The identity of the intestinal stem niche has long remained elusive. Early evidence suggested that Paneth cells, a secretory cell type of the intestinal epithelium, functioned as a key component of the intestinal stem cell niche.1 Situated in the crypt base, Paneth cells intercalate between intestinal stem cells and express multiple signaling molecules that could promote stem cell activity. However, subsequent studies have shown that Paneth cells are dispensable for intestinal stem cell maintenance, proliferation, and function,2,3 thereby raising doubt over the cellular identity of the intestinal stem cell niche. In recent years, our laboratory, along with others, has shown the important role played by mesenchymal cell populations in maintaining intestinal stem cell function.4, 5, 6, 7, 8 In particular, we identified FoxL1-expressing (FoxL1+) subepithelial telocytes as a critical component of the intestinal stem cell niche and the essential source of Wnt signaling, without which intestinal stem cells cannot survive.4,7,9
To study FoxL1+ telocytes and understand their contribution to gastrointestinal health, we must rely on effective mouse models that enable genetic tracing and cell type–specific gene ablation. Our laboratory previously generated a transgenic FoxL1CreER mouse line that uses the Cre-loxP system to induce reporter expression and conditional gene ablation specifically within FoxL1+ telocytes. However, this mouse model relied on random integration into the mouse genome and therefore is subject to transgene silencing. In addition, this FoxL1CreER mouse line does not provide permanent fluorescent labeling of FoxL1+ telocytes; fluorescent labeling of FoxL1+ telocytes therefore can be accomplished only by crossing this mouse to a loxP-dependent reporter line and treating the animals with tamoxifen to activate the reporter allele.
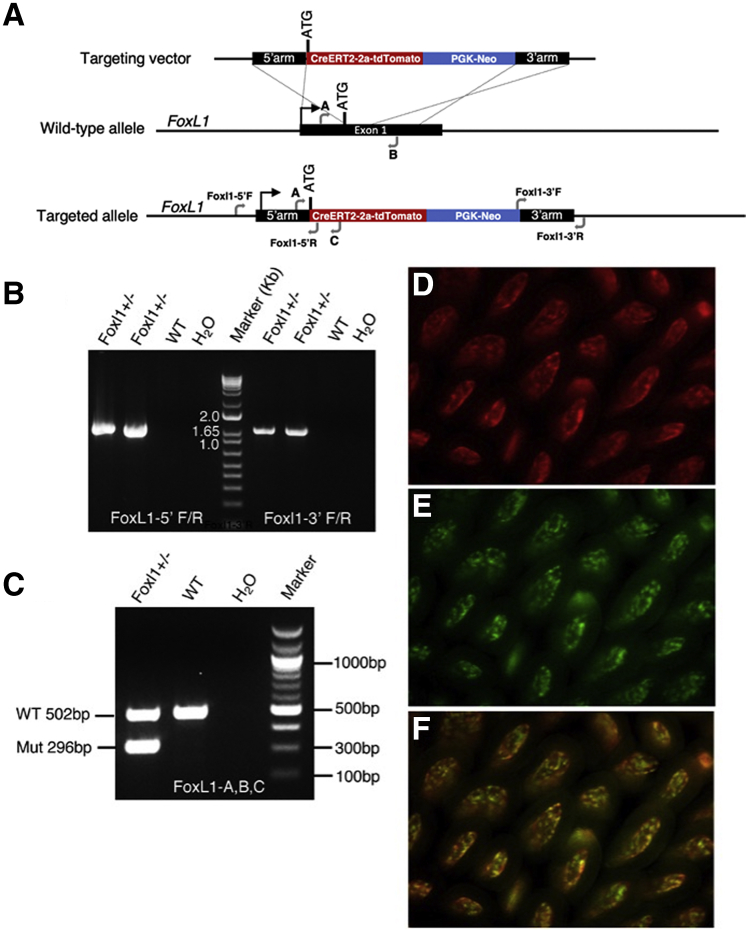
To overcome these limitations, we generated a novel FoxL1-CreERT2-2A-tdTomato (FoxL1CreER-tdTom) mouse line in which the endogenous FoxL1 promoter drives expression of both a tdTomato fluorescent reporter and the tamoxifen-dependent Cre (Causes recombination) recombinase. An ATG-less CreERT2-2A-tdTomato cassette harboring a neomycin-resistance gene was targeted into the FoxL1 locus directly after the endogenous translational start site of this single-exon gene (Figure 1A and B). Targeting was performed in mouse V6.5 embryonic stem cells, and a properly targeted clone was injected into blastocysts to establish the FoxL1CreER-tdTom mouse strain. Subsequent genotyping of the strain was performed with a 3-primer polymerase chain reaction enabling simultaneous detection of both the targeted and wild-type alleles (Figure 1C). This strategy retains all endogenous FoxL1 promoter and 5’ untranslated regions and thus should provide high concordance with endogenous FoxL1 expression.
