Abstract

Real-time (quantitative) polymerase chain reaction (qPCR) has been widely applied in molecular diagnostics due to its immense sensitivity and specificity. qPCR multiplexing, based either on fluorescent probes or intercalating dyes, greatly expanded PCR capability due to the concurrent amplification of several deoxyribonucleic acid sequences. However, probe-based multiplexing requires multiple fluorescent channels, while intercalating dye-based multiplexing needs primers to be designed for amplicons having different melting temperatures. Here, we report a single fluorescent channel-based qPCR duplexing method on a model containing the sequence of chromosomes 21 (Chr21) and 18 (Chr18). We combined nonspecific intercalating dye EvaGreen with a 6-carboxyfluorescein (FAM) probe specific to either Chr21 or Chr18. The copy number (cn) of the target linked to the FAM probe could be determined in the entire tested range from the denaturation curve, while the cn of the other one was determined from the difference between the denaturation and elongation curves. We recorded the amplitude of fluorescence at the end of denaturation and elongation steps, thus getting statistical data set to determine the limit of the proposed method in detail in terms of detectable concentration ratios of both targets. The proposed method eliminated the fluorescence overspilling that happened in probe-based qPCR multiplexing and determined the specificity of the PCR product via melting curve analysis. Additionally, we performed and verified our method using a commercial thermal cycler instead of a self-developed system, making it more generally applicable for researchers. This quantitative single-channel duplexing method is an economical substitute for a conventional rather expensive probe-based qPCR requiring different color probes and hardware capable of processing these fluorescent signals.
1. Introduction
Polymerase chain reaction (PCR) is a powerful technique to amplify a small number of specific sequences of deoxyribonucleic acid (DNA) molecules up to a detectable level.1 Since its invention in 1986, variants of PCR-based methods have been developed,2 such as standard PCR (end-point PCR),3 real-time/quantitative PCR (qPCR),4 and digital PCR (dPCR).5 qPCR is capable of monitoring the PCR progress in real-time and determining the initial concentration of the target genes. It is currently a gold standard for nucleic acid (NA) detection, especially during the coronavirus disease 2019 (COVID-19) pandemic.6 It is performed using either nonspecific intercalating fluorescent dyes, such as SYBR Green I or EvaGreen,7 or specific oligonucleotides (probes) with fluorophores,8 such as 6-carboxyfluorescein (FAM)9 and 2′-chloro-7′phenyl-1,4-dichloro-6-carboxy-fluorescein (VIC).10
Multiplexing of any PCR expands its detection capability as more than one target gene is simultaneously amplified and detected.11−14 qPCR multiplexing is often used for the detection of a DNA component with minor content with a background of another more abundant DNA, such as gene mutations with wild types in oncological research and diagnostics.15,16 The infectious agents in the background of human sequences are often determined in microbiology and virology.17 The genes of genetically modified organisms are searched in food control.18 Paternally inherited disease-associated sequences of DNA are investigated in the background of cell-free DNA in the plasma of pregnant females, and they are used in noninvasive prenatal diagnostics.19,20 The correct quantification of both mixture components by qPCR is complicated once there are large differences in their representation in the mixture.
Probe-based qPCR multiplexing utilizes probes attached to fluorophores with different emission wavelengths. The fluorescent signals are then split and detected using different optical channels. The number of target genes for the concurrent detection is theoretically unlimited as long as there is no emission wavelength overlap between the different probes. However, it is limited by the optical detection system as the increasing number of detectable colors makes the system complicated and costly. Additionally, the design of the probes and primers for each target gene should also be done carefully otherwise the interaction between them inhibits the PCR multiplexing efficiency. Expression analysis using qPCR highly depends on the quantification of all of the simultaneously analyzed targets. It is assumed that qPCR multiplexed amplification does not alter each other’s efficiency.21,22 The prerequisites for the successful interpretation of duplexed qPCR experiments were extensively studied, which resulted in a generally accepted methodology of 2–ΔΔCt, which is considered as the gold standard for the relative quantification of mixtures.23
An intercalating dye-based qPCR multiplexing method simplifies the optical detection system as only one fluorescent channel is required, and there is no need to design different probes. The genes are typically distinguished based on different melting temperatures (TM) during the melting curve analysis (MCA) after temperature cycling.24,25 However, it is an end-point detection, and primers specific to each target gene must be designed to have a different TM. Alternatively, it can be performed using the continuous fluorescence monitoring (CFM) method allowing monitoring the PCR progress and optimizing it.26 It also enables to perform dynamic MCA during the transition from the elongation to denaturation step and to construct the PCR amplification curves of two27 or three genes,28 or perhaps more. Another CFM-based method was proposed combining the FAM probe with the EvaGreen dye for simultaneous detection as both FAM and EvaGreen share the same emission wavelength,29 which effectively doubles the PCR throughput. Unfortunately, both of these CFM-based methods are restricted from being further applied as they only allow one experiment at a time and require a customized setup. Recently, a novel single fluorescent channel-based multiplexing method was proposed using intercalating dyes, and the result was processed based on multidimensional standard curves.30−32 Moreover, the data set was achieved by commercial qPCR instruments, and the absolute quantification was determined, thus extending the use of these devices.
