR kit (Boehringer Mannheim) and a Progene thermal cycler, from Techne (Cambridge) Ltd. The RT stage was performed at 55°C for 30 min. After 1 min at 94°C to denature the DNA, amplification was carried out as follows: (i) 10 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C; (ii) 15 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, the elongation time of each cycle being increased by 5 s; and (iii) a final elongation step of 7 min at 72°C. The primers used to amplify a 606-bp subfragment of the ARO9 gene situated 84 bp downstream from the start codon were 5′-CCGTTCAGTAGTCAGGTCGCT-3′ and 5′-GGATCCCCCAGTGAGCTACCCATT-3′. For the ARO10 (YDR380w) gene, the 745-bp amplicon is located 60 bp downstream from the start codon, and the primers used were 5′-CCATACCTATCAGTCAGTACGTCTCC-3′ and 5′-CGATCAGCAACAATTCCGTTTGGTG-3′. The ACT1 amplicon was produced with the primers 5′-GACTCCTACGTTGGTGATGA-3′ and 5′-CTGGAGGAGCAATGATCTTG-3′.
RESULTS
Detection of a UAS element required for induction of the ARO9 gene.
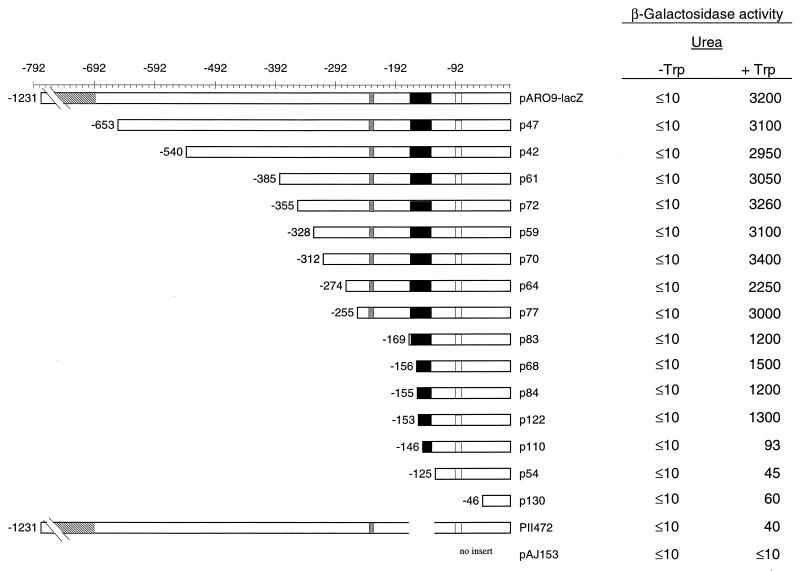
To roughly locate the cis-acting sequences involved in ARO9 induction, we examined the effects of a series of 5′ deletions produced in the ARO9 promoter region (Fig. 2). The starting plasmid was YCpARO9-lacZ, containing 1,231 bp of upstream region and the first 11 codons of the ARO9 gene fused to a promoterless lacZ gene (34). Our previous work had shown that this fusion product responds to the same regulatory signals as the chromosomal ARO9 gene. In particular, it is very weakly expressed in the absence of inducer, induced by aromatic amino acids, and subject to the ammonia effect (34). The effect on induction of removing different promoter sequences was monitored by measuring the β-galactosidase activity in plasmid-bearing yeast strain 23346c (ura3) grown on minimal urea medium in the presence or absence of inducer. Deletions extending from −1231 through −255 had no major effect on gene induction. Further removal of sequences up to −169 resulted in a two- to threefold decrease in the level of induction, and no further change was observed with deletions extending to −153. This segment of the promoter thus seems to contribute modestly to activation. It contains several sequences of potential interest: the sequence −234-TTGCCGCCGA-225 almost perfectly matches (9 of 10 nucleotides) the URS1 element (44), the binding site of Car80p/Ume6p, a key regulator of nitrogen metabolism and early meiosis gene expression (74). The imperfect palindrome −231-CCGCCGACGGCCG-219 overlaps with this element. Further deletion of sequences up to −145 and beyond caused almost complete loss of tryptophan-induced expression, indicating that an element essential to activation is present in the promoter near position −145. This region contains, from −168 to −133, a remarkable sequence consisting of four CCG repeats regularly spaced by 7 bp (Fig. 1A and B). An internal deletion removing this 36-bp sequence was produced in plasmid pII472. The deletion abolished induction of the gene (Fig. 2), showing that this 36-bp promoter fragment is necessary for ARO9 induction.
FIG. 2.
Effects of 5′-nested deletions on induced ARO9-lacZ expression. Strain 23346c (ura3) transformed with a low-copy-number plasmid carrying the different promoter deletion constructs was grown on minimal urea medium with (+ Trp) or without (− Trp) tryptophan at 500 μg/ml. β-Galactosidase activities, expressed in nanomoles per minute per milligram of protein, are averages of two independent experiments. β-Galactosidase activity variations were ≤15%. The black box indicates the position of UASaro. The grey box indicates the position of the sequence similar to URS1. The hatched box represents the position of the SPL2 (YHR136c) gene (20). Vertical lines represent the positions of two TATA consensus sequences (TATAAA).
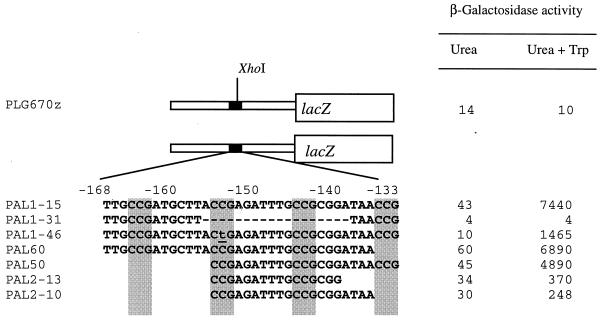
We constructed a hybrid promoter (28) to demonstrate that the 36-bp fragment is also sufficient for specific transcriptional activation. We used a heterologous CYC1-lacZ gene, present on multicopy plasmid pLG670-Z (28), possessing the transcriptional and translational start sites and TATA boxes of CYC1 but lacking its UAS elements. As expected, the CYC1-lacZ fusion was very weakly expressed and did not respond to the presence of tryptophan in the growth medium (Fig. 3). In contrast, plasmid PAL1-15 carrying the putative UAS of ARO9 in front of the CYC1-lacZ gene displayed high activation levels in response to tryptophan (Fig. 3). Furthermore, mutations introduced in vitro into the 36-bp fragment severely affected or abolished tryptophan-induced expression (Fig. 3). Induction was completely abolished by removal of 18 central bp of the sequence in plasmid PAL1-31. It was fivefold reduced by single substitution of a T for a C in the second CCG triplet (PAL1-46). Derived constructs truncated at either the 3′ or the 5′ end and containing only three CCG triplets displayed high induction levels (PAL50 and PAL60). Simultaneous elimination of both external CCG triplets led to 20- to 30-fold-decreased induction but did not totally suppress induction (PAL2-13 and PAL2-10). Taken together, these results demonstrate that the 36-bp sequence contains a UAS element necessary and sufficient for induced expression of the fusion gene in response to tryptophan. The entire 36-bp sequence will be called UASaro throughout this work.
