Abstract
PB125® is a phytochemical composition providing potent Nrf2 activation as well as a number of direct actions that do not involve Nrf2. Nrf2 is a transcription actor that helps maintain metabolic balance by providing redox-sensitive expression of numerous genes controlling normal day-to-day metabolic pathways. When ordinary metabolism is upset by extraordinary events such as injury, pathogenic infection, air or water pollution, ingestion of toxins, or simply by the slow but incessant changes brought about by aging and genetic variations, Nrf2 may also be called into action by the redox changes resulting from these events, whether acute or chronic. A complicating factor in all of this is that Nrf2 levels decline with aging, leaving the elderly less able to maintain proper redox balance. The dysregulated gene expression that results can cause or exacerbate a wide variety of pathological conditions, including susceptibility to viral infections. This review examines the characteristics desirable in Nrf2 activators that have therapeutic potential, as well as some of the patterns of dysregulated gene expression commonly observed during pulmonary infections and the normalizing effects possible by judicious use of phytochemicals to increase the activation level of available Nrf2.
Keywords: Nrf2, PB125, Carnosic acid, SARS-CoV-2, Coronavirus, COVID-19, Cytokines, Interferon, Endothelium
Graphical abstract
1. Introduction
Nrf2, encoded by the NFE2L2 gene, is a transcription factor that is sequestered in the cytoplasm by a redox-sensing protein called Keap1, which binds to Nrf2 and targets it for proteasomal degradation. Nrf2 regulates a number of the genes that defend directly against oxidative stress, including superoxide dismutases, catalase, numerous peroxidases, and the enzymes that synthesize glutathione as well as regulate its redox poise. It may be surprising to some that Nrf2 also regulates literally hundreds of genes involved in metabolic pathways that seem to have little if anything to do with oxidative stress or redox balance [1]. By evolution, these pathways have come to be controlled by Nrf2 because when a cell undergoes an adverse shift in redox balance numerous pathways may need to be adjusted. Some of these pathways may be able to correct a transient imbalance to return to a state of homeostasis, while others may repair damage incurred. Many pathways may have to adjust to a “new normal” if the situation is a chronic one such as aging, or any age-related disease. For example, there may be more metabolic demand for pathways to produce reducing equivalents (NADH, NADPH, glutathione) to neutralize oxidizing species at the expense of energy production. Unfortunately, Nrf2 production declines steadily with age in many tissues and cell types [[2], [3], [4]] which may reflect increased production of a miRNA (miR-146a) that impedes translation of the Nrf2 mRNA [5]. This age-related decline in Nrf2 is not seen consistently in all tissues, and sometimes Nrf2 basal expression is actually elevated [6]. One possible explanation for this phenomenon might be that in some aging organs age-related diseases may have increased ROS production, and the organ response is to increase its “basal” rate of Nrf2 production, which may in fact be an “induced” or partially compensated level of Nrf2. In any event, most aging cells and tissues show a decreased ability for electrophiles to activate the Nrf2 that they have [6]. This loss of active Nrf2 is associated with frailty [7] and results in diminished resistance to stress [8]. In a youthful cell experiencing little oxidative stress most of the Nrf2 produced is never released to enter the nucleus. Rather, it is ubiquitinylated and sent for degradation [9]. Maintaining a reserve of Nrf2 in the cytosol and a sufficient rate of production to rapidly replenish it is vital for dealing with a rapidly changing state of oxidative stress. Nrf2 is also quickly retrieved from the nucleus by Keap1 and sent for degradation by the proteosome. This rapid production and destruction may appear to be inefficient, but it is the price paid for rapid response in either direction. Failure to respond rapidly when a corrective measure is needed and overshooting the return-to-balance are both unacceptable when precise control is needed. In an aging or stressed cell, however, the reserve supply of Nrf2 may become limiting due to the demand for Nrf2 in the nucleus exceeding the available supply. More and more frequently a transient exposure to oxidative stress may exhaust the dwindling reserve, leaving the aging cell in periods of slow recovery, or unable to bring about a complete recovery at all-- a condition of chronic oxidative stress. To help maintain healthy Nrf2 signaling during aging we have sought to develop phytochemical combinations that potently activate the Nrf2 pathway at multiple synergistic control points in the pathway as previously described while minimizing electrophilic toxicity [[10], [11], [12], [13]]. PB125 consists of three plant extracts in a 15:5:2 ratio by mass: the first, an extract from Rosmarinus officinalis, is standardized to contain 15% carnosic acid and 6% carnosol (Flavex, Rehlingen, Germany); the second, an extract from Withania somnifera is standardized to contain 2% withaferin A (Verdure Sciences, Noblesville, IN, USA); the third, an extract from Sophora japonica is standardized to contain 98% luteolin (Jiaherb, Pine Brook, NJ, USA). These active ingredients were chosen for very specific reasons as previously described [10]. The Nrf2 activation pathway is controlled by