In this work, we performed a comprehensive study of single-channel multiplexing based on the combination of EvaGreen and a FAM probe. We used a commercial qPCR instrument with 96 wells, which allowed us to study multiplexing using robust, statistically significant data sets. We showed the strengths and weaknesses of the proposed method and also compared it with the dual-channel multiplexing method, thereby making this PCR duplexing universal. Although the proposed work is based on the same principle as our previous one,29 we performed a quantitative study of the method, having statistically significant data set using a commercial thermocycler instead of a home-made system to determine the limit of this method in detail in terms of detectable concentration ratios of both targets. The proposed methodology was more generally applicable for system optimization compared with the previous one, as researchers could perform it using any commercial qPCR instruments. We show fully quantitative duplexing capable to be performed by a conventional thermocycler and thus available to anyone.
2. Principle of the Proposed Multiplexing Method
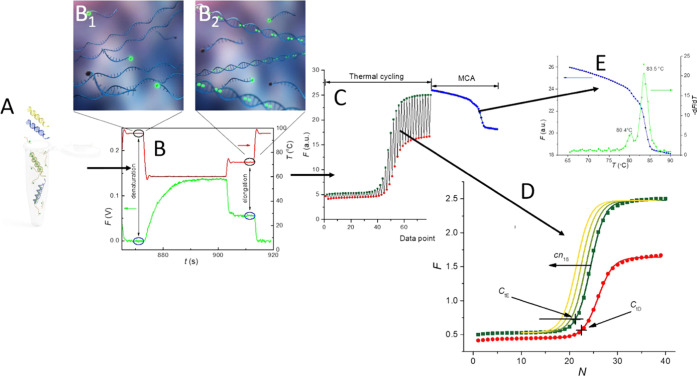
We assumed that we prepared PCR master mix using a fragment of chromosome 21 (Chr21) specific to a FAM probe with a fixed number of copies (cn21) and a fragment of chromosome 18 (Chr18) with a fixed number of copies (cn18) as the parameter (Figure 1A). We then performed the PCR in the presence of EvaGreen while collecting the fluorescent amplitude (F) at the end of the denaturation as well as the elongation in each PCR cycle (Figure 1B), followed by an MCA (Figure 1C). We split the collected data and formed two amplification curves, the denaturation and elongation curves (Figure 1D). The denaturation curve only consists of the FAM probe as all of the double-stranded DNA molecules melted into single-stranded DNAs, and EvaGreen’s contribution to F is negligible. Changing cn18 has no influence on the denaturation curve. The elongation curves represent EvaGreen intercalated with both Chr21 and Chr18 amplicons in combination with the FAM probe linked to Chr21. Changing cn18 causes the PCR curve shift. The MCA extracted at the end of the PCR proved the presence of both amplicons in the PCR product (Figure 1E).
Figure 1.
Principle of the proposed qPCR multiplexing method. (A) We prepared the PCR master mix containing two DNA templates, two sets of relevant forward and reverse primers, a FAM probe specific to one DNA template, and EvaGreen intercalating dye. (B) We performed the PCR and captured the F (green curve) during each cycle at the end of the denaturation and elongation steps, while the red curve represents the heater temperature. Contributions to the F at the denaturation step are only from the FAM probe (inset B1). F at the elongation step consists of both the FAM probe linked to Chr21 as well as the EvaGreen intercalating dye with both targets (inset B2). (C) All of the collected data during the PCR followed by the MCA were split (D) into two sets and plotted separately, which represents the denaturation amplification curve (red) and the elongation amplification curves in gradient colors from dark to light green for different cn18. (E) We also plotted an MCA as well as the negative derivation of F showing the presence of two different amplicons in the PCR master mix.
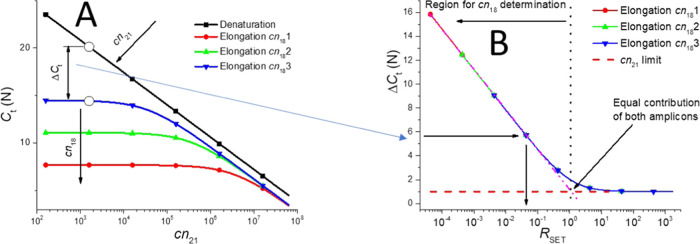
The threshold of the denaturation curve (CtD) of the mixture is only determined by the value of cn21 as the only FAM probe presence, and its concentration contributes to the F value at TD (Figure 2A, black line with squares)
| 1 |
where AD is an offset and BD is a PCR curve slope determined by the reaction efficiency of Chr21.
Figure 2.
Determination of the copy numbers of both targets based on calculation. (A) Values of CtD and CtE as a function of cn21 with cn18 as the parameter. The black lines with squares are three denaturation curves overlapping each other that have been calculated using eq 1. The blue curve with downward triangles, the green with upward triangles, and the red with circles are elongation curves calculated by eq 2. Each of them represents different values of cn18. (B) ΔCt values as a function of RSET with cn18 as the parameter that has been extracted from Figure 2A. All three curves overlap, which shows that it is the RSET value determining the ΔCt instead of an absolute value of cn18.