FIG. 3.
UASaro renders the UASaro-CYC1-lacZ gene inducible by aromatic amino acids. Strain 23346c (ura3) transformed with a high-copy-number plasmid carrying the wild-type form (PAL1-15) or a mutated form of UASaro was grown on minimal urea medium with (+ Trp) or without tryptophan at 500 μg/ml. β-Galactosidase activities, expressed in nanomoles per minute per milligram of protein, are averages of two independent experiments. β-Galactosidase activity variations were ≤15%. The underlined lowercase “t” in the PAL1-46 plasmid indicates substitution of a thymine for a cytosine.
Isolation of mutants impaired in ARO9 induction.
Cells of strain 30332d (ura3) transformed by multicopy plasmid PAL1-15 bearing the UASaro-CYC1-lacZ fusion gene form blue colonies during growth on urea plus tryptophan medium supplemented with X-Gal. To isolate mutants affected in ARO9 induction through the UASaro element, we mutagenized transformed 30332d cells and analyzed clones forming white or pale blue colonies. Among 17 analyzed mutants, strains IHI602, IHI606, and IHI614 displayed no induction of either β-galactosidase or tryptophan-phenylpyruvate aminotransferase activity. Strain IHI611 was only partially deficient (Table 2). The same induction deficiencies were observed in mutant cells cured of the plasmid and back-transformed with PAL1-15 (data not shown). The mutants were defective in induction of both the chromosomal ARO9 gene and the multiple plasmid-borne copies of the UASaro-CYC1-lacZ gene. The mutations most likely affect trans-acting elements involved in UASaro-mediated induction.
TABLE 2.
Enzyme activities in mutant isolates defective in ARO9 inductiona
| Strain | Sp act of enzyme:
|
|
|---|---|---|
| β-Galactosidase | Trp-phenylpyruvate aminotransferase | |
| 30332d | 8,143 | 90 |
| IHI602 | 5 | 20 |
| IHI606 | 1 | 13 |
| IHI611 | 4,641 | 40 |
| IHI614 | 4 | 4 |
Strain 30332d (ura3) and mutant strains bearing the PAL1-15 plasmid were grown on minimal urea medium containing 500 μg of tryptophan per ml. Specific activities are expressed in nanomoles per minute per milligram of protein.
ARO9 induction is impaired in mutants lacking the Gap1p and Wap1p permeases.
Mutant strain IHI611 failed to grow on minimal medium containing tryptophan, phenylalanine, tyrosine, or citrulline as the sole nitrogen source and grew at a very reduced rate on isoleucine. Genetic analyses indicated that two mutations were responsible for this growth phenotype (Table 3). One of them affected the GAP1 locus and was called gap1-92. The second mutation did not by itself produce a particular phenotype, but in double mutants it markedly worsened the growth deficiencies caused by the gap1-92 mutation (Table 3). To identify the altered genetic locus, we searched a wild-type genomic library for fragments complementing the growth deficiencies of the double mutant strain 32102c (see Materials and Methods). Subsequent subcloning experiments indicated that the second mutation affected the YCL025c gene on chromosome III (56). On the basis of sequence similarities, this gene had been postulated to encode an amino acid permease (2, 56). We temporarily named the second mutation ycl025-1 and constructed a ycl025Δ-1 deletion mutant strain. Growth phenotype analysis, complementation tests, and analysis of 21 tetrads derived from the cross between strain IHI726 (gap1-92 ycl025Δ-1 ura3) and strain 32102c (gap1-92 ycl025-1 ura3) showed that the recessive mutations ycl025-1 and ycl025Δ-1 are allelic.
TABLE 3.
Growth phenotype of segregants bearing gap1-92 and wap1-1 mutationsa
| Strain | Genotype | M Am | M Trp | M Phe | M Tyr | M Ile | M Cit |
|---|---|---|---|---|---|---|---|
| 30622a | wap1-1 gap1-92 ura3 | ++++ | − | − | − | + | − |
| 30622b | gap1-92 ura3 | ++++ | + | ++++ | +++ | ++++ | − |
| 30622c | ura3 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| 30622d | wap1-1 ura3 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| 23346c | ura3 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| 32102c | wap1-1 gap1-92 ura3 | ++++ | − | − | − | + | − |
Tetratype tetrad 30622 was derived from a cross between strains 32102c (wap1-1 gap1-92 ura3) and 23346c (ura3). Strain 32102c is a segregant from the backcross of the mutant isolate IHI611 with the wild type. The genotypes of 30622c and 30622d were determined by measuring Wap1p permease activity. All media were supplemented by uracil at 50 μg/ml. Abbreviations: M, minimal medium; Am, ammonia; Cit, citrulline. ++++ to −, most to least growth, respectively.
The growth deficiencies of gap1-92 ycl025-1 double mutants suggested that aromatic amino acids are transported not only by Gap1p (26) but also by the putative permease Ycl025p. We conducted uptake experiments with gap1 deletion mutants to avoid massive interference of the general permease with uptake. The cells were grown on urea medium to avoid both the NCR and NCI effects produced by preferred nitrogen sources on the synthesis and activity of permeases (see the introduction). We added radiolabeled tryptophan (final concentration, 20 μM) to cultures of exponentially growing gap1Δ single mutants or gap1Δ ycl025Δ-1 double mutants and measured the rate at which the amino acid accumulated in the cells (data not shown). The two strains displayed markedly different accumulation profiles. In gap1Δ ycl025Δ-1 cells, the import rate remained very low throughout the experiment. In gap1Δ cells, uptake activity gradually developed upon addition of the amino acid, in a manner reminiscent of γ-aminobutyric acid (GABA)-triggered induction of the UGA4 gene encoding the high-affinity GABA permease (3, 27). Similar results have been obtained for the accumulation of phenylalanine and tyrosine (33). Our interpretation is that Ycl025p can indeed transport aromatic amino acids and that these act as inducers of YCL025c gene expression. A detailed study of YCL025c expression and of the properties of the new permease is presented elsewhere (33), showing that YCL025c does encode an inducible amino acid permease with broad substrate specificity. Consequently YCL025c was named WAP1, for wide-specificity amino acid permease. An independent analysis of YCL025c function has also appeared during the preparation of the manuscript (65). To investigate the effects of the gap1Δ and wap1Δ mutations on tryptophan-dependent ARO9 induction, we determined steady-state ARO9-lacZ expression levels in cells grown in the presence or absence of inducer at a high concentration (500 μg/ml) (Table 4). Elimination of both permeases did not significantly reduce the level of ARO9 expression on tryptophan-supplemented minimal ammonia medium. On tryptophan-supplemented urea medium, elimination of Gap1p or Wap1p did not decrease the level of expression, but simultaneous loss of both permeases led to a 75% decline, similar to that observed with the original IHI611 mutant. This decline probably reflects poor inducer uptake, but the very low inducer uptake rates observed in the double mutant remain compatible with a moderate level of ARO9 induction. Individual loss of Gap1p or Wap1p led to a twofold increase of ARO9 expression, an aspect which remains unexplained. Overall, these results show that Gap1p and Wap1p are the main transporters involved in aromatic amino acid utilization. They suggest also that cellular inducer concentration limits ARO9 induction in gap1 wap1 double mutant strains grown on urea plus tryptophan medium.