more than a dozen different actions occurring between the transcription of the Nrf2-encoding NFE2L2 gene and the eventual proteasomal destruction of the Nrf2 protein. All of these actions participate in controlling the quantity, intensity and duration of Nrf2's gene regulatory actions. The complexity of the pathway makes it unlikely that safe and satisfactory control of Nrf2 can be obtained by any single compound, whether pharmaceutical or phytochemical, acting at only one of these multiple control points. By far, the most studied Nrf2 activators rely on electrophilic attack on Keap1, which is a necessary step to release Nrf2 from its cytoplasmic storage site, but which may result in excessive electrophilic toxicity and rapid depletion of the short-lived Nrf2 transcription factor. Luteolin was chosen in part because it was found to nearly double Nrf2 mRNA production, not by gene induction but by relieving epigenetic silencing of the Nrf2 promoter through reducing CpG methylation of the Nrf2 promoter region [14]. A second action of luteolin that increases Nrf2 activity is its ability to potently inhibit GSK3β [15,16], the enzyme responsible for activation of the Fyn kinase that phosphorylates nuclear Nrf2 at tyrosine 586 [17] causing Nrf2 to be ejected from the nucleus, effectively shutting down Nrf2-dependent transcription. GSK3β activation has been shown in SARS-CoV-2 infected cells and GSK3β inhibitors have been suggested as potential therapeutic interventions [18]. By preventing the activation of Fyn, the limited supply of Nrf2 available in aging cells may remain in the nucleus for a longer time, possibly preventing Nrf2 depletion. Withaferin A is a component of PB125 because it activates Nrf2 in a Keap1-independent manner via the PTEN/PI3K/Akt pathway [19], unlike the prototypical Nrf2 inducers sulforaphane and CDDO-Im, and importantly, different as well from carnosic acid/carnosol. The rationale for pharmacological or phytochemical Nrf2 activators is that increasing the amount of Nrf2 released by Keap1 or extending its duration in the nucleus may restore redox homeostasis at times when Nrf2 reserves are low. A phenomenon seen in some but not all tissues and cell lines is that Nrf2 can induce itself (unpublished observations). Thus, when oxidative stress sends Nrf2 into the nucleus, it increases or replenishes the cytosolic reserve by transcriptional upregulation of its own promoter. When the stress causing nuclear translocation subsides, the self-induction would similarly go away.
2. Are all Nrf2 activators equivalent?
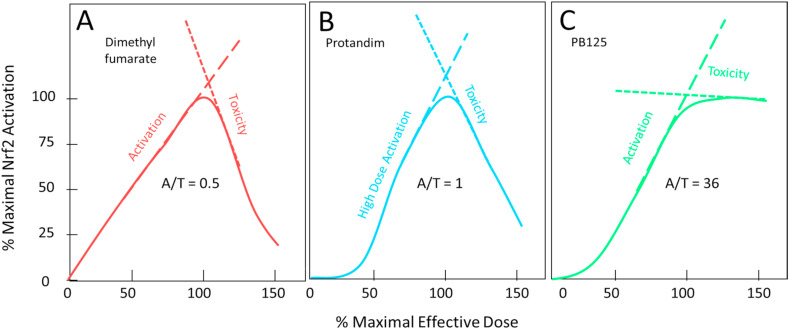
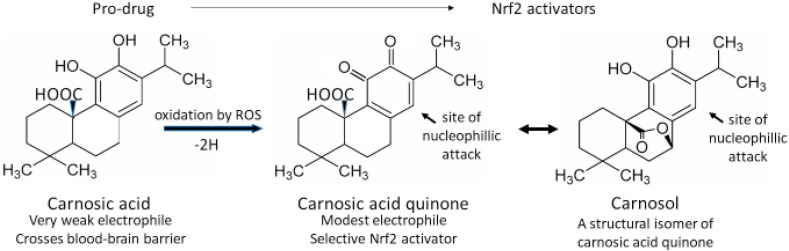
Most Nrf2 activators act by electrophilic attack on three cysteine residues (Cys-151, -273, and -288) which serve as “redox sensors” on the surface of the Keap1 protein causing S-alkylation of these residues and inducing conformational changes in Keap1. Once S-alkylated, the Keap1 releases its bound Nrf2 to enter the nucleus rather than to be tagged for degradation [20]. Because electrophiles react with the ionized thiolate form of cysteine (‾S-Cys), these “sensor” residues are situated on the protein surface near positively charged residues, thereby stabilizing their ionized form and increasing their likelihood of reacting with electrophiles. Satoh and Lipton [21] have introduced the terms “S-alkylation” and “S-alkylation for redox signaling” to distinguish between random electrophilic attack on thiol groups (considered a toxic pathological event) and targeted electrophilic modification of specially sensitized protein thiol groups (such as those on Keap1) which result in changes of biological activity of the modified protein causing protective or reparative measures to be initiated within the cell. At first glance it may appear ironic that the action required to activate Nrf2 is a potentially toxic electrophilic attack within the cell. As an example, electrophilic lipid-peroxidation products containing α,β-unsaturated carbonyls are generated under conditions of oxidative stress. These compounds are toxic to the cell in a number of ways, but they can also S-alkylate Keap1 causing it to liberate Nrf2, which will enter the nucleus and strongly induce the AKR1B10 gene, which encodes an aldo-ketoreductase that will safely detoxify the α,β-unsaturated carbonyls [22]. So, in fact, it makes perfect sense that the compounds that activate the sensor are among the compounds that Nrf2 ultimately eliminates by the genes that it induces. This does, however, pose a bit of a problem when selecting electrophilic phytochemicals or synthetic compounds to potentially serve as therapeutic Nrf2 activators. The desirable candidates should have electrophilicities that are sufficient to react efficiently with the sensitized sensor sulfhydryl groups of Keap1, but that are not so strongly electrophilic as to react with random protein sulfhydryl groups, with low molecular weight thiols such as reduced glutathione, or with the nucleophilic guanine bases of DNA [23]. In other words, the ideal Nrf2 activator should be a “silver bullet” exhibiting a high degree of discrimination between toxic or random “S-alkylation” and “S-alkylation for redox signaling.” Satoh et al. [24] have described a way to analyze the dose behavior of various Nrf2 activators to assess the broadness of their therapeutic indices by examining the ratio of their rates of Nrf2 activation and of toxicity vs dose. Fig. 1 shows two Nrf2 activators that we have previously examined [1] compared to PB125 which derives most of its Nrf2 activation from its carnosol and carnosic acid