The threshold of the elongation curve (CtE) is affected by both cn18 and cn21, and we can create a plot of CtE as a function of cn21 with cn18 as the parameter
| 2 |
where AE is an offset and BE is the overall PCR efficiency (Figure 2A, blue, red, and green lines). Equation 2 can be split into two limiting situations, cn18≫ cn21
| 3 |
and cn18 ≪ cn21
| 4 |
where A1 and A2 are the offsets and B1 and B2 are the individual PCR efficiencies of Chr18 and Chr21 amplifications, respectively.
We then extracted the differences from the corresponding Ct values between the elongation and denaturation curves (ΔCt) and plotted them as a function of the ratio between the cn of the amplicon with and without a link to FAM (RSET) with log cn18 as the parameter. Here, it would be RSET = cn18/cn21. It shows that the ΔCt value is a function of RSET independent of the cn absolute values (Figure 2B). The ΔCt can be expressed as
| 5 |
where A is intercept with the Y-axis and B is a slope, respectively. We can then, with eq 4, simplify the relationship as
| 6 |
to show its independence on an absolute value of cn21 and cn18 (Figure 2B). Parameter B defines the precision of RSET and thus cn18 determination.
Thus, once we form the PCR standard curve, as shown in Figure 2A, we can determine the cn21 of an unknown sample from the denaturation curve (marked by a black arrow). Then, we calculate the ΔCt value and determine the cn18 based on the extracted RSET values from Figure 2B. Note that the cn18 can only be determined from the left part of the graph starting at the point where both amplicons’ contribution is equal; otherwise, cn21 dominates the change of CtE.
3. Results and Discussion
3.1. Preliminary Experiments
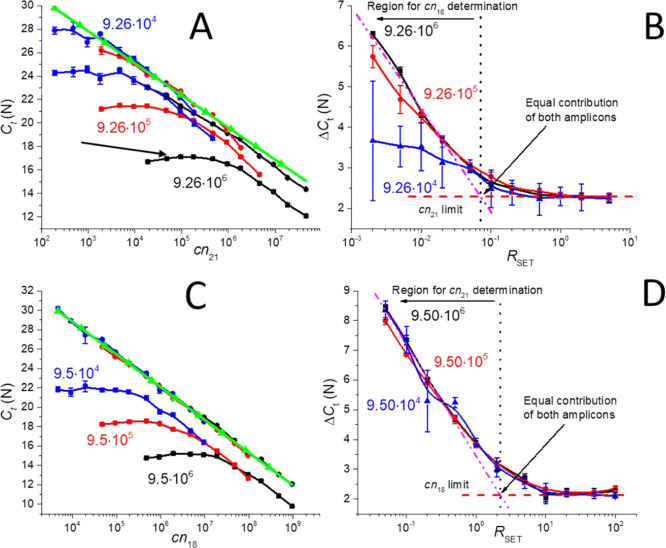
We first performed the PCR and extracted elongation and denaturation standard curves of Chr21 with cn21 from 9.5 × 101 to 9.5 × 106 with and without a FAM probe to determine the Ct shift from the elongation to the denaturation as well as the PCR efficiency for both curves. Each experiment was repeated four times to suppress any random error. Then, the same experiment was conducted using Chr18 from cn18 between 9.26 × 102 and 9.26 × 106 with a VIC probe (Supporting Information, Section S1). We used a VIC specific to Chr18 as a reference to the single-channel multiplexing method. We found that the PCR efficiency was very similar for all combinations, and its value is (−3.46 ± 0.03) N ×(log cn)−1 (mean ± fitting error from four measurements), which demonstrated the correct experiment setting as well as the minimal pipetting error (Supporting Information, Section S1).
3.2. Multiplexing Using the Target cn18 Constant and the cn21 Variable with FAM Specific to Chr21
We then mixed Chr21 with a FAM probe and Chr18 with the ratio RSET with three different cn18 values of 9.26 × 104, 9.26 × 105, and 9.26 × 106 and a cn21 variable (Supporting Information, Section S2). We performed the PCR and extracted the denaturation and elongation standard curves (Figure 3A). The denaturation curves (black, red, and blue line with the squares) were solely determined by cn21, but they were nevertheless influenced by the abundance of Chr18, as exhibited by the curve deviation from the straight line.
Figure 3.
Ct values extracted from the PCR with EvaGreen intercalating dye and the FAM probe. (A) PCR standard curves of the variable cn21 values with three different cn18 values as the parameter with the denaturation curves marked with circles and the elongation with squares. The cn21 values could be determined in an entire range from the denaturation standard curves. The arrow pointed to the curved region where minor Chr21 clearly influences Chr18 amplification. The green line marked with triangles was formed by the curve fitting of an average value from all three denaturation curves. (B) Values of ΔCt plotted as a function of RSET showing the importance of the RSET value and not the individual cn21 or cn18. (C) Equivalent of (A) with cn18 as the variable and cn21 as the parameter with elongation curves marked by squares and the denaturation by circles. (D) Equivalent of (B) with cn18 as the variable and cn21 as a parameter.