TABLE 4.
Tryptophan-induced expression of the ARO9-lacZ gene in gap1 and wap1 deletion mutant strainsa
| Strain | Genotype | β-Galactosidase activity
|
|||
|---|---|---|---|---|---|
| Ammonia
|
Urea
|
||||
| − Trp | + Trp | − Trp | + Trp | ||
| 23346c | Wild type | 6 | 129 | 3 | 2,046 |
| 30629c | gap1Δ | 2 | 281 | 3 | 1,900 |
| 30637a | wap1Δ | 2 | 300 | 3 | 3,085 |
| 30633c | wap1Δ gap1Δ | 2 | 102 | 3 | 478 |
Wild-type and mutant cells transformed with the YCpARO9-lacZ plasmid were grown on ammonia or urea medium in the presence (+ Trp) or absence (− Trp) of the inducer tryptophan at 500 μg/ml. Specific β-galactosidase activities are expressed in nanomoles per minute per milligram of protein. The reported activities are averages of the values from at least two independent experiments. Variations were ≤15%.
The ARO80 gene is essential to induced expression of ARO9.
Mutant strains IHI602, IHI606, and IHI614 failed to grow on minimal tryptophan medium; their growth rate was reduced on minimal phenylalanine or tyrosine medium and unaffected on other sources of nitrogen. Genetic analyses showed that all three mutations segregate 2:2; are recessive; complement the aro8, aro9, gap1, and wap1 mutations; and belong to the same complementation class, which we have named ARO80. When an aro80 mutation was associated with an aro8 mutation affecting aromatic aminotransferase I (77), the aro80 aro8 double mutant cells grew very slowly on ammonia medium unsupplemented with phenylalanine or tyrosine. This confirmed the absence of inducible aromatic aminotransferase II activity in the double mutants, our previous studies having shown that the simultaneous absence of aromatic aminotransferases I and II results in phenylalanine and tyrosine auxotrophy (77).
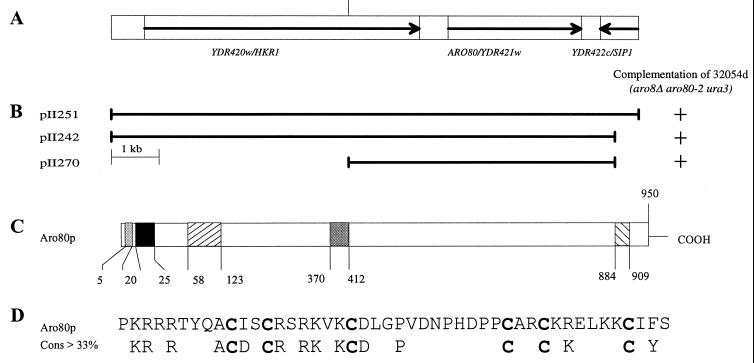
The ARO80 gene was isolated by seeking genomic fragments complementing the double auxotrophy of strain 32054d (aro8Δ1-1 aro80-2 ura3) (see Materials and Methods). Complementing fragments carried the YDR421w gene (Fig. 4A and B) of chromosome IV (37). YDR421w also complemented the growth deficiencies of aro80 single mutants and was thus postulated to be identical to the ARO80 gene. To confirm this, we deleted YDR421w from the chromosome of strain IHI708 (ydr421wΔ-1::Kanr ura3). Growth phenotype analyses, complementation tests, and analysis of 28 tetrads from a cross between strains 32054b (aro80-2 ura3) and IHI708 (ydr421wΔ-1::Kanr ura3) demonstrated that the ydr421wΔ-1 and aro80-2 mutations are allelic. Deletion mutant ydr421wΔ-1 was named aro80Δ-1.
FIG. 4.
(A) Map of chromosome IV region around the ARO80 gene. (B) Schematic drawing of the DNA fragments that complement the auxotrophy of the aro8Δ aro80-2 double mutation. (C) Schematic diagram of predicted Aro80p domains: putative bipartite nuclear targeting sequence ( ), C6 zinc finger domain (▪), coiled-coil region (▨), middle homology region (
), C6 zinc finger domain (▪), coiled-coil region (▨), middle homology region ( ), and acidic domain (▧). (D) Alignment of the C6 zinc finger sequence of the ARO80 gene and the consensus sequence of the domain (64).
), and acidic domain (▧). (D) Alignment of the C6 zinc finger sequence of the ARO80 gene and the consensus sequence of the domain (64).
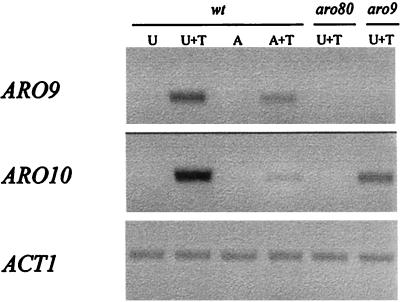
To confirm the role of Aro80p in ARO9 regulation, we compared expression of the ARO9 gene in the wild-type and aro80Δ genetic backgrounds. Table 5 shows that tryptophan-dependent induction of both plasmid-borne ARO9-lacZ and chromosomal ARO9 was abolished in extracts of the aro80Δ deletion mutant. Table 5 further shows that inducible aminotransferase activity was restored in the mutant after transformation with a centromeric plasmid carrying the cloned ARO80 gene. Steady-state ARO9 transcript levels were also measured semiquantitatively by RT-PCR (Fig. 5). A PCR product of the correct size was obtained with ARO9-specific primers and RNA prepared from wild-type cells grown on urea medium supplemented with tryptophan. A much less abundant amplification product was obtained with RNA from wild-type cells grown on tryptophan-supplemented ammonia medium. No perceptible ARO9 amplification signal was obtained with RNA preparations from wild-type cells grown on inducer-free medium or from aro80Δ cells grown on inducer-supplemented ammonia or urea medium. There is thus complete agreement among the regulation pattern of the native enzyme, the reporter gene expression data, and the results of RT-PCR transcript analysis. We conclude that induction of aromatic aminotransferase II results from Aro80p-dependent transcriptional activation of the ARO9 gene.