Figure 1.
Targeting a CreER-2A-tdTomato reporter to the endogenous FoxL1 locus. (A) Targeting vector for inserting a CreER-2A-tdTomato cassette at the translational start site of the endogenous FoxL1 locus. Primer locations for validating insertion at the 5’ and 3’ insertion sites are indicated, as are primers used for genotyping of FoxL1CreER-2A-Tomato mice (A, B, and C). (B) Polymerase chain reaction assays spanning the 5’ and 3’ arms of homology validate proper gene targeting. (C) Genotyping assay for the FoxL1CreER-2A-Tomato mouse strain using a 3-primer (A, B, and C) polymerase chain reaction enables resolution of wild-type (WT) and targeted (Mut) FoxL1 alleles. (D) Endogenous tdTomato expression in the subepithelial telocyte plexus in the jejunum of FoxL1CreER-2A-Tomato mice. (E) Sun1GFP expression in a FoxL1CreER-tdTomRosa26Sun1GFP/+ mouse after 5 doses of tamoxifen . (F) Overlap of endogenous tdTomato and Sun1GFP expression. PGK, phosphoglycerate kinase I.
To validate Cre activity, FoxL1CreER-tdTom mice were crossed to the Rosa26LSL-SUN1-GFP reporter mouse line,10 in which the nuclear membrane protein, SUN1, is labeled with 2 tandem copies of superfolder (sf) green fluorescent protein (GFP). The Sun1-sfGFP-myc cassette is expressed from the ubiquitously active Rosa26 locus and is preceded by a CAG promoter and a loxP-3x polyA-loxP cassette, which blocks downstream transcription. Cre-expressing cells remove this transcriptional stop cassette to allow expression of Sun1-sfGFP-myc. Whole-mount direct fluorescence imaging of FoxL1CreER-tdTomRosa26Sun1GFP/+ mice showed endogenous tdTomato expression driven by the FoxL1 promoter, which localized to the subepithelial plexus of the jejunum and overlapped extensively with Sun1GFP reporter expression (Figure 1D–F).