The elongation curves (black, blue, and red with circles) show two distinguished regions, one determined by cn18 and the other by cn21, as predicted by eqs 2–6, and their graphical representation is shown in Figure 2A. We performed the curve fitting (green line in Figure 3A) of the average of all three denaturation curves using eq 1 and obtained AD and BD as (36.03 ± 0.56) N and (−2.74 ± 0.11) N ×(log cn21)−1 (both mean ± fitting error), respectively.
The saturation at the elongation curve determined by the cn18 value exhibited a curved shape instead of a flat line parallel to the X-axis (pointed by an arrow in Figure 3A), a clear sign of cn21 influence. With a lower value of cn21, the CtE was also lowered, demonstrating that the influence of Chr21 was diminishing. All of the PCR experiments were accompanied with MCAs, and there was a trend showing the transition from the region dominated by Chr21 to a region dominated by Chr18 with an increased number of amplicons of Chr18, while the product of Chr21 dominated regardless of cn21 (Supporting Information, Section S3).
We then calculated the values of ΔCt and plotted them as a function of RSET (Figure 3B). We found that each amplicon had different contributions to the CtE value as the point of the equal contribution of both the amplicons was not for RSET = 1. We then modified the original elongation curves per eq 2 as
| 7 |
where the critical ratio (RC) was a coefficient reflecting the different contributions of each amplicon to the CtE value. Then, eq 6 would convert to
| 8 |
We extracted the slope B from the curves as (−2.40 ± 0.03) N × (log RSET)−1 (mean ± fitting error), thereby defining the sensitivity of RSET determination as well as the RC value of ≈0.07 RSET. The low RC value in comparison with RSET showed strong dominance of Chr21 contribution to the CtE values. As predicted by eq 7, the ΔCt was a function of RSET instead of an absolute number of either cn21 or cn18.
We determined the absolute value of cn21 in the entire range from the denaturation curve (curves in Figure 3A marked with circles) with a precision of (−2.74 ± 0.11) N × (log cn21)−1 (mean ± fitting error). The cn18 value was identified by extracting the RSET value from Figure 3B and multiplying it with cn21.
We found that the precision of the cn18 determination is (−2.40 ± 0.03) N × (log RSET)−1 (mean ± fitting error) for RSET values smaller than RC ≈ 0.07. The lower values of cn18 deviated from the described behavior because the presence of Chr21 probably inhibited the PCR of Chr18, as shown by the MCAs (Supporting Information, Section S3).
3.3. Multiplexing Using a Target cn21 Constant and a cn18 Variable with FAM Specific to Chr18
We repeated the same measurement with a FAM probe specific to Chr18 and Chr21 with cn21 fixed at the three different values with the cn18 variable and plotted the results into Figure 3C (Supporting Information, Section S4). Here, the RSET was cn21/cn18. We performed the curve fitting as it was done in the previous section and found that the transition RC changed from 0.07 to 2.26, which showed the dominance of the Chr21 regardless of the FAM probe specificity. We also calculated the ΔCt and plotted it as a function of RSET (Figure 3D). The results verified our original hypothesis that ΔCt was determined by the value of RSET instead of the absolute values of cn18 and cn21.
The absolute value of cn18 couldbe determined in an entire range from the denaturation curve (curves in Figure 3C marked with squares) with a precision of (−3.38 ± 0.03) N × (log cn21)−1 (mean ± fitting error). The cn21 value could be extracted from Figure 3D using the RSET values and a value of cn18 with a precision of (−3.48 ± 0.26) N × (log RSET)−1 (mean ± fitting error) for the RSET values smaller than RC ≈ 2.26.
3.4. Multiplexing with Target cn21 Having a Chr21-Specific FAM Probe and cn18 with a Chr18-Specific VIC Probe
We performed a PCR using Chr21 and Chr18 specific to FAM and VIC probes, respectively, with either a cn18 constant and a cn21 variable or vice versa (Supporting Information, Section S5).
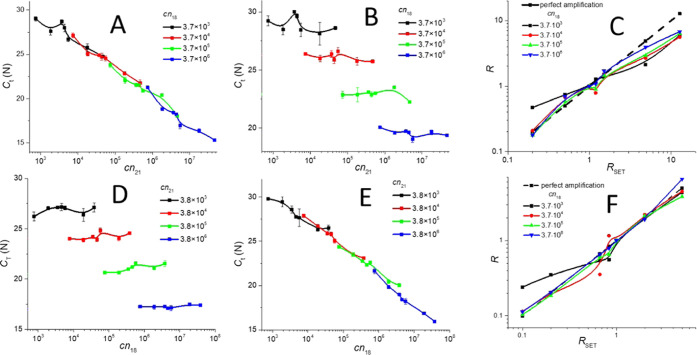
Each experiment was conducted five times. We then extracted the Ct values using eq 11 and plotted Ct as a function of cn21 from the FAM and VIC channels, as shown in Figure 4A,B, respectively.