TABLE 5.
The ARO80 gene is required for transcriptional induction of ARO9a
| Enzyme and strain | Genotype | Plasmid | Gene on plasmid | Sp act
|
|
|---|---|---|---|---|---|
| Urea | Urea + Trp | ||||
| β-Galactosidase | |||||
| 23346c | ura3 | YCpARO9-lacZ | ARO9-lacZ | ≤10 | 2,500 |
| 30701a | aro80Δ ura3 | YCpARO9-lacZ | ARO9-lacZ | ≤10 | ≤10 |
| Tryptophan-phenylpyruvate aminotransferase | |||||
| 23346c | ura3 | pFL38 | 10 | 98 | |
| 30701a | aro80Δ ura3 | pFL38 | 10 | 8 | |
| 30701a | aro80Δ ura3 | pII271 | ARO80 | 7 | 88 |
Strains 23346c (ura3) and 30701a (aro80Δ ura3) bearing the YCpARO9-lacZ, pII271, or pFL38 plasmid were grown on minimal urea medium with or without tryptophan at 500 μg/ml. Specific activities are expressed in nanomoles per minute per milligram of protein. The reported activities are averages of the values from at least two independent experiments. Variations were ≤15%.
FIG. 5.
RT-PCR analysis of ARO9 and ARO10 transcripts. Strains 23346c (ura3), 30701a (aro80Δ ura3), and 30312d (aro9Δ ura3) were grown on minimal medium containing the following nitrogen sources: urea (U), urea plus tryptophan (U+T), ammonia (A), or ammonia plus tryptophan (A+T). Total RNA was used to perform the analysis (see Materials and Methods). wt, wild type.
Aro80p is a member of the Zn2Cys6 transcription factor family.
The ARO80 (YDR421w) open reading frame encodes a 950-residue polypeptide with a calculated molecular mass of 118 kDa. Its codon bias index is 0.022 (7), and its codon adaptation index is 0.12 (67), both values being indicative of a low expression level, typical of yeast transcriptional activators. Two poor matches to the TATA consensus sequence lie at positions −49 and −72 from the putative initiation codon. This upstream region also contains a cluster of three 5′-GATA-3′ sequences from positions −137 to −114, raising the possibility that GATA factors control ARO80 expression. The deduced amino acid sequence of Aro80p contains several motifs (Fig. 4C and D) that characterize the Gal4p class of transcription factors, as already analyzed by Schjerling and Holmberg (64) in their study of the 56 Gal4p family members present in the entire yeast genome: the most salient feature is an N-terminal segment highly similar to the consensus sequence of the Zn2Cys6-type zinc finger motif (Fig. 4D), an established DNA-binding domain (40). The Aro8p motif has all the features found in the other cloverleaf-like Zn2Cys6 binuclear clusters (57), but the region of variable length linking the two cysteine-rich substructures comprises 12 amino acids instead of 6 to 9 as commonly found in other proteins of the family; it shares this property with the putative yeast transcription factor Yml076p of unknown function; several clusters of acidic residues are present in the predicted Aro80p. The cluster with the highest negative charge (−9) is located at the C-terminal end spanning positions 884 to 909. Similar C-terminal acidic regions have been shown to mediate protein-protein interactions and to contribute to the ability of the protein to activate transcription (46). In Aro80p, this C-terminal acidic segment is followed by an asparagine-rich region; several coiled-coil structures with a potential to form heptad repeats of an amphipathic α-helix are predicted by the Pepcoil algorithm (45). It is common for this family of factors to act as homodimers, and crystal structures have revealed that coiled-coil structures mediate formation of Gal4 and Ppr1 homodimers (50, 51). Other noteworthy structural features of Aro80p have not been reported previously: (i) a recognizable part of the middle homology region (64) spans positions 370 to 412; (ii) a putative bipartite nuclear targeting sequence (61) spans amino acids 5 to 20; (iii) an ATP-GTP binding site spans residues 647 to 654; and (iv) a putative PEST region extends from residue 836 to residue 854.
The ARO80 gene is also essential to induced expression of the ARO10 (YDR380w) gene encoding a putative indole-3-pyruvate decarboxylase.
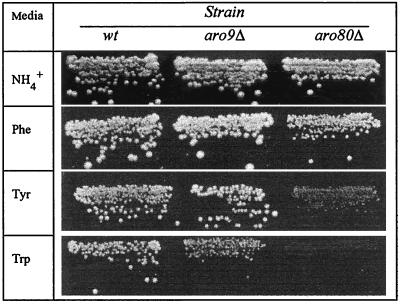
On media containing tryptophan, phenylalanine, or tyrosine as the sole nitrogen source, growth of the aro80Δ mutant was clearly affected more strongly than that of the aro9Δ mutant (Fig. 6). This indicates that Aro80p activates other genes besides ARO9 in the aromatic amino acid utilization pathway. Genes involved in aromatic amino acid transport were obvious candidates for this regulation. Yet the tryptophan, phenylalanine, and tyrosine uptake rates were unaffected by the aro80Δ mutation, and the ARO80 gene proved not to be involved in induction of the new Wap1p permease (data not shown). We searched the genome for sequences similar to UASaro. The pattern 5′-CCG(7X)CCG(7X)CCG(7X)CCG-3′ was found, spanning bp −342 to −310 upstream from the open reading frame YDR380w. The deduced Ydr380p sequence is highly similar to those of yeast pyruvate decarboxylases Pdc6p, Pdc5p, and Pdc1p and to the recently described α-ketoisocaproate decarboxylase Kid1p (15). It is also highly similar to the three bacterial indolepyruvate decarboxylase sequences present in databases. In fact, together with pyruvate decarboxylases and Kid1p, Ydr380p is the yeast protein most similar to them, as established by sequence comparisons (data not shown). Thus, Ydr380p may be the yeast indole-3-pyruvate decarboxylase catalyzing conversion of indole-3-pyruvic acid to indole-3-acetaldehyde, the second reaction involved in tryptophan degradation to tryptophol. In line with this hypothesis, we found the YDR380w and ARO9 genes to be regulated in a very similar manner. RT-PCR transcript analysis (Fig. 5) showed weak expression of YDR380w on both urea and ammonia medium. On both media, YDR380w transcription was induced upon cell exposure to tryptophan, the activation level being markedly higher on urea medium. In both cases, induction of transcription was abolished in aro80Δ mutant cells, showing that ARO80 is also essential to YDR380w induction by tryptophan. These facts highlight a second target of Aro80p and integrate the YDR380w gene into tryptophan metabolism. We have thus given YDR380w the name ARO10. Further characterization of this gene is under way.
FIG. 6.