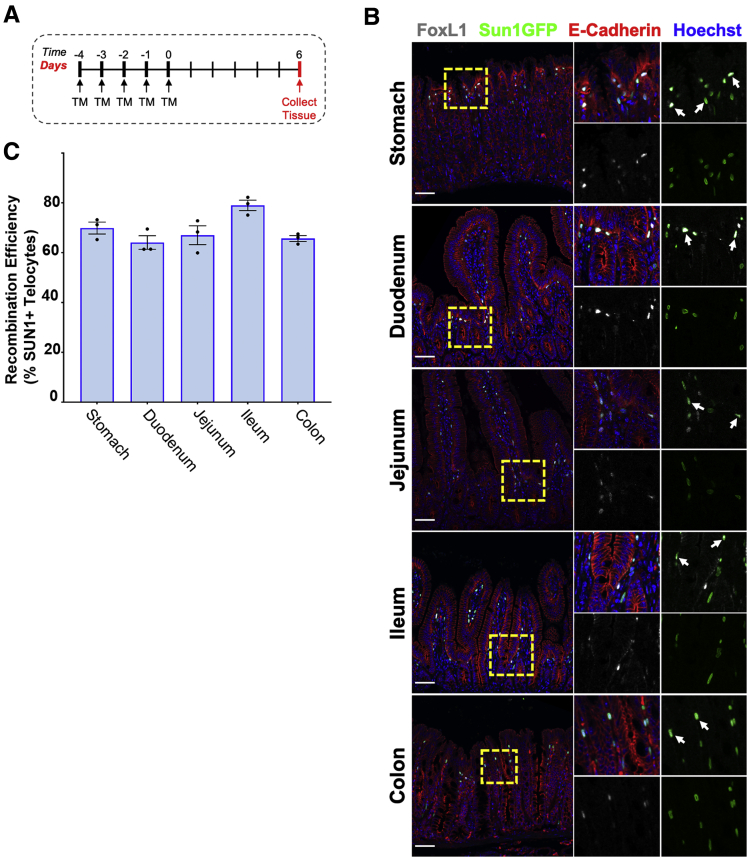
Using FoxL1CreER-tdTomRosa26Sun1GFP/+ mice, we next assessed recombination efficiency in tissues collected along the proximal-to-distal axis of the gastrointestinal tract. To induce Cre activity, adult FoxL1CreER-tdTomRosa26Sun1GFP/+ mice were treated once daily with tamoxifen for 5 consecutive days; tissues were analyzed 6 days after the final tamoxifen dose (Figure 2A). Segments of the stomach, duodenum, jejunum, ileum, and colon were surveyed for FoxL1 and Sun1GFP protein expression by immunohistochemistry (Figure 2B). Recombination efficiency was calculated as the number of FoxL1+Sun1GFP+ nuclei normalized to the total number of FoxL1+ cells. Recombination efficiency varied along the gastrointestinal tract, increasing slightly from the duodenum (64.1%) to the ileum (79.0%), with a decrease in efficiency in the colon (65.7%) (Figure 2C). The highest recombination efficiency was observed in the ileum, while the lowest was observed in the colon. Recombination efficiency in the stomach was 69.0%. In other studies using 3 consecutive days of tamoxifen treatment, we observed similar recombination frequencies ranging from 66.5% to 73.7%, depending on tissue (data not shown).
Figure 2.
The FoxL1 promoter drives Cre recombinase and reporter expression in tissues of the gastrointestinal tract. (A) Timeline of tamoxifen injection and tissue-harvesting scheme. (B) Immunohistochemistry of gastrointestinal regions from FoxL1CreER-tdTomRosa26Sun1GFP/+ mice (n = 3). Cre activity induces Sun1GFP expression in the nuclear envelopes of subepithelial mesenchymal cells. Boxed regions are shown at higher magnification (right). Co-localization of FoxL1 protein and Sun1GFP is observed (arrows). Scale bars: 50 μm. (C) Recombination efficiency was quantified as the percentage of Sun1GFP+FoxL1+ cells normalized by the total number of FoxL1+ cells. Recombination efficiency increased slightly along the proximal-to-distal axis of the small intestine, with the lowest recombination efficiency observed in the duodenum and the highest observed in the ileum. Error bars represent means with SEM (n = 3). TM, tamoxifen.