Figure 4.
Ct values extracted from the PCR with FAM and VIC probes. (A) FAM channel with a cn21 variable with cn18 as the parameter showing the problems with the dual-channel amplification as the standard curves with different values of cn18 should be collinear and form a single line regardless of the cn18 value. (B) Same PCR mixtures using a VIC channel also showed the influence of cn21 on cn18 as the results should be independent of cn21 since it was Chr18 with the VIC probe. (C) Implementation of the 2–ΔΔCt method showing its shortcoming with the noncompensated data for fluorescence spillover. (D–F) Similar results for a mixture with the cn18 value variable and the cn21 as the parameter.
We calculated the average errors in Ct determination (Table 1), and they were rather small—between 2.4 and 1.5% for the lowest and highest cn18. The determination of the individual Ct values varied much more, especially for the lowest value of cn18 (black line in Figure 4B). We used the 2–ΔΔCt method23 with the mixture having RSET = 1 as the calibrator. We plotted the extracted 2–ΔΔCt values as a function of RSET (Figure 4C). The results showed that even the widely accepted method of 2–ΔΔCt had a problem with determining R precisely when applied to the data without any compensation for fluorescence spillover.33
Table 1. Results from Two Fluorescence Channels Showing Ct Determination Errors.
| cn | intercept (N) | slope (log cn) | mean value (N) | |
|---|---|---|---|---|
| cn21 variable | ||||
| FAM channel | 38.60 ± 0.43 | –3.05 ± 0.08 | ||
| VIC channel | 3.7 × 103 | 29.03 ± 0.70 | ||
| 3.7 × 104 | 26.13 ± 0.33 | |||
| 3.7 × 105 | 22.92 ± 0.35 | |||
| 3.7 × 106 | 19.55 ± 0.30 | |||
| cn18 variable | ||||
| FAM channel | 3.8 × 103 | 26.89 ± 0.37 | ||
| 3.8 × 104 | 24.16 ± 0.35 | |||
| 3.8 × 105 | 20.99 ± 0.42 | |||
| 3.8 × 106 | 17.22 ± 0.16 | |||
| VIC channel | 39.59 ± 0.39 | –3.07 ± 0.07 | ||
We repeated the same experiment with the cn18 variable and cn21 as the parameter and extracted the results in the same fashion as for the first experiment (Figure 4D–F), and the extracted data and errors were shown in Table 1.
3.5. Comparison of the Single-Channel Duplexing Method with the Method Employing Two Probes with Different Fluorophores
We performed the single-channel duplexing qPCR method using Chr21 with a FAM-specific probe and Chr18 both in the presence of EvaGreen intercalating dye. We also alternated the mixture composition having a FAM-specific probe to the Chr18 sequence instead of Chr21. We found that we could determine the value of the cn of the amplicon with the FAM probe in the entire range of the values. The second amplicon, cn, could only be determined once the RSET value was smaller than the RC, which could vary for each individual pair of targets. In comparison, we performed standard multiplexing using FAM and VIC probes. The multichannel technique exhibited large error especially for the lower values of cns, where the Ct value varied in a range between 27 and 30 (black line in Figure 4B). We then processed the extracted data using the 2–ΔΔCt method23 without performing channel overspilling compensation. It showed that even these widely accepted methods did not produce precise data (Figure 4C,F). Single-channel multiplexing produced more precise data, but it could determine both amplicons only for RSET < RC in half of the total range of cn.
4. Conclusions
We proposed and verified a method of quantitative polymerase chain reaction (qPCR) duplexing using a single fluorescein isothiocyanate (FITC) fluorescent channel with a commercially available qPCR system with a 96-well platform. We amplified the following two sequences: one from chromosome 21 (Chr21) and one from chromosome 18 (Chr18) in the presence of the EvaGreen intercalating dye. We added a 6-carboxyfluorescein (FAM) probe specific to either Chr21 or to Chr18 and extracted the PCR curves after the denaturation and elongation steps, and then, we plotted the PCR standard curves, one for denaturation and the second one for elongation. We were able to determine the copy number (cn) of an amplicon with a FAM-specific probe in the entire range of the cn values from the denaturation standard curve with precision values of (−2.74 ± 0.11) N × (log cn21)−1 and (−3.38 ± 0.03) N × (log cn18)−1 (both mean ± fitting error) for Chr21 and Chr18, respectively.
The cn value of the other amplicon was extracted from the difference of the critical thresholds between the denaturation and elongation curves with precision values of (−2.40 ± 0.03) N × (log RSET)−1 and (−3.48 ± 0.26) N × (log RSET)−1 (both mean ± fitting error) for Chr21 and Chr18, respectively.