Growth phenotypes of strains 23346c (ura3), 30312d (aro9Δ ura3), and 30701a (aro80Δ ura3) on minimal medium containing ammonia, phenylalanine, tyrosine, or tryptophan as the nitrogen source. All media were supplemented by 50 μg of uracil per ml. wt, wild type.
Nitrogen regulation of ARO9 expression.
Like many other nitrogen catabolic genes, ARO9 is subject to nitrogen source regulation (see the introduction). A systematic study was undertaken to discover the mechanisms underlying this control. Wild-type and appropriate mutant strains transformed with the ARO9-lacZ plasmid were grown under conditions favoring repression (ammonia medium) or derepression (urea medium) and in the presence or absence of the inducer tryptophan. The transformed strains each carried one or several mutations affecting factors involved in either NCR or NCI. β-Galactosidase activity was taken as a measure of ARO9 expression.
(i) Role of the NCR general negative factors.
In the absence of inducer, wild-type cells grown on either ammonia or urea medium displayed very low, similar levels of ARO9 expression; hence, no ARO9 derepression was observed on urea medium (Table 6, row 1). In the absence of inducer, likewise, we observed no increased expression of ARO9 in cells harboring, separately or in combination, the mutations ure2, uga43Δ, and gzf3Δ, disabling factors that mediate nitrogen repression (Table 6, rows 2 to 8). This contrasts with the behavior of the inducible genes CAR1, GDH2, DUR1, DUR2, and DAL1 (47, 80); PUT1, PUT2, and UGA1 (81); and UGA4 (4), whose basal expression in inducer-free media is subject to nitrogen repression. Yet despite the apparent insensitivity of ARO9 to nitrogen repression, once wild-type cells were exposed to the inducer, much higher levels of ARO9 transcription were observed on urea medium than on ammonia medium (Table 6, row 1). This ammonia effect can hardly be attributed to nitrogen repression exerted by Ure2p, by GATA factor Gzf3p or Uga43p, or by a combination of these, since expression increased very little in NH4+-grown cells lacking these factors (Table 6, rows 2 to 8). Furthermore, the threefold increase observed in cells carrying the ure2 mutation, alone or associated with the gzf3Δ and uga43Δ mutations (Table 6, rows 2, 5, and 7 and 8), was lost in ure2 aro80Δ cells (Table 6, row 9), showing that it is dependent on induction.
TABLE 6.
Nitrogen regulation of ARO9 expressiona
| Strain | Genotype | β-Galactosidase activity
|
|||
|---|---|---|---|---|---|
| Ammonia
|
Urea
|
||||
| − Trp | + Trp | − Trp | + Trp | ||
| 23346c | Wild type | ≤10 | 175 | ≤10 | 2,500 |
| 26854a | ure2Δ | ≤10 | 505 | ≤10 | 3,830 |
| 30078c | uga43Δ | ≤10 | 230 | ≤10 | 2,975 |
| SBS10 | gzf3Δ | ≤10 | 296 | ≤10 | 2,740 |
| 26992c | ure2 uga43Δ | ≤10 | 250 | ≤10 | 2,670 |
| 50036c | uga43Δ gzf3Δ | ≤10 | 264 | ≤10 | 2,255 |
| 50000b | ure2 gzf3Δ | ≤10 | 770 | ≤10 | 4,120 |
| 50054a | ure2 gzf3Δ uga43Δ | ≤10 | 600 | ≤10 | 4,500 |
| 32173c | ure2 aro80Δ | ≤10 | ≤10 | ≤10 | ≤10 |
| 30505b | gln3Δ | ≤10 | 720 | ≤10 | 2,125 |
| SBS21 | nil1Δ | ≤10 | 280 | ≤10 | 3,625 |
| 32164c | gln3Δ nil1Δ | 40 | 300 | 46 | 390 |
| 30629c | gap1Δ | ≤10 | 292 | ≤10 | 1,984 |
| 33089d | gap1pgr | ≤10 | 715 | ≤10 | 1,988 |
| 27038a | npi1 | ≤10 | 1,976 | ≤10 | 3,701 |
| 30831b | npi1 ure2 | ≤10 | 2,450 | ≤10 | 3,642 |
| 30768d | gap1Δ npi1 | ≤10 | 1,325 | ≤10 | 3,289 |
The table shows the role of Ure2p and negative GATA factors Uga43p and Gzf3p (top nine rows), of positive GATA factors Gln3p and Nil1p (middle three rows), and effect of mutations affecting NCI (bottom five rows). All strains carried the ura3 marker mutation and bore the YCpARO9-lacZ plasmid. Cells were grown on minimal ammonia or urea medium with (+ Trp) or without (− Trp) tryptophan at 500 μg/ml. Specific activities are expressed in nanomoles per minute per milligram of protein. The reported activities are averages of the values from at least two independent experiments. Variations were ≤15%.
(ii) Role of the positive factors Gln3p and Nil1p.
Nitrogen-regulated genes are typically under the positive control of the GATA family factors Gln3p and Nil1p (47, 52). On tryptophan-supplemented urea medium, ARO9 transcription was unaffected in cells lacking one of these factors (Table 6, rows 10 and 11). Surprisingly, the activity in the gln3Δ mutant is higher than that in the wild-type strain after growth in minimal ammonia plus tryptophan (Table 6, rows 10 and 1; Table 7). This unexpected result has also been observed in the case of the arginine induction of the CAR1 gene encoding arginase (18). ARO9 expression was greatly reduced upon elimination of both activators (Table 6, row 12), indicating that they both can contribute to the amplification of induction observed under conditions of nitrogen derepression. Yet under conditions of nitrogen repression, elimination of both activators had no depressing effect on ARO9 induction (Table 6, row 12), in contrast to what happens with other nitrogen-regulated genes (9, 71, 73). Furthermore, Gln3p and Nil1p activate expression of nitrogen-sensitive genes via 5′-GATA-3′ UASs. The ARO9 promoter contains several dispersed GATA motifs (Fig. 1A) but no GATA element similar to either UASNTR (58, 59), UASN (55), or UASGATA (4). Moreover, we failed to obtain evidence that Gln3p and Nil1p exert their stimulatory effect at a site different from UASaro: none of our various ARO9 promoter constructs enabled us to distinguish Gln3p and Nil1p target elements from the induction element UASaro. First, we introduced the constructs obtained by exonuclease III treatment (Fig. 2) into wild-type and gln3Δ nil1Δ strains and compared their expression in both genetic backgrounds. In the set of deletion mutants, loss of induction closely paralleled loss of the amplifying action of the two GATA activators (data not shown). Removal of the sequence between −255 and −169 resulted in a two- to threefold decrease in the level of induction in the double gln3Δ nil1Δ mutant from in the wild-type strain. Next, we confirmed that the amplifying effect of Gln3p and Nil1p requires the integrity of the UASaro element, as judged from the effect of deleting the element in the ARO9-lacZ fusion construct (Table 7). Finally, we observed that UASaro alone suffices to drive a much higher level of tryptophan-induced transcription on urea medium than on ammonia and that this amplification is no longer observed in gln3Δ nil1Δ double mutant cells (Table 7). Hence, Gln3p and Nil1p appear to amplify induction of ARO9 mainly through UASaro. The low constitutive activity measured in the gln3Δ nil1Δ double mutant (Table 6, row 12) is possibly due to a particular phenomenon connected to its very low growth in minimum medium.