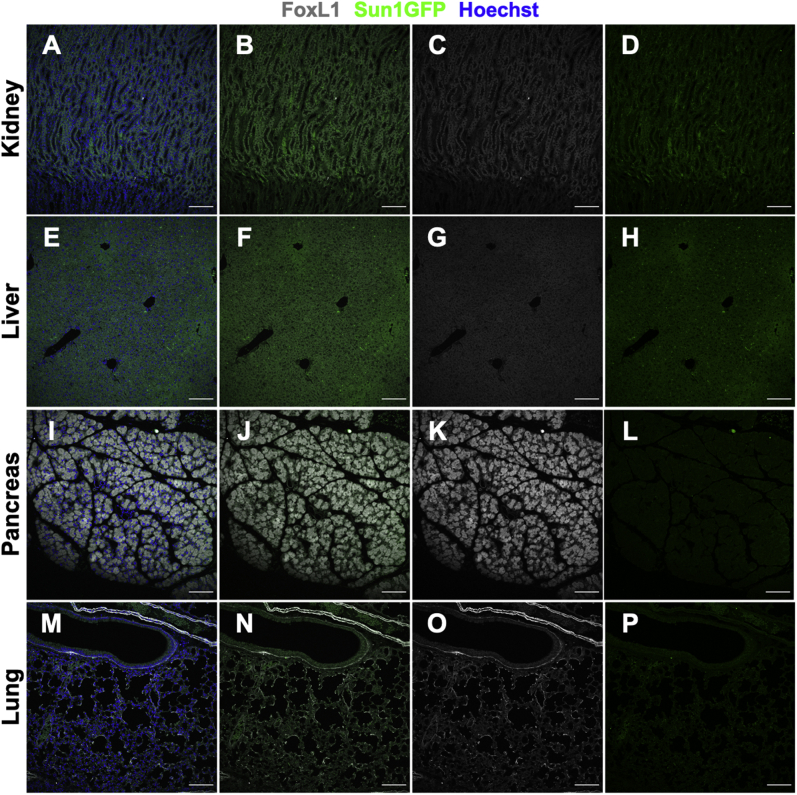
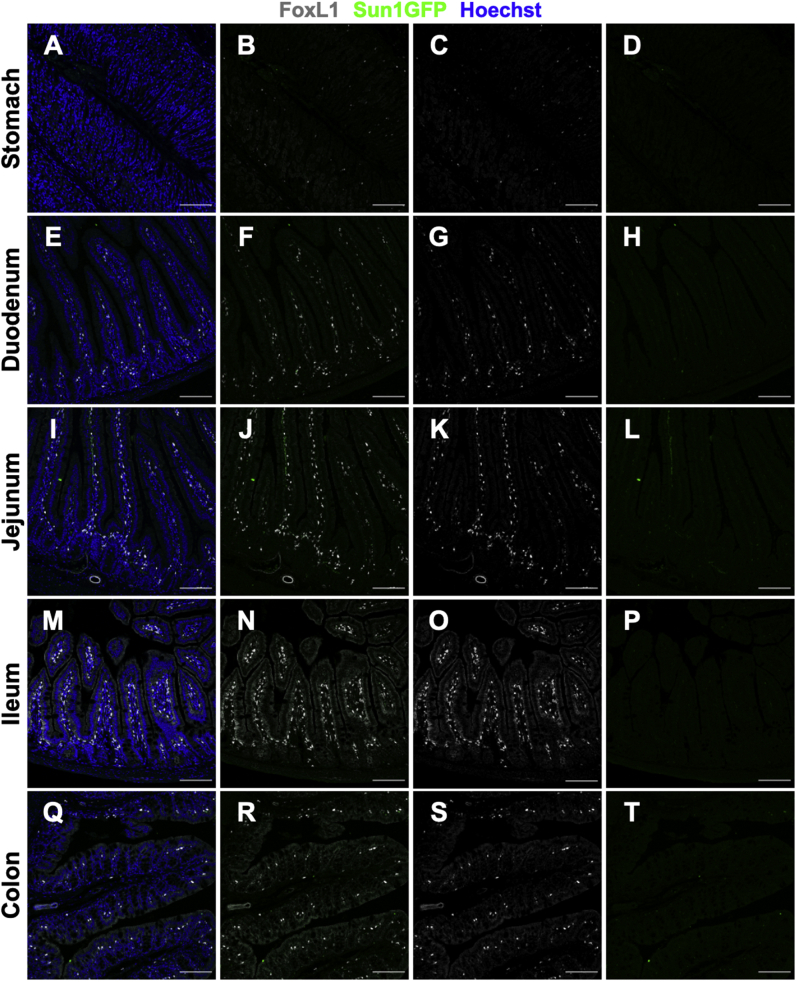
To determine the extent of extraintestinal Cre expression, the kidney, liver, pancreas, and lung of FoxL1CreER-tdTomRosa26Sun1GFP/+ mice were surveyed for Sun1GFP reporter expression (Supplementary Figure 1). Neither FoxL1 nor Sun1GFP expression was observed in the nuclei of any of these tissues. Furthermore, significant (>2%) Sun1GFP reporter expression was not observed in tissues collected from FoxL1CreER-tdTomRosa26Sun1GFP/+ mice that were not induced with tamoxifen (Supplementary Figure 2).