This proposed method has better precision than the standard technique of working with differently labeled probes, which eliminates the problem of overspilling between fluorescent channels. However, this method is not capable of operating in the entire range of possible ratios of mixture components as the RSET value has to be greater than the RC to determine the cn of the target without the FAM probe. Nevertheless, if the determination of the value with RSET < RC is required, it is possible to change the system and use a FAM probe that is specific to the second target. The combination of the probe-based and intercalating dye-based methods brings the potential drawbacks of both such as the high cost for probe-based qPCR and the low specificity for intercalating dye-based qPCR. However, it is not necessary to have two expensive probes and a complex fluorescence system; thus, the proposed method can lower the cost as only one probe is applied. Additionally, it eliminates the fluorescence overspilling, which exists in probe-based qPCR multiplexing. Moreover, the melting curve analysis conducted immediately once the thermal cycling is completed will determine the amplicon specificity.
This proposed duplexing method using a single FITC fluorescent channel can be implemented for a wide variety of examinations where mixture analysis is desired once either multifluorescence channel is not a viable option or to lower the test cost. This qPCR duplexing thus opens new horizons for practical applications, providing an original, easily applicable technical solution.
5. Material and Methods
5.1. DNA Sequences and PCR Master Mix
We used the synthesized target fragments of Chr21 and Chr18 as well as primers and probes for both, as shown in Table 2. The chosen amplicons, designed primers, and the lock nucleic acids (LNA) probes were used beforehand for the noninvasive prenatal diagnostics using dPCR, and they were optimized to perform in multiplex reactions.34 We then prepared different types of PCR master mixes based on the following designed experiments by adjusting the volume of probes for both targets (Supporting Information, Section S6). The final volume of the PCR master mix was ≈10 μL by adding sterilized deionized H2O.
Table 2. Information of Target Sequences and the Corresponding Primers and Probes.
| target gene | gene locationa | primer type | primer sequence (5′–3′) | amplicon size (bp) | amplicon TM (°C) |
|---|---|---|---|---|---|
| Chr21 | 14099141-14099218 | forward | ctaggagactgtccctgagctt | 78 | 83.5 |
| reverse | agggggaacatagaggcttg | ||||
| LNA probe | FAM-ccctgcctct-BHQ1 | ||||
| Chr18 | 1248496-1248575 | forward | ccatctccataacccaaatacc | 80 | 80.4 |
| reverse | ccttgcaaacctcatgttga | ||||
| LNA probe | VIC-cccacctcca-BHQ1 | ||||
| FAM-cccacctcca-BHQ1 |
Gene location according to the human genome assembly version hg19/Genome Reference Consortium Human Build 37.
5.2. qPCR Protocol
We performed the qPCR with the protocol set in the following way: a hot start at 95 °C for 30 s to activate the polymerase followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 52 °C for 30 s, and elongation at 72 °C for 10 s using a commercial 96-well real-time PCR system collecting two F values during each PCR cycle at the end of the elongation and denaturation steps. We then conducted an MCA from 65 to 95 °C with a ramping rate set to 0.5 K•s–1.
5.3. Data Processing
The collected data were processed using a MATLAB script by first splitting them into two blocks. The first 80 points contained the PCR amplification curves, and the rest represented the MCA. During each cycle, we collected two data points corresponding to the two adjacent sample points, F values were collected at the end of the denaturation and elongation steps. Then, we split the amplification curve data according to their sequence. The data whose serial number was odd (even) were selected to form the denaturation (elongation) amplification curve. The elongation curve was the same as the conventional PCR amplification curve. The denaturation curve was different since the PCR cycle started with denaturation and ends with elongation. We skipped the first point of the denaturation curve as it had no meaning since no PCR amplification had occurred by that point in time. As a result, the denaturation curve had only 39 points, while the elongation had typical 40 points resulting from 40 cycles. After that, the amplification curves were processed independently.
5.4. Data Normalization
We normalized the amplification curves according to the following formula
| 9 |
where FN is the normalized F amplitude value and Fmax is the maximal amplitude value of the F data. Fmin is manually selected for each curve due to the unpredictable fluctuation in the F value of the first few data. If the variation range (Fmax – Fmin) of a curve is judged to be lower than 50 000 arbitrary units (a.u.), this curve is regarded as a nonamplification curve, and the FN value is set to 0 a.u.
5.5. Ct Value Extraction
We performed the curve fitting using a Boltzmann function similar to the methods published earlier35,36
| 10 |
where FN(i) is the normalized amplitude of the F value of cycle number i, i0 is an inflexion point of the Boltzmann curve, and dF determines the Boltzmann curve slope at F(i0) by  . In previous work,36 the author defined Ct as a value of N at a fluorescence
inflection point. Here, we calculated
the Ct value as an N value
for a 10% increase of the F value above its baseline
. In previous work,36 the author defined Ct as a value of N at a fluorescence
inflection point. Here, we calculated
the Ct value as an N value
for a 10% increase of the F value above its baseline
| 11 |
This method is similar to the one conventionally used for calculating the Ct defining the Ct value as a cycle number for 10% of F increased above the baseline. The Boltzmann function-based Ct value determination was then implemented using the MATLAB script.