TABLE 7.
Amplification by Gln3p and Nil1p of ARO9 expression requires the UASaro elementa
| Strain | Genotype | Plasmid | Gene on plasmid | β-Galactosidase activity
|
|||
|---|---|---|---|---|---|---|---|
| Ammonia
|
Proline
|
||||||
| − Trp | + Trp | − Trp | + Trp | ||||
| 23346c | ura3 | YCpARO9-lacZ | ARO9-lacZ | ≤10 | 248 | ≤10 | 2,260 |
| 32164c | gln3Δ nil1Δ ura3 | YCpARO9-lacZ | ARO9-lacZ | 54 | 384 | 46 | 450 |
| 23346c | ura3 | pII472 | ARO9Δ-UASaro-lacZ | ≤10 | 20 | 17 | 47 |
| 32164c | gln3Δ nil1Δ ura3 | pII472 | ARO9Δ-UASaro-lacZ | 12 | 20 | 19 | 25 |
| 23346c | ura3 | PAL1-15 | UASaro-CYC1-lacZ | 27 | 2,755 | 30 | 6,480 |
| 32164c | gln3Δ nil1Δ ura3 | PAL1-15 | UASaro-CYC1-lacZ | 227 | 2,020 | 335 | 1,808 |
Transformed strains 23346c (ura3) and 32164c (gln3Δ nil1Δ ura3) were grown on minimal ammonia or proline medium with (+ Trp) or without (− Trp) tryptophan at 500 μg/ml. Specific activities are expressed in nanomoles per minute per milligram of protein. The reported activities are averages of the values from at least two independent experiments. Variations were ≤15%.
(iii) Role of inducer exclusion.
The above results suggest that NH4+ and the GATA factors Gln3p and Nil1p may affect ARO9 induction indirectly, by inducer exclusion. Inducer uptake as a limiting step in ARO9 induction is indeed consistent with the observation that ARO9 expression is severely reduced in gap1 wap1 double mutants which take up tryptophan at a very reduced rate (Table 4). To test this hypothesis, we monitored induction of ARO9 in gap1pgr and npi1 mutants grown on ammonia medium. These strains are defective in NCI and incorporate tryptophan faster than do wild-type cells on ammonia medium (see the introduction). The gap1pgr mutation protects only the Gap1p permease from inactivation, whereas the npi1 mutation prevents inactivation of other NCI-sensitive permeases as well (23). On tryptophan-supplemented ammonia medium, gap1pgr mutant cells displayed four-times-higher ARO9 expression levels than wild-type cells (Table 6, row 14). In npi1 cells grown on the same medium, ARO9 expression was increased more than 10-fold, reaching 80% of the level found in wild-type cells grown on urea plus tryptophan (Table 6, row 15). ARO9 can thus be expressed to near-maximum levels under conditions of nitrogen repression in mutants released from NCI. Simultaneous release from NCR and NCI in a ure2 npi1 double mutant caused but a slight further increase in expression compared to the effect of the npi1 mutation alone (Table 6, row 16). These results strongly indicate that the ammonium effect on ARO9 expression is produced mainly by inducer exclusion through permease inactivation. The Gap1p permease is neither the only nor the principal tryptophan permease involved in exclusion, since (i) the ARO9 induction level was only slightly affected in gap1Δ cells compared to that in wild-type cells (Table 6, row 13) and (ii) the npi1 gap1Δ strain (Table 6, row 17) attains about 60% of the induction level reached by the npi1 single mutant. The experiments for Table 6 were also carried out with phenylalanine or tyrosine as an alternative inducer, yielding very similar results (data not shown).
DISCUSSION
The ARO9 gene encodes aromatic aminotransferase II, the enzyme catalyzing the first reaction of aromatic amino acid catabolism (34, 41, 77). Transcription of the ARO9 gene is induced by aromatic amino acids and subject to nitrogen regulation. In this work, we have characterized cis- and trans-acting elements mediating this induction. We have also shown that nitrogen source regulation of ARO9 expression is due mainly to inducer exclusion, in contrast to the situation observed with many genes involved in secondary nitrogen source utilization. We further show, in this report, that the two main uptake systems for utilization of aromatic amino acids are Gap1p, the general amino acid permease, and Wap1p, a newly discovered inducible amino acid permease with wide substrate specificity. Finally, we present evidence that transcription of the ARO10 gene, which very likely encodes the second enzyme of the aromatic catabolic pathway, indolepyruvate decarboxylase, is subject to the same new induction mechanism.
Induction-specific regulatory elements.
Induction of ARO9 by aromatic amino acids requires UASaro, a 36-bp element, and the trans-acting factor Aro80p. UASaro and Aro80p behave functionally like the pathway-specific elements ArgRIIp (53), Put3p (48), Uga3p (1), and Cha4p (31) and their cognate UASs, which control induction of the arginine, proline, GABA, serine, and threonine catabolic pathways in yeast, respectively. Like ArgRIIp, Put3p, Uga3p, and Cha4p, Aro80p is a member of the Zn2Cys6 family of transcriptional activators. Its deduced sequence displays all the features that characterize this protein family. Aro80p is required for specific induction of ARO9 expression, and its function depends on sequences present in the UASaro element. This 36-bp element, on the other hand, is necessary and sufficient to mediate transcriptional activation of a reporter gene in response to aromatic amino acids. UASaro could thus be the DNA element to which Aro80p binds. Its structural features support this hypothesis: binding sites for Zn2Cys6 proteins often comprise two symmetrically placed CGG elements, separated by a characteristic spacing (64). UASaro possesses one CGG triplet which could form an imperfect palindrome with any of several CCG triplets in the element (Fig. 1B). The CCGCGG core of one such palindrome, for example, is identical to the core of the Pdr3p binding site (39). Yet not all zinc cluster proteins bind to sites with inverted CGG repeats. Hap1p binding sites, for instance, are composed of two direct CGG repeats, and their spacer displays no symmetry (82). The most obvious feature of the UASaro sequence is the pattern of four regularly spaced direct CCG repeats. A similar pattern was found in the ARO10 (YDR380w) promoter. When the putative UAS of ARO10 is aligned with UASaro, the consensus sequence of the motif is extended to T(T/A)(G/A)CCG followed by four variable nucleotides (Fig. 1C). ARO10 is also inducible by tryptophan, and its transcriptional activation is Aro80p dependent. The ARO9 and ARO10 genes are thus coregulated by aromatic amino acids.