content [10]. Because all the currently useful Nrf2 activators have two opposing electrophilic actions resulting in both protective and detrimental outcomes, the compounds show inverse U-shaped dose-response curves where the upslope reflects activation of Nrf2 (usually measured by production of a Nrf2-induced reporter gene) and the downslope reflects undesirable toxic adduct formation that may damage enzymes, structural protein, or DNA. This downslope manifests as diminishing production of the reporter gene but has been documented more broadly as diminishing ATP content in the cells [25]. The toxicities become apparent at doses beyond the point where all available Nrf2 has been activated. In Fig. 1 it may be seen that dimethyl fumarate shows strong activation (A), but even stronger toxicity (T), yielding an A/T ratio of 0.5. This ratio is related to therapeutic index, which may be defined in a variety of ways, but which reflects a ratio of the dose that produces the desired effect relative to the dose that produces adverse effects. The larger the ratio the safer the drug. Protandim®, a mixture of five phytochemical extracts, shows a high activation rate at doses greater than 40% of the dose needed for maximal activation. At doses below that amount, which include the physiologically attainable plasma and tissue levels activation is quite low. Song et al. [26] found low bioavailability and poor tissue distribution by three of the Protandim ingredients when orally administered to rats, especially for skin and brain where the active components were barely detectable. Protandim's in vitro toxicity rate, however, is about half that seen with dimethyl fumarate, making its A/T ratio about 1. By contrast, Fig. 1, Panel C, shows PB125 where carnosic acid/carnosol provide the predominant Nrf2 activation. Here there is very strong activation of Nrf2, but after the maximum is reached the rate of toxicity is markedly slower resulting in an A/T ratio of 36. There are several important reasons for this difference. Carnosic acid acts as a pro-drug, not being electrophilic itself and therefore possessing very low toxicity. Carnosic acid is, however, very easily oxidized by reactive oxygen species such as are produced at pathophysiological sites by the inflammatory system, producing carnosic acid quinone, a structural isomeric form of carnosol [24], as shown in Fig. 2 . The creation of an quinone-type structure (ortho or para) creates an electrophilic site on the ring which can readily react with the activated thiolate on Keap1 or on other proteins that are regulated by S-alkylation [27]. This property of carnosic acid allows administration of a non-toxic pro-drug that self-activates upon arriving at sites of oxidative stress to yield a mild and targeted electrophile with efficient Nrf2-activation. This would seem to go a long way in alleviating the legitimate issue of indiscriminate electrophilic toxicity associated with traditional Nrf2 activators intended to act via “S-alkylation for redox regulation.” Both pharmaceutical products Bardoxolone® and Tecfidera® have the problem of electrophilic toxicity [25,28]. To avoid it some are seeking Nrf2 activators that can inhibit BACH1, a transcriptional repressor of Nrf2 [29]. One such small molecule BACH1 inhibitor suppressed RANKL-mediated osteoclastogenesis and bone destruction following local injection in mice by increasing Nrf2, suggesting that BACH1 inhibitors may have potential utility in bone destructive diseases, such as osteoporosis, periodontitis, and rheumatoid arthritis [30]. Jo et al. have described a series of bis-sulfone Antioxidant Response Element (ARE) activator molecules with good efficacy at low micromolar concentrations and little or no electrophilic cellular toxicity at 20 μM [31], but they are yet to be assessed in vivo. Others are seeking compounds that can physically block the Keap1 binding site for Nrf2 [32,33]. It remains to be seen whether these approaches to Nrf2 activation will provide better therapeutic efficacy, or whether they will bring a new set of toxicity problems. Another requirement for a therapeutically successful Nrf2 activator is that it may be orally dosed with sufficient bioavailability to reach effective but non-toxic levels in most tissues, especially brain. Here, carnosic acid does well with multiple studies showing that carnosic acid from Rosemary extract is quickly absorbed and bioavailable to multiple organs, including brain [[34], [35], [36], [37], [38], [39]]. In an interesting in vivo mouse model examining brain injury following sublethal six-day cyanide poisoning (designed to mimic a human situation of potential bioterrorism) oral pretreatment with carnosic acid significantly protected from cyanide-induced brain damage, showing up to 90% protection in some behavioral and grip/strength tests and a very significant reduction in delayed mortality [37]. The dosage and oral delivery of carnosic acid were considered consistent with a human taking a daily oral supplement of Rosemary extract.
Fig. 1.
Panels A and B are adapted from Hybertson et al. (see reference 1) and normalized to maximal Nrf2 activation at their respective maximally effective doses in cultured cells and the results analyzed according to Satoh et al. (see reference 17). Panel C represents the mean of 4 replicates. A/T reflects the ratio of the slope of the activation response to the slope of the toxic response, with a higher number being desirable.
Fig. 2.
Conversion of the pro-drug carnosic acid at physiological sites of inflammation or other causes of oxidative stress.