Supplementary Figure 1.
FoxL1CreER-tdTom recombination was not observed in extraintestinal tissues. FoxL1CreER-tdTomRosa26Sun1GFP/+ mice were induced with 3 consecutive days of tamoxifen, and tissues were collected 6 days after the final tamoxifen dose (n = 1). Tissues from the (A–D) kidney, (E–H) liver, (I–L) pancreas, and (M–P) lung were stained for FoxL1 and Sun1GFP. (B, F, J, and N) Overlay of FoxL1 and Sun1GFP channels is shown. (C, G, K, and O) Nuclear staining of FoxL1 was not observed in these extraintestinal tissues. (D, H, L, and P) Accordingly, Sun1GFP reporter expression was not observed. Scale bars: 100 μm.
Supplementary Figure 2.
Sun1GFP expression was not observed in FoxL1CreER-tdTomRosa26Sun1GFP/+ mice that were not induced with tamoxifen. Gastrointestinal tissues were collected from FoxL1CreER-tdTomRosa26Sun1GFP/+ mice that were not treated with tamoxifen. Segments of the (A–D) stomach, (E–H) duodenum, (I–L) jejunum, (M–P) ileum, and (Q–T) colon were stained for FoxL1 and Sun1GFP. (B, F, J, N, and R) Overlay of FoxL1 and Sun1GFP channels is shown. (C, G, K, O, and S) FoxL1 protein is shown in the nuclei of subepithelial mesenchymal cells. (D, H, L, P, and T) Significant Sun1GFP expression was not observed, showing the efficacy of reporter activity. Scale bars: 100 μm.
In sum, we report the derivation and characterization of a novel FoxL1CreER-tdTom mouse line that drives tdTomato and Cre expression directly from the FoxL1 locus. Because of the inclusion of a constitutively expressed marker, this model can be used to isolate and study telocytes without prior treatment with tamoxifen, which can have deleterious effects in the mouse stomach.11 When crossed with a Rosa26LSL-SUN1-GFP reporter mouse line, we observed Cre activity and subsequent fluorescent reporter expression in the subepithelial mesenchymal cells of gastrointestinal tissues. In using these mice, it should be noted that the FoxL1CreER-tdTomRosa26Sun1GFP allele is a Foxl1 null allele. Mice heterozygous for FoxL1, previously known as Fkh6, are phenotypically normal; however, mice null for FoxL1 show developmental delays in gut formation and abnormal proliferation and architecture of the gastrointestinal epithelium.12 Thus, FoxL1CreER-tdTomRosa26Sun1GFP/+ mice should not be homozygosed unless analysis of the FoxL1 null phenotype is intended. This novel mouse model will be made available via the Mutant Mouse Resource and Research Centers (mmrrc.org) and hopefully will become a valuable tool for the investigation of FoxL1+ telocytes and their contribution to the maintenance and functioning of the intestinal stem cell niche.
All authors had access to the study data and approved the final manuscript.
Acknowledgments
The authors would like to thank the members of the Kaestner and Lengner laboratories for their helpful discussions throughout this project. The authors thank Alanis Perez for her help with animal husbandry, and Dr Avital Swisa for help with tissue collections. The authors also thank the Cell and Developmental Biology Microscopy Core for use of their microscopy services, as well as Kate Bennett and Nahyun Son of the Center for Molecular Studies in Digestive and Liver Diseases (P30 DK050306) for the use of the Molecular Pathology and Imaging Core.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R37 DK053839 (K.H.K.) and R01 DK106309 (C.J.L.).
Contributor Information
N. Li, Email: ningli@upenn.edu.
K.H. Kaestner, Email: kaestner@pennmedicine.upenn.edu.