Our data clearly show that Aro80p is essential to UASaro-mediated induction by tryptophan, phenylalanine, and tyrosine. Although aro80 cells are unable to grow on tryptophan medium, their growth is slowed down only on medium containing tyrosine or phenylalanine as the sole nitrogen source (Fig. 6). Similarly, aro9 cells are unable to grow on tryptophan but grow normally on phenylalanine or tyrosine (77). Thus, ARO9, ARO80, and UASaro are essential to utilization of tryptophan as a nitrogen source. The growth phenotype of aro80 cells suggests that other genes involved in phenylalanine and tyrosine catabolism and their regulatory elements remain to be discovered.
Nitrogen regulation of ARO9 is exerted mainly through inducer exclusion.
ARO9 is less actively transcribed in ammonia-grown than in urea-grown cells, indicating that the gene is subject to nitrogen source regulation. Nitrogen catabolic genes are commonly regulated by nitrogen repression, i.e., by the competitive action at upstream 5′-GATA-3′ elements of positive and negative GATA family factors and by the inhibition exerted by Ure2p on Gln3p (47, 52). Our results strongly suggest that ARO9 is an exception. Nitrogen regulation of ARO9 transcription would appear to be achieved mainly or even exclusively by inducer exclusion, via regulation of permease synthesis and activity. This conclusion is based on the fact that mutations relieving nitrogen repression only slightly affect ARO9 expression, compared with mutations relieving NCI of permeases. After growth on ammonia medium containing tryptophan (or phenylalanine or tyrosine), npi1 cells display a high level of ARO9 transcription, reaching 80% of the level displayed by wild-type cells grown on tryptophan-supplemented urea medium. Under growth conditions promoting nitrogen repression, ARO9 expression can thus reach near-maximum levels provided that permease inactivation is prevented. The CAR2 gene encoding ornithine aminotransferase in the arginine catabolic pathway (14) is likewise subject to an ammonia effect but apparently insensitive to nitrogen repression. In this case, it was possible to correlate closely the amount of enzyme synthesized with the cellular pool of arginine, the inducer (14). The hypothesis that the nitrogen source regulates ARO9 expression principally by modulating inducer availability is consistent with other observations presented here, notably the fact that ARO9 expression is induction dependent under all examined conditions. The very low level of ARO9 expression in cells grown on inducer-free medium is in line with the very small cellular pools of aromatic amino acids (13, 19, 41). The negative effect of the gln3 nil1 double mutation can also be attributed to reduced inducer uptake. We have shown that Gap1p and Wap1p are the main transporters of aromatic amino acids and that gap1 wap1 cells display severely reduced levels of ARO9 expression on tryptophan-supplemented urea medium. It is known that the GAP1 transcription level is very low in gln3 nil1 cells (71, 73). We show elsewhere (33) that WAP1 responds in a very similar manner, a result also published recently by another group (65). Thus, the marked negative effect of the double gln3 nil1 mutation is most likely a consequence of poor permease synthesis. The inducer exclusion hypothesis also seems to best explain why the gln3 nil1 double mutation does not affect induced ARO9 expression under conditions of nitrogen repression. Analysis of the npi1 mutant has indeed shown that permease inactivation, and not permease synthesis, limits inducer availability under these growth conditions.
In a separate paper (33), we show that induction of WAP1 and other amino acid permease genes is controlled by Apf7p, a sensor of external amino acids and the likely head of a new signaling pathway in which Grr1p and Uga35p also intervene. An interesting corollary is that external amino acids also influence ARO9 expression via the control exerted on WAP1 and possibly other aromatic amino acid permease genes.
ACKNOWLEDGMENTS
We are grateful to S. Soussi-Boudekou for the gift of strains prior to publication, to K. Broman for help in writing the manuscript, and to the members of the laboratory for fruitful discussions.
This work was supported by Medical Scientific Research Fund grant 3.4602.94 (FRSM, Belgium) and by a research grant from the Université Libre de Bruxelles. During this work, I.I. was the recipient of a predoctoral fellowship from the Communauté Française de Belgique and then from the Fondation Universitaire David et Alice Van Buuren.
REFERENCES
- 1.André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990;220:269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- 2.André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 3.André B, Hein C, Grenson M, Jauniaux J C. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:17–25. doi: 10.1007/BF00282779. [DOI] [PubMed] [Google Scholar]
- 4.André B, Talibi D, Soussi Boudekou S, Hein C, Vissers S, Coornaert D. Two mutually exclusive regulatory systems inhibit UASGATA, a cluster of 5′-GAT(A/T)A-3′ upstream from the UGA4 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:558–564. doi: 10.1093/nar/23.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 6.Béchet J, Grenson M, Wiame J M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 7.Bennetzen J L, Hall B D. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 8.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 9.Coffman J A, Rai R, Cunningham T, Svetlov V, Cooper T G. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman J A, Rai R, Loprete D M, Cunningham T, Svetlov V, Cooper T G. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1981. pp. 39–99. [Google Scholar]
- 12.Coornaert D, Vissers S, André B, Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet. 1992;21:301–307. doi: 10.1007/BF00351687. [DOI] [PubMed] [Google Scholar]
- 13.Delforge J, Messenguy F, Wiame J M. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR− mutations and the general control of amino-acid biosynthesis. Eur J Biochem. 1975;57:231–239. doi: 10.1111/j.1432-1033.1975.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 14.Deschamps J, Dubois E, Wiame J M. l-Ornithine transaminase synthesis in Saccharomyces cerevisiae: regulation by inducer exclusion. Mol Gen Genet. 1979;174:225–232. doi: 10.1007/BF00267794. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson J R, Lanterman M M, Danner D J, Pearson B M, Sanz P, Harrison S J, Hewlins M J. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1997;272:26871–26878. doi: 10.1074/jbc.272.43.26871. [DOI] [PubMed] [Google Scholar]
- 16.Drillien R, Lacroute F. Ureidosuccinic acid uptake in yeast and some aspects of its regulation. J Bacteriol. 1972;109:203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubois E, Grenson M, Wiame J M. The participation of the anabolic glutamate dehydrogenase in the nitrogen catabolite repression of arginase in Saccharomyces cerevisiae. Eur J Biochem. 1974;48:603–616. doi: 10.1111/j.1432-1033.1974.tb03803.x. [DOI] [PubMed] [Google Scholar]
- 18.Dubois E, Messenguy F. Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p, and Ume6p. Mol Gen Genet. 1997;253:568–580. doi: 10.1007/s004380050359. [DOI] [PubMed] [Google Scholar]
- 19.Fantes P A, Roberts L M, Huetter R. Free tryptophan pool and tryptophan biosynthetic enzymes in Saccharomyces cerevisiae. Arch Microbiol. 1976;107:207–214. doi: 10.1007/BF00446842. [DOI] [PubMed] [Google Scholar]
- 20.Flick J S, Thorner J. An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:33–47. doi: 10.1093/genetics/148.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grenson M. The utilization of exogenous pyrimidines and the recycling of uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur J Biochem. 1969;11:249–260. doi: 10.1111/j.1432-1033.1969.tb00767.x. [DOI] [PubMed] [Google Scholar]