Acknowledgments
H.Z., Z.Y., X.W., and P.N. were supported by the Ministry of Science and Technology of the P.R. China [grant no. 2018YFE0109000]. M.K. and S.L. were supported by the Ministry of Education, Youth, and Sports of the Czech Republic [grant no. LTACH19005] and by a grant from the Ministry of Health of the Czech Republic [grant no. RVO-VFN 64165].
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02971.
Standard curves of single genes, amplicon ratios of Chr21 and Chr18 with Chr21 variable, MCAs between Chr21 with the FAM probe and Chr18 having cn18 with three different values and cn21 variable, amplicon ratios of Chr21 and Chr18 with Chr18 variable and MCA after PCR, amplicon ratios of Chr21 and Chr18, and PCR master mix (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Mullis K.; Faloona F.; Scharf S.; Saiki R.; Horn G.; Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. Quant. Biol. 1986, 263–273. 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Zhang H.; Xu Y.; Laššáková S.; Korabečná M.; Neužil P. PCR past, present and future. BioTechniques 2020, 69, 317–325. 10.2144/btn-2020-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B.; Zhang C.; Xing D. Segmented continuous-flow multiplex polymerase chain reaction microfluidics for high-throughput and rapid foodborne pathogen detection. Anal. Chim. Acta 2014, 826, 51–60. 10.1016/j.aca.2014.04.017. [DOI] [PubMed] [Google Scholar]
- Kubista M.; Andrade J. M.; Bengtsson M.; Forootan A.; Jonák J.; Lind K.; Sindelka R.; Sjöback R.; Sjögreen B.; Strömbom L.; Ståhlberg A.; Zoric N. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 27, 95–125. 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Vogelstein B.; Kinzler K. W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9236–9241. 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Fohlerová Z.; Pekárek J.; Basova E.; Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoute L. C.; Loppnow G. R. Characterization of the binding interactions between EvaGreen dye and dsDNA. Phys. Chem. Chem. Phys. 2018, 20, 4772–4780. 10.1039/C7CP06058K. [DOI] [PubMed] [Google Scholar]
- Lehmusvuori A.; Karhunen U.; Tapio A.-H.; Lamminmäki U.; Soukka T. High-performance closed-tube PCR based on switchable luminescence probes. Anal. Chim. Acta 2012, 731, 88–92. 10.1016/j.aca.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Zimmermann B.; Holzgreve W.; Wenzel F.; Hahn S. Novel real-time quantitative PCR test for trisomy 21. Clin. Chem. 2002, 48, 362–363. 10.1093/clinchem/48.2.362. [DOI] [PubMed] [Google Scholar]
- Farzan V. M.; Kvach M. V.; Aparin I. O.; Kireev D. E.; Prikazchikova T. A.; Ustinov A. V.; Shmanai V. V.; Shipulin G. A.; Korshun V. A.; Zatsepin T. S. Novel homo Yin-Yang probes improve sensitivity in RT-qPCR detection of low copy HIV RNA. Talanta 2019, 194, 226–232. 10.1016/j.talanta.2018.10.043. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. S.; Gibbs R. A.; Rainer J. E.; Nguyen P. N.; Thomas C. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988, 16, 11141–11156. 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaňová M.; Zhang H.; Zhu H.; Korabečná M.; Neužil P. Multiplexed digital polymerase chain reaction as a powerful diagnostic tool. Biosens. Bioelectron. 2021, 181, 113155 10.1016/j.bios.2021.113155. [DOI] [PubMed] [Google Scholar]
- DuVall J. A.; Le Roux D.; Thompson B. L.; Birch C.; Nelson D. A.; Li J.; Mills D. L.; Tsuei A.-c.; Ensenberger M. G.; Sprecher C.; Storts D. R.; Root B. E.; Landers J. P. Rapid multiplex DNA amplification on an inexpensive microdevice for human identification via short tandem repeat analysis. Anal. Chim. Acta 2017, 980, 41–49. 10.1016/j.aca.2017.04.051. [DOI] [PubMed] [Google Scholar]
- Li Y.; Li Y.; Zheng B.; Qu L.; Li C. Determination of foodborne pathogenic bacteria by multiplex PCR-microchip capillary electrophoresis with genetic algorithm-support vector regression optimization. Anal. Chim. Acta 2009, 643, 100–107. 10.1016/j.aca.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Arnold L.; Alexiadis V.; watanaskul T.; Zarrabi V.; Poole J.; Singh V. Clinical validation of qPCR Target Selector assays using highly specific switch-blockers for rare mutation detection. J. Clin. Pathol. 2020, 73, 648–655. 10.1136/jclinpath-2019-206381. [DOI] [PubMed] [Google Scholar]
- Emaus M. N.; Anderson J. L. Allelic discrimination between circulating tumor DNA fragments enabled by a multiplex-qPCR assay containing DNA-enriched magnetic ionic liquids. Anal. Chim. Acta 2020, 1124, 184–193. 10.1016/j.aca.2020.04.078. [DOI] [PubMed] [Google Scholar]