Supplementary Materials and Methods
Derivation of FoxL1-CreERT2-2A-tdTomato Gene Replacement Allele
FoxL1CreERT2-2A-tdTomato mice were derived by inserting a CreERT2-2A-tdTomato-phosphoglycerate kinase I-NeomycinR cassette into the endogenous FoxL1 locus via homology-directed repair in mouse V6.5 embryonic stem cells (129/sv × C57Bl/6 F1 hybrid male). The targeting vector was generated by In-Fusion Cloning (Takara Bio Inc, Shiga, Japan), and arms of homology were amplified from genomic DNA of the V6.5 cells to be targeted. All components were verified by Sanger sequencing. Two hundred base pairs of the FoxL1 coding sequence downstream of the ATG was deleted, rendering this reporter allele null for FoxL1 protein expression. The linearized targeting vector was introduced by nucleofection (Amaxa; Lonza, Basel, Switzerland) into V6.5 cells, after which the cultures were subjected to G418 selection for 7 days and drug-resistant colonies were subcloned and screened for proper insertion. Insertion was validated at both the 5’ and 3’ insertion sites using primers forward: 5’-AGCCAGACCCAAGGACCTTGTTG, 5’R-TGCGAACCTCATCACTCGTT and 3’F-TCCCCAACTGGGGTAACCTT, 3’R-TTCCGGTGGCTGGTTAAGTC. Several correctly targeted clones were identified, and one was injected into C57Bl/6 blastocysts, resulting in the generation of several high-contribution male chimeras. Chimeras were backcrossed to C57Bl/6 females to establish the FoxL1CreERT2-2A-tdTomato strain. FoxL1CreERT2-2A-tdTomato mice subsequently were routinely genotyped using a 3-primer polymerase chain reaction (Figure 1C) enabling resolution of both wild-type and FoxL1CreERT2-2A-tdTomato alleles. Primer FoxL1A: AGCCATGAAGAAGGGACAAAGCG, FoxL1B: GGCGGGCTTGGGCTTCCTCTTCCTG, FoxL1C: CGACCGGCAAACGGACAGAAGCA.
Mouse Studies
All animal experiments performed in this study were reviewed and approved by the Institutional Animal Care and Use Committee of the Office of Animal Welfare at the University of Pennsylvania. Housed mice were subjected to 12-hour light-dark cycles, fed standard rodent chow, and had access to water at all times. Two- to 3-month-old male and female mice were used for all experiments. Rosa26Sun1GFP/+ mice have been described previously10 and were generously gifted by Dr Jeremy Nathans (Johns Hopkins University, Baltimore, MD). To induce Cre activity, animals were treated by intraperitoneal injection with 1.6 mg/30 g body weight of tamoxifen (T5648; Sigma-Aldrich, St. Louis, MO) dissolved in sunflower seed oil. Animals were injected with tamoxifen once daily for 3 or 5 consecutive days. Mice were euthanized by cervical dislocation 6 days after the final tamoxifen dose.
Tissue Isolation and Processing
The entire gastrointestinal tract, from stomach to distal colon, was removed. The stomach was isolated, opened along the greater curvature, rinsed in cold phosphate-buffered saline, and placed in a cassette for fixation. The small intestine was split into thirds, and the most proximal 4 cm of tissue was isolated as duodenum. The next 5 cm was isolated as jejunum and made into a Swiss roll. From the final third of intestine, the most proximal 1 cm of tissue was isolated as the ileum. All small intestine pieces were opened longitudinally and rinsed in phosphate-buffered saline before fixation. From the colon, the proximal 3 cm of tissue was isolated, opened longitudinally, rinsed in phosphate-buffered saline, and made into a Swiss roll. The pancreas, liver, lung, and kidney were removed, rinsed in phosphate-buffered saline, and placed in individual cassettes for fixation. All tissue pieces were fixed in 4% paraformaldehyde overnight at 4°C, rinsed in phosphate-buffered saline, and embedded in paraffin. Tissue sections (5 μm) were obtained from paraffin blocks.