- 23.Grenson M. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur J Biochem. 1983;133:135–139. doi: 10.1111/j.1432-1033.1983.tb07438.x. [DOI] [PubMed] [Google Scholar]
- 24.Grenson M. Amino acid transporters in yeast: structure, function and regulation. In: De Pont J J L L M, editor. Molecular aspects of transport proteins. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 219–245. [Google Scholar]
- 25.Grenson M, Dubois E, Piotrowska M, Drillien R, Aigle M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol Gen Genet. 1974;128:73–85. doi: 10.1007/BF00267295. [DOI] [PubMed] [Google Scholar]
- 26.Grenson M, Hou C, Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol. 1970;103:770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grenson M, Muyldermans F, Broman K, Vissers S. 4-Aminobutyric acid (GABA) uptake in baker’s yeast Saccharomyces cerevisiae is mediated by the general amino acid permease, the proline permease and a GABA-specific permease integrated into the GABA-catabolic pathway. Life Sci Adv Ser C. 1987;6:35–39. [Google Scholar]
- 28.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 29.Hein C, André B. A C-terminal di-leucine motif and nearby sequences are required for NH4+-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol Microbiol. 1997;24:607–616. doi: 10.1046/j.1365-2958.1997.3771735.x. [DOI] [PubMed] [Google Scholar]
- 30.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 31.Holmberg S, Schjerling P. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics. 1996;144:467–478. doi: 10.1093/genetics/144.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iraqui I, Vissers S, André B, Urrestarazu A. Characterization of cis- and trans-acting elements responsible for transcriptional induction of the ARO9 gene encoding aromatic aminotransferase II in Saccharomyces cerevisiae. Arch Physiol Biochem. 1998;106:B8. [Google Scholar]
- 33.Iraqui I, Vissers S, Bernard F, De Craene J-O, Boles E, Urrestarazu A, André B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- 35.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs P, Jauniaux J C, Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980;139:691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- 37.Jacq C, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature. 1997;387:75–78. [PubMed] [Google Scholar]
- 38.Jauniaux J C, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other baker’s yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- 39.Katzmann D J, Burnett P E, Golin J, Mahe Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986;231:699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- 41.Kradolfer P, Niederberger P, Hütter R. Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases. Arch Microbiol. 1982;133:242–248. doi: 10.1007/BF00415010. [DOI] [PubMed] [Google Scholar]
- 42.Lingens F, Goebel W, Uesseler H. Regulation of the biosynthesis of aromatic amino acids in Saccharomyces cerevisiae. 2. Repression, induction and activation. Eur J Biochem. 1967;1:363–374. doi: 10.1111/j.1432-1033.1967.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 43.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 44.Luche R M, Sumrada R, Cooper T G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990;10:3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 46.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 47.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 283–317. [Google Scholar]
- 48.Marczak J E, Brandriss M C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991;11:2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marini A M, Vissers S, Urrestarazu A, André B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994;13:3456–3463. doi: 10.1002/j.1460-2075.1994.tb06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 51.Marmorstein R, Harrison S C. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994;8:2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- 52.Marzluf G A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messenguy F, Dubois E. The yeast ARGRII regulatory protein has homology with various RNases and DNA binding proteins. Mol Gen Genet. 1988;211:102–105. doi: 10.1007/BF00338399. [DOI] [PubMed] [Google Scholar]
- 54.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 55.Miller S M, Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliver S G, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 57.Pan T, Coleman J E. GAL4 transcription factor is not a “zinc finger” but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci USA. 1990;87:2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rai R, Daugherty J R, Cooper T G. UASNTR functioning in combination with other UAS elements underlies exceptional patterns of nitrogen regulation in Saccharomyces cerevisiae. Yeast. 1995;11:247–260. doi: 10.1002/yea.320110307. [DOI] [PubMed] [Google Scholar]
- 59.Rai R, Genbauffe F S, Sumrada R A, Cooper T G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reece R J, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 61.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 62.Rowen D W, Esiobu N, Magasanik B. Role of GATA factor Nil2p in nitrogen regulation of gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3761–3766. doi: 10.1128/jb.179.11.3761-3766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 64.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schreve J L, Sin J K, Garrett J M. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J Bacteriol. 1998;180:2556–2559. doi: 10.1128/jb.180.9.2556-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.SentheShanmuganathan S. The mechanism of the formation of higher alcohols from amino acids by Saccharomyces cerevisiae. Biochem J. 1960;74:568–576. doi: 10.1042/bj0740568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharp P M, Li W H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 69.Shin M, Shinguu T, Sano K, Umezawa C. Metabolic fates of l-tryptophan in Saccharomyces uvarum (Saccharomyces carlsbergensis) Chem Pharm Bull (Tokyo) 1991;39:1792–1795. doi: 10.1248/cpb.39.1792. [DOI] [PubMed] [Google Scholar]
- 70.Siddiqui A H, Brandriss M C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989;9:4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux J C, André B. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1997;23:1157–1168. doi: 10.1046/j.1365-2958.1997.3021665.x. [DOI] [PubMed] [Google Scholar]
- 71a.Soussi-Boudekou, S., and B. André. Unpublished data.
- 72.Springael J Y, André B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanbrough M, Rowen D W, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 75.Svetlov V V, Cooper T G. Review: compilation and characteristics of dedicated transcription factors in Saccharomyces cerevisiae. Yeast. 1995;11:1439–1484. doi: 10.1002/yea.320111502. [DOI] [PubMed] [Google Scholar]
- 76.Talibi D, Grenson M, André B. Cis- and trans-acting elements determining induction of the genes of the gamma-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:550–557. doi: 10.1093/nar/23.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urrestarazu A, Vissers S, Iraqui I, Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet. 1998;257:230–237. doi: 10.1007/s004380050643. [DOI] [PubMed] [Google Scholar]
- 78.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 79.Webb A D, Ingraham J L. Fusel oil. Adv Appl Microbiol. 1963;5:317–353. [Google Scholar]
- 80.Wiame J M, Grenson M, Arst H N J. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv Microbiol Physiol. 1985;26:1–87. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- 81.Xu S, Falvey D A, Brandriss M C. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2321–2330. doi: 10.1128/mcb.15.4.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang L, Guarente L. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 1994;8:2110–2119. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]