- Muenchhoff M.; Mairhofer H.; Nitschko H.; Grzimek-Koschewa N.; Hoffmann D.; Berger A.; Rabenau H.; Widera M.; Ackermann N.; Konrad R.; et al. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Eurosurveillance 2020, 25, 2001057 10.2807/1560-7917.ES.2020.25.24.2001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottenet G.; Blancpain C.; Sonnard V.; Chuah P. F. Two FAST multiplex real-time PCR reactions to assess the presence of genetically modified organisms in food. Food Chem. 2019, 274, 760–765. 10.1016/j.foodchem.2018.09.050. [DOI] [PubMed] [Google Scholar]
- Pazourkova E.; Zednikova I.; Korabecna M.; Kralova J.; Pisacka M.; Novotna M.; Calda P.; Horinek A.. Optimization of diagnostic strategy for non-invasive cell-free foetal RHD determination from maternal plasma Vox Sang. 2021, 13099. 10.1111/vox.13099 [DOI] [PubMed]
- Suwannakhon N.; Pangeson T.; Seeratanachot T.; Mahingsa K.; Pingyod A.; Bumrungpakdee W.; Sanguansermsri T. Noninvasive prenatal screening test for compound heterozygous beta thalassemia using an amplification refractory mutation system real-time polymerase chain reaction technique. Hematol. Rep. 2019, 11, 8124 10.4081/hr.2019.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J. F.; The dMIQE Group; Whale A. S.; et al. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- Bustin S. A.; Benes V.; Garson J. A.; Hellemans J.; Huggett J.; Kubista M.; Mueller R.; Nolan T.; Pfaffl M. W.; Shipley G. L.; et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Gubala A. J. Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 2006, 65, 278–293. 10.1016/j.mimet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Waggoner J. J.; Abeynayake J.; Sahoo M. K.; Gresh L.; Tellez Y.; Gonzalez K.; Ballesteros G.; Pierro A. M.; Gaibani P.; Guo F. P.; et al. Single-reaction, multiplex, real-time rt-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Neglected Trop. Dis. 2013, 7, e2116 10.1371/journal.pntd.0002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Li H.; Zhu H.; Pekárek J.; Podešva P.; Chang H.; Neužil P. Revealing the secrets of PCR. Sens. Actuators, B 2019, 298, 126924 10.1016/j.snb.2019.126924. [DOI] [Google Scholar]
- Ahrberg C. D.; Manz A.; Neuzil P. Single fluorescence channel-based multiplex detection of avian influenza virus by quantitative PCR with intercalating dye. Sci. Rep. 2015, 5, 11479 10.1038/srep11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Zhang H.; Ni S.; Korabečná M.; Yobas L.; Neuzil P. The vision of point-of-care PCR tests for the COVID-19 pandemic and beyond. TrAC, Trends Anal. Chem. 2020, 130, 115984 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrberg C. D.; Neužil P. Doubling throughput of a real-time PCR. Sci. Rep. 2015, 5, 12595 10.1038/srep12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzano J.; Moniri A.; Malpartida-Cardenas K.; Dronavalli J.; Davies F.; Holmes A.; Georgiou P. Simultaneous single-channel multiplexing and quantification of carbapenem-resistant genes using multidimensional standard curves. Anal. Chem. 2019, 91, 2013–2020. 10.1021/acs.analchem.8b04412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniri A.; Rodriguez-Manzano J.; Malpartida-Cardenas K.; Yu L.-S.; Didelot X.; Holmes A.; Georgiou P. Framework for DNA quantification and outlier detection using multidimensional standard curves. Anal. Chem. 2019, 91, 7426–7434. 10.1021/acs.analchem.9b01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniri A.; Miglietta L.; Malpartida-Cardenas K.; Pennisi I.; Cacho-Soblechero M.; Moser N.; Holmes A.; Georgiou P.; Rodriguez-Manzano J. Amplification curve analysis: data-driven multiplexing using real-time digital PCR. Anal. Chem. 2020, 92, 13134–13143. 10.1021/acs.analchem.0c02253. [DOI] [PubMed] [Google Scholar]
- Wang C.; Gao D.; Vaglenov A.; Kaltenboeck B. One-step real-time duplex reverse transcription PCRs simultaneously quantify analyte and housekeeping gene mRNAs. Biotechniques 2004, 36, 508–519. 10.2144/04363RN06. [DOI] [PubMed] [Google Scholar]
- Tan C.; Chen X.; Wang F.; Wang D.; Cao Z.; Zhu X.; Lu C.; Yang W.; Gao N.; Gao H. J. A.; et al. A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst 2019, 144, 2239–2247. 10.1039/C8AN02018C. [DOI] [PubMed] [Google Scholar]
- Liu W.; Saint D. A. Validation of a quantitative method for real time PCR kinetics. Biochem. Biophys. Res. Commun. 2002, 294, 347–353. 10.1016/S0006-291X(02)00478-3. [DOI] [PubMed] [Google Scholar]
- Rutledge R. Sigmoidal curve-fitting redefines quantitative real-time PCR with the prospective of developing automated high-throughput applications. Nucleic Acids Res. 2004, 32, e178 10.1093/nar/gnh177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.