Histology, Immunohistochemistry, and Whole-Mount Imaging
Tissue sections were deparaffinized with xylene and rehydrated by serial incubation of 100%, 95%, 80%, and 75% ethanol followed by phosphate-buffered saline. Antigen retrieval was performed in Tris-EDTA-Tween buffer (10 mmol/L Tris Base, 1 mmol/L EDTA, 0.05% Tween 20, pH 9) in a pressure cooker (2100 Antigen Retriever; Aptum Biologics, Ltd, Southampton, UK) and slides were cooled to room temperature. Slides then were washed in phosphate-buffered saline and endogenous peroxidases were quenched in 3% hydrogen peroxide for 15 minutes at room temperature. After additional phosphate-buffered saline washes, tissue sections were blocked with blocking buffer (008120, CAS-Block; Invitrogen, Carlsbad, CA) for 1 hour, followed by overnight incubation of primary antibodies diluted in blocking buffer at 4°C in a humidified chamber. Primary antibodies included guinea pig anti-FoxL1 (1:1500), goat anti-GFP (ab6673, 1:500; Abcam, Cambridge, UK), and mouse anti–E-cadherin (610181, 1:500; BD Biosciences, Franklin Lakes, NJ). The following day, the FoxL1 signal was amplified using biotinyl tyramide (NEL700A001KT; PerkinElmer, Waltham, MA). Four phosphate-buffered saline/tween washes were performed, followed by blocking with avidin and biotin (SP-2001; Vector Laboratories, Burlingame, CA). A biotinylated secondary antibody (BA-7000, 1:200; Vector Laboratories) targeting guinea pig epitopes then was applied to tissue sections, followed by additional phosphate-buffered saline/tween washes. Streptavidin-conjugated peroxidase then was applied for 15 minutes, followed by phosphate-buffered saline/tween washes and subsequent incubation with biotinyl tyramide for 10 minutes (NEL700A001KT; PerkinElmer). After additional phosphate-buffered saline/tween washes, tissue sections were incubated in secondary antibodies for 2 hours at room temperature in the dark. Secondary antibodies included TSA-conjugated Cy5 (NEL745001KT; PerkinElmer) to visualize the amplified FoxL1 signal, Cy3 anti-mouse, and Cy2 anti-goat antibodies. Nuclei were labeled using Hoechst stain (H3570, 1:10,000; Molecular Probes, Eugene, OR). Fluorescent images were obtained using a KEYENCE BZ-X800 microscope (KEYENCE, Osaka, Japan) and a Leica laser scanning confocal microscope at thePenn Cell and Developmental Biology Microscopy Core (TCS SP8; Leica, Wetzlar, Germany). Images were processed and brightness and contrast were enhanced using FIJI (ImageJ, version 2.1.0; National Institutes of Health, Bethesda, MD).
For whole-mount imaging, the jejunum was dissected and fixed overnight at 4°C in zinc formalin fixative, pH 6.25 (21516-3.75; Polysciences, Warrington, PA). After washing with phosphate-buffered saline, the specimens were cut longitudinally and placed on a glass-bottom dish (no. 1 coverslip) (P35G-1.0-14-C; MatTek, Ashland, MA), mounted in fluorescent mounting medium (71-00-16; SeraCare, Milford, MA), and imaged using an inverted Leica DMi8 microscope equipped with an harmonic compound system, positive low, fluorite abberation correction (HC PL FLUOTAR) 10×/0.30 dry objective and Leica Application Suite X software. Images were analyzed with Leica Application Suite X software and FIJI (ImageJ, version 2.1.0) software.
Supplementary Material
References
- 1.Sato T. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durand A. Proc Natl Acad Sci U S A. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T.H. Proc Natl Acad Sci U S A. 2012;109:3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki R. Cell Mol Gastroenterol Hepatol. 2016;2:175–188. doi: 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degirmenci B. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 6.Greicius G. Proc Natl Acad Sci U S A. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoshkes-Carmel M. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stzepourginski I. Proc Natl Acad Sci U S A. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaestner K.H. Cell Mol Gastroenterol Hepatol. 2019;8:111–117. doi: 10.1016/j.jcmgh.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mo A. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning E.H. Cell Mol Gastroenterol Hepatol. 2020;10:655–657 e651. doi: 10.1016/j.jcmgh.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaestner K.H. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]