3. Carnosic-acid based Nrf2 activation and antiviral protection
We recently examined the effects of PB125 on lipopolysaccharide (LPS)-stimulated primary human pulmonary artery endothelial cells (HPAEC) as a model of microbial-induced gene expression [11]. We reported that 36 cytokines, including interleukins, interferon-related genes, tumor necrosis factor-related genes, and cell adhesion genes were strongly upregulated by LPS, but were significantly downregulated by PB125. As these genes are also strongly upregulated by many respiratory viruses, including the SARS-CoV-2 virus responsible for the current COVID-19 pandemic, we also examined host genes (in liver-derived HepG2 cells) that have been identified as specifically involved with SARS-CoV-2 attachment (ACE2) and spike protein activation (TMPRSS2), finding them downregulated as well. These results strongly suggested that Nrf2 activation by host or inactivation by virus might play some important roles in the development of COVID-19, as others have demonstrated for respiratory syncytial virus [[40], [41], [42]], dengue virus [43], herpes simplex virus-1 [44], zika virus [45], and as we and others have shown for the human immunodeficiency virus HIV [12,13], and which has now been demonstrated by Olagnier et al. for SARS-CoV-2 [46]. We have continued to “mine” the microarray data showing the effects of PB125 on LPS-stimulated HPAEC (experimental details previously published [11]) to seek out additional effects of PB125 that may modulate COVID-19-associated events such as coagulopathy and alternative modes of cell entry.
4. Inflammatory and antiviral responses: how much is too much?
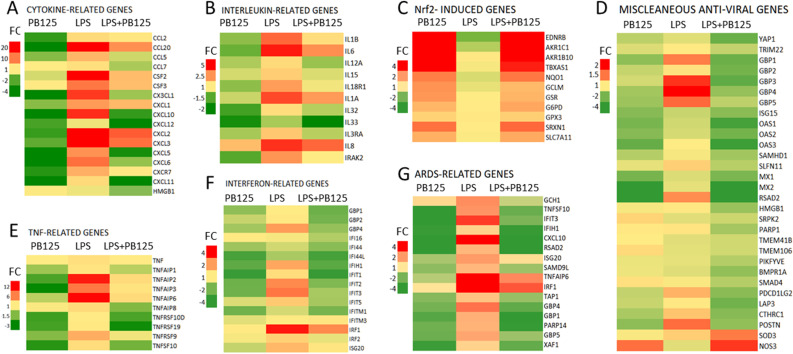
A general theme has come into focus regarding the pathology that accompanies COVID-19. In several ways the host responses to SARS-CoV-2 seem to be overly exuberant, turning around host responses aimed at limiting pathogen-induced damage such that they cause more damage to the host than they do to the virus. The first example of this is the so-called “cytokine storm” [47,48], a phenomenon that has been observed in some other viral infections [49], in graft-versus-host disease [50], in macrophage activation syndrome [51], in sepsis [52], and in acute respiratory distress syndrome (ARDS) [47]. Indeed, this exuberant upregulation of cytokines is also apparent in LPS-treated HPAEC. Fig. 3 is a heatmap representation of the cytokines we first reported to be strongly downregulated by PB125 along with 79 additional genes. The left column of each panel shows the effect of overnight treatment with 5 μg/ml PB125 on non-LPS-stimulated HPAEC vs untreated control cells. Center columns show expression in HPAEC following 5 h of exposure to LPS vs control. Columns on the right show expression of HPAEC pretreated overnight with PB125, then stimulated for 5 h with LPS vs control. Panel A shows 17 chemokines that are commonly seen in hyperinflammatory scenarios. One addition is high mobility group box 1 protein (HMGB1), a newly recognized pro-inflammatory cytokine marker for neuroinflammation that is elevated about 4.5-fold in multiple sclerosis patients versus healthy controls [53]. Wei et al. found HMGB1 regulates ACE2 expression and is critical for SARS-CoV-2 entry [54]. PB125 reduced HMGB1 expression by about -1.7-fold in LPS stimulated HPAEC. All of the genes in panels A (cytokine-related), B (interleukin-related), and E (tumor necrosis factor-related) were downregulated by PB125. Panel C shows a few of the characteristic canonical genes that are expected to be upregulated by a Nrf2 activator, and indeed they are very similarly upregulated in cells with or without exposure to LPS, although the average induction was diminished by about 15% in the LPS-treated cells. This probably reflects generalized metabolic stress caused by the cellular responses to LPS. An observation that was unexpected, and perhaps somewhat surprising, is seen in Panels F (interferon-related genes) and D (miscellaneous antiviral genes). Here genes that are sometimes considered to be antiviral are consistently downregulated by Nrf2 activation. Sauter and Kirchhoff have recently reviewed an under investigated aspect of host antiviral responses—their potential adverse effects on host metabolism [55]. They review the diverse spectrum of antiviral proteins, carefully considering the costs and benefits from the host perspective of each evolutionary step, and they conclude that antiviral actions come at a price. So, is the over exuberant induction of antiviral proteins another clever trick that some viruses have mastered to turn another of our primary defense systems against us? Is there an “interferon storm”? There is evidence suggesting there is [56,57]. There are some examples that suggest that antiviral genes that evolved early in mammals may have reached the point where evolutionary pressure is now pushing for their removal. The interferon induced IFIH1 gene that encodes the MDA5 protein is a good example of this possibility. MDA5 functions as a pattern recognition receptor capable of detecting double stranded RNA (dsRNA) which is involved in the replicative stage of RNA viruses such as the coronaviruses [58]. MDA5 binds to double-stranded RNA in the cytosol and signals through mitochondrial antiviral-signaling protein (MAVS) to activate expression of both type I and type III interferons and to induce inflammation by the production of the proinflammatory IL-1 family of cytokines, including IL-1β and IL-18 [59]. The protection afforded by IFIH1/MDA5, however, comes at a steep price. A mutant mouse line bearing a constitutively active IFIH1 gene developed lupus-like autoimmune symptoms without virus infection [60]. Spontaneous inflammation developed in multiple organs, especially in the kidney. The mice showed growth retardation and 60% died at 24 weeks of age. In the kidney, production of inflammatory cytokines and chemokines including IFN-β, IL-6, CXCL10 and tumor TNF-α was significantly upregulated [60]. Thus, an ultimately lethal hyperinflammatory state can be induced by constitutive activation of this single “antiviral” gene IFIH1. Zhang et al. recently investigated a panel of 36 genes associated with severity of acute respiratory distress syndrome (ARDS) to identify which of those genes also play roles in macrophage polarization to the inflammatory M1 state [61]. Five genes were identified as hub genes. IFIH1 showed the strongest connection with ARDS, and they confirmed that IFIH1 is a novel regulator promoting M1 macrophage polarization. In our LPS/HPAEC study all five of the hub genes identified by Zhang et al. were downregulated by PB125. Fig. 3, Panel G, represents 16 of the genes that were found to be upregulated in ARDS patients that were also upregulated by LPS in HPAEC. All five of the identified hub genes were downregulated by PB125 treatment, including IFIH1 (downregulated -4.4-fold). Other studies have found that reduced expression if the IFIH1 gene protects against the development of type 1 diabetes (T1D) [62,63]. IFIH1 ± mice express roughly half the level of MDA5 protein as wild-type mice. When infected with coxsackievirus serotype B4, a clinically relevant stimulator of T1D, the IFIH1 heterozygous non-obese diabetic (NOD) mice were protected from T1D, underscoring the ability of IFIH1/MDA5 to augment autoimmunity and T1D [63]. Concern has been voiced that new-onset T1D is being seen in COVID-19 patients [64]. An interesting evolutionary observation is that pangolins have lost the functionality of their IFIH1 gene by acquiring several frame-shift mutations [65]. This unique loss of a gene important to the innate immune response to RNA-viruses has resulted in pangolins becoming tolerant to SARS-CoV-2, and becoming suspects for carrying the virus to humans [66]. Another group of RNA-sensing antiviral proteins, OAS1, OAS2, OAS3, provide protection against viral infection but can also be activated by host RNAs. Carey et al. have reported that loss-of-function alleles of OAS1 are pervasive among non-human primates, possibly providing a way to mitigate the costs associated with OAS1/RNase L signaling [67]. These losses of antiviral mechanisms imply that, on the whole, pangolins and non-human primates are better off without these particular antiviral mechanisms in the ever-shifting ongoing battle between mammals and microbes, and it raises the question of whether pharmaceutical (or phytochemical) restraints might be indicated when a microbe generates an overly exuberant and potentially damaging immune response from us. Clearly, more work needs to be done to understand fully how these delicate balances might be maintained. It is also intriguing to consider whether there may be mechanisms that permit SARS-CoV-2-induced changes in gene expression to linger for months or longer, giving rise to the myriad symptoms associated with “long Covid,” some of which resemble autoimmune diseases.
Fig. 3.
Heatmaps showing groups of related genes expressed in HPAEC. Gene expression in each of the three treatment groups is expressed relative to untreated control cells. Experimental details have been previously described (see reference 10).
5. Effects of Nrf2 activation on genes involved in coagulation
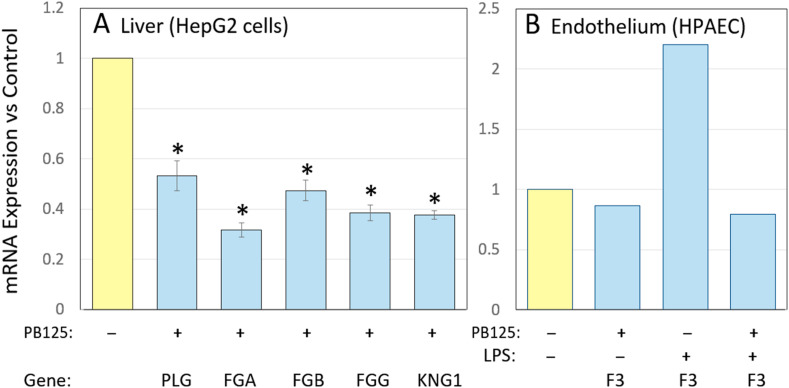
Hypercoagulation can create life-threatening conditions in a number of diseases, but clearly manifests in age-related diseases such as atherosclerosis, coronary artery disease, and thrombotic stroke. A number of genes involved in coagulation pathways are expressed more highly in the elderly. Key components of the coagulation cascade such as tissue factor (F3) and fibrinogen are epidemiologically and functionally linked with inflammatory disease [[68], [69], [70]]. Furthermore, plasma concentration of fibrinogen was found to increase by 57% in 70 year old vs 25 year old subjects [71]. Elevated fibrinogen has been shown to place males at higher risk of later life mortality [72]. Fig. 4 , Panel A, shows the effect of PB125 on the expression levels of plasminogen (PLG) and the three genes encoding subunits of fibrinogen (FGA, FGB, and FGG) in HepG2 cells. Most coagulation factors are synthesized in the liver and secreted into the plasma to be systemically available. Plasminogen expression was decreased 47% by PB125. At first glance a decrease in plasmin, the fibrinolytic product of plasminogen, might appear be a pro-coagulative change, but note that the actual clot-forming protein, fibrin, would be decreased by 68% based on FGA expression. Furthermore, Ji et al. [73] found elevated plasminogen to be a common risk factor for COVID-19 susceptibility and discussed a number of possible mechanisms. Activated plasmin plays a role in many inflammatory conditions including hereditary angioedema that produces aggressive tissue swelling due to overproduction of bradykinin [74] and macrophage activation syndrome [51]. Decreasing plasminogen in a mouse model of Alzheimer disease dramatically improved the pathology, and decreased glial cell activity in the brain [75]. Tissue factor (encoded by the F3 gene) is the primary coagulation factor acting locally and is released at sites of tissue injury by the affected tissue. Fig. 4, Panel B, shows that in our model of LPS-treated HPAEC [11] LPS induced tissue factor by more that 2-fold, and that this increase was completely prevented by PB125. Eslamifar et al. have concluded that tissue factor plays the major role in the activation of the coagulation cascade during a number of types of viral infection, including SARS-CoV-2 [76]. Tissue factor has been shown capable of triggering disseminated intravascular coagulation in LPS-treated rabbits [77]. Finally, we also noted that the kininogen 1 gene (KNG1) was repressed 60% by PB125 in HepG2 cells (Fig. 4, Panel A). This gene encodes high-molecular-weight kininogen (HMWK), a circulating plasma protein giving rise to products such as [des-Arg9] bradykinin which participate in the initiation of blood coagulation, vasodilation and microvascular leakage, pain, and fibrosis. COVID-19 increases bradykinin and its metabolites, which facilitate lung inflammation, fever, coagulation and complement activation. Colarusso et al. [78] and van de Veerdonk [79] have proposed that blocking the conversion of HMWK to its metabolites might relieve some of COVID-19's worst symptoms. We suggest that similar effects might be achieved by the ability of PB125 to downregulate expression of KNG1/HMWK -2.5-fold (Fig. 4, Panel A).
Fig. 4.
Effects of PB125 on coagulation-related genes in liver HepG2 cell (Panel A) and in HPAEC with and without LPS stimulation (Panel B). PLG encodes plasminogen; FGA, FGB, and FGG encode the three subunits of fibrinogen; KNG1 encodes high-molecular weight kininogen. Results in Panel A were determined by RNA-Seq; * indicates < 0.001 by Student t-test, n=3. Results in Panel B were by a single Affymetrix GeneChip where each sample was three pooled replicates as previously described (see reference 10).
6. Additional potential effects of PB125 on viral entry
Early in the COVID-19 pandemic Hoffman et al. [80], Itawa-Yoshikawa et al. [81] and other investigators identified a very important role of the cell surface-bound serine-protease TMPRSS2 in activating the SARS-CoV-2 spike protein on virus particles bound by ACE2 on the surface of a soon-to-be infected lung cell. Experiments showed TMPRSS2 was both necessary and sufficient for spike protein activation and entry in the cell line investigated, but that obviously did not preclude different modes of entry in other tissues throughout the body which show great diversity with regard to TMPRSS family member expression. Other members of the TMPRSS family expressed in other organs and cell types have been implicated as possible facilitators of entry including TMPRSS4, TMPRSS11A, TMPRSS11B, TMPRSS11E, and TMPRSS12 [[82], [83], [84], [85], [86]]. Fig. 5 shows that several TMPRSS genes expressed in HepG2 cells are similarly repressed by PB125, suggesting that Nrf2 activation might be effective at impeding viral entry in organs other than lung that may activate spike proteins via alternative proteases.
Fig. 5.
PB125 inhibits gene expression of other members of the TMPRSS family in HepG2 cells. (* indicates p < 0.05) by Student t-test, n=4. Results were determined by RNA-Seq as previously described (see reference 10).
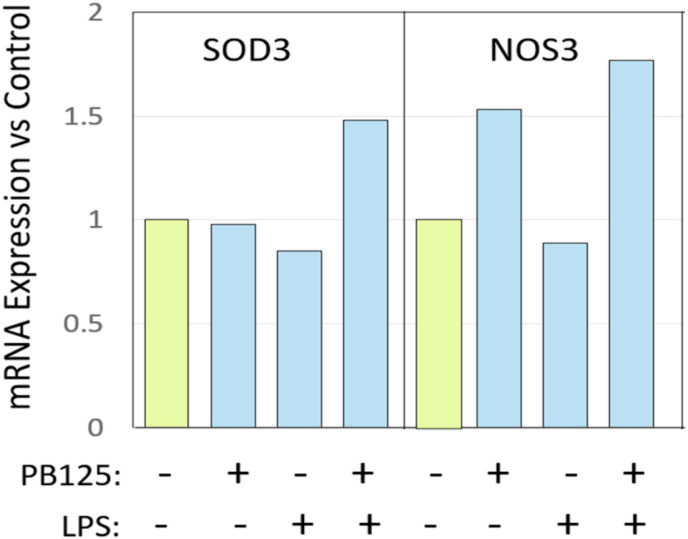
A recent study by Abouhashem et al. [87] examined differently expressed genes in alveolar type II cells from the healthy lungs of elderly versus young human subjects. In the elderly subjects 263 genes were found to be downregulated while 95 were upregulated. Surprisingly, the extracellular superoxide dismutase (SOD3) was the top-ranked, most downregulated gene in the elderly alveolar type II cells. The study was performed in the context of asking why COVID-19 might be so much more severe in the elderly than in the young. It prompted us to examine SOD3 expression in the results of our HPAEC study [11]. We saw no effect of PB125 on the expression of SOD3 in the absence of LPS stimulation. LPS produced a small 15% downregulation of SOD3, but PB125 in the presence of LPS raised it by 74% (Fig. 6 , left panel). Various forms of SOD have been shown to be therapeutic against pulmonary disease including emphysema and fibrosis [[88], [89], [90], [91]]. Also shown in Fig. 6, right panel, is an induction by PB125 at 5 μg/ml of endothelial nitric oxide synthase, eNOS, encoded by the NOS3 gene. The question has been raised as to whether eNOS-derived nitric oxide may be a factor that protects the young from severe COVID-19 complications [92,93]. The amount of nitric oxide produced by eNOS decreases with aging in rats [94]. The physiology and chemical reactivity of NO• are rather complex and depend greatly on precisely where, when, and how much is generated [95,96]. It can be involved, constructively or destructively, at almost every level of the inflammatory process [97], but the induction of eNOS by PB125 is noteworthy and interesting.
Fig. 6.
Effects of PB125 on two additional genes of interest in HPAEC with and without LPS stimulation. Results were determined by Affymetrix GeneChip as previously described (see reference 10). Each sample represents three pooled replicates.
7. Antiviral effects of PB125 independent of Nrf2 activation
PB125 contains a withaferin A enriched extract of Withania somnifera [10]. Potentially significant antiviral effects have recently been reported for these phytochemicals that reflect direct binding actions to host or viral proteins related to cell entry and viral infectivity. Kumar et al. [98] used molecular docking and molecular dynamics simulations to examine the binding potential of Withaferin A for the host protease activator of the spike protein, TPMRSS2, using a known inhibitor, Camostat mesylate [80] for comparison. They found that withaferin A and a structurally similar component, withanone, could bind and stably interact at the catalytic site of TMPRSS2 with binding parameters very similar to those calculated for Camostat mesylate. They predicted that the two phytochemicals could probably function as drugs to block S-protein priming of SARS-CoV-2. Interestingly, these investigators also noted the ability of withanone to downregulate the expression of TMPRSS2 in the MCF7 human breast carcinoma-derived cell line by as much as 2-fold [98]. We suggest that this is by the same Nrf2-dependent mechanism as we demonstrate for PB125's repression of TMPRSS2 in HepG2 cells, shown in Fig. 5. Kumar et al. [99] have also used the same computer modeling techniques to predict that withanone, but not withaferin A, will show tight binding to the Main Protease, Mpro, of SARS-CoV-2, at the substrate binding pocket.
PB125 also contains 9% luteolin by weight from Sophora japonica as one of its three active ingredients [10]. Shawan et al. [100] employed similar molecular docking and molecular dynamics simulations to examine the potential binding affinities of 43 different flavonoid compounds including luteolin, for three important targets involved in SARS-CoV-2 infection: two viral proteases Mpro/3CLpro and PLpro, and the host protein ACE2. The 43 flavonoid phytochemicals were scanned for drug likeness/pharmacophore features along with 3 drugs used for control purpose: remdesivir; hydroxychloroquine; and Camostat mesylate. Two of the flavonoids, abyssinone II and luteolin, emerged as superior or comparable to remdesivir, the best of the three control drugs against the three target proteins in terms of free energy of binding and lowest inhibition constants (Ki). Furthermore, all three of the control drugs failed ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) profile analysis, raising flags for remdesivir (tumorigenic, irritant, reproductive system injury) as well as for hydroxychloroquine and Camostat mesylate (AMES positive, carcinogenic and mutagenic), while the two flavonoids were considered harmless [100]. The ADMAT analysis predicts safety/toxicity for pharmaceutical candidate compounds [101].
8. Summary and conclusions
Effective cellular defenses and immune function are critical components of health, and evidence points to a key role for the Nrf2 transcription factor in maintaining those systems. A wide variety of chemical compounds, including some dietary phytochemicals and some synthetic drug compounds, have been shown to activate the Nrf2 signaling pathway. Many of these activators are electrophiles that react with Keap1 protein in the cytoplasm which allows Nrf2 to migrate to the nucleus. The optimal electrophiles for Nrf2 activation have electrophilicity sufficient to react with sensor sulfhydryl groups on Keap1, but not to react with other important intracellular molecules and cause toxicity. One such candidate includes carnosic acid, a phytochemical in Rosmarinus officinalis. Carnosic acid has low electrophilicity but it is readily oxidized to become the potent Nrf2 activator carnosol at cellular locations that are under oxidative stress. One straightforward goal during aging is to position the body and its immune system to remain as healthy and resilient as possible. For that reason, we are examining dietary supplement formulations based on phytochemical combinations including carnosic acid/carnosol, withaferin A, and luteolin. We believe that appropriate augmentation of Nrf2 activation supports the body's resilience in the face of health challenges, and our cell culture studies have shown PB125-induced regulation of genes relevant to the protection against oxidative, viral, inflammatory, and coagulopathic insults. Rather than experiencing “death by a thousand cuts” we believe that judicious activation of Nrf2 as the body ages may provide “health and longevity by a thousand little remedies.”
Author contributions
Conceptualization J.M.M., B.M.H., B.G., and A.C.-G.; methodology B.M.H., B.G., and A.C.-G.; investigation A.C.-G and B.G.; data analysis J.M.M., A.C.-G., and B.G.; manuscript preparation J.M.M., manuscript review and editing J.M.M., B.M.H., A.C.-G., and B.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by grant number 1R43AG053128 from the National Institutes of Health (NIH) of the United States to A.C.-G.
Declaration of competing interest
B.M.H., B.G., and J.M.M. are cofounders of Pathways Bioscience, which owns and markets the PB125 dietary supplement. A.C.-G.. has no potential conflicts of interest. The NIH funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
The authors thank Ken Jones and Wenhua Ren for their assistance in analyzing the genearray and RNA-seq data. This work utilized the Genomics Shared Resource of the University of Colorado Cancer Center (P30CA046934). The authors dedicate this work to the memory of Kara P. Geraci.
Footnotes
This is part of a Special Issue - The effects of diet on human in vivo redox state
Guest Editors: Dana R. Crawford, Young Joon Surh
References
- 1.Hybertson B.M., Gao B., Bose S.K., McCord J.M. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol. Aspect. Med. 2011;32:234–246. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L., Zhang H., Davies K.J.A., Forman H.J. Aging-related decline in the induction of Nrf2-regulated antioxidant genes in human bronchial epithelial cells. Redox Biol. 2018;14:35–40. doi: 10.1016/j.redox.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh J.H., Shenvi S.V., Dixon B.M., Liu H., Jaiswal A.K., Liu R.M., Hagen T.M. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang D.D., Fan S.D., Chen X.Y., Yan X.L., Zhang X.Z., Ma B.W., Yu D.Y., Xiao W.Y., Zhuang C.L., Yu Z. Nrf2 deficiency exacerbates frailty and sarcopenia by impairing skeletal muscle mitochondrial biogenesis and dynamics in an age-dependent manner. Exp. Gerontol. 2019;119:61–73. doi: 10.1016/j.exger.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Smith E.J., Shay K.P., Thomas N.O., Butler J.A., Finlay L.F., Hagen T.M. Age-related loss of hepatic Nrf2 protein homeostasis: potential role for heightened expression of miR-146a. Free Radic. Biol. Med. 2015;89:1184–1191. doi: 10.1016/j.freeradbiomed.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Davies K.J.A., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015;88:314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Assar M., Angulo J., carnicero J.A., walter S., garcia-garcia F.J., lopez-hernandez E., sanchez-puelles J.M., rodriguez-manas L. Frailty is associated with lower expression of genes involved in cellular response to stress: results from the toledo study for healthy aging. J. Am. Med. Dir. Assoc. 2017;18:734.e731–734.e737. doi: 10.1016/j.jamda.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Lomeli N., Bota D.A., Davies K.J.A. Diminished stress resistance and defective adaptive homeostasis in age-related diseases. Clin. Sci. (Lond.) 2017;131:2573–2599. doi: 10.1042/cs20160982. [DOI] [PubMed] [Google Scholar]
- 9.Sun Z., Zhang S., Chan J.Y., Zhang D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hybertson B.M., Gao B., Bose S., McCord J.M. Phytochemical combination PB125 activates the Nrf2 pathway and induces cellular protection against oxidative injury. Antioxidants. 2019;8 doi: 10.3390/antiox8050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCord J.M., Hybertson B.M., Cota-Gomez A., Geraci K.P., Gao B. Nrf2 activator PB125® as a potential therapeutic agent against COVID-19. Antioxidants. 2020;9 doi: 10.3390/antiox9060518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simenauer A., Assefa B., Rios-Ochoa J., Geracia K., Hybertson B., Gao B., McCord J., Elajaili H., Nozik-Grayck E., Cota-Gomez A. Repression of Nrf2/ARE regulated antioxidant genes and dysregulation of the cellular redox environment by the HIV Transactivator of Transcription. Free Radic. Biol. Med. 2019;141:244–252. doi: 10.1016/j.freeradbiomed.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kukoyi A.T., Fan X., Staitieh B.S., Hybertson B.M., Gao B., McCord J.M., Guidot D.M. MiR-144 mediates Nrf2 inhibition and alveolar epithelial dysfunction in HIV-1 transgenic rats. Am. J. Physiol. Cell Physiol. 2019;317:C390–C397. doi: 10.1152/ajpcell.00038.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo Q., Wu R., Xiao X., Yang C., Yang Y., Wang C., Lin L., Kong A.N. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem. 2018;119:9573–9582. doi: 10.1002/jcb.27275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reudhabibadh R., Binlateh T., Chonpathompikunlert P., Nonpanya N., Prommeenate P., Chanvorachote P., Hutamekalin P. Suppressing Cdk5 activity by luteolin inhibits MPP+-Induced apoptotic of neuroblastoma through Erk/Drp1 and Fak/Akt/GSK3β pathways. Molecules. 2021;26:1307,. doi: 10.3390/molecules26051307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jing Z., Wang C., Yang Q., Wei X., Jin Y., Meng Q., Liu Q., Liu Z., Ma X., Liu K., et al. Luteolin attenuates glucocorticoid-induced osteoporosis by regulating ERK/Lrp-5/GSK-3β signaling pathway in vivo and in vitro. J. Cell. Physiol. 2019;234:4472–4490. doi: 10.1002/jcp.27252. [DOI] [PubMed] [Google Scholar]
- 17.Kaspar J.W., Jaiswal A.K. Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression. Faseb. J. 2011;25:1076–1087. doi: 10.1096/fj.10-171553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rana A.K., Rahmatkar S.N., Kumar A., Singh D. Glycogen synthase kinase-3: a putative target to combat severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Cytokine Growth Factor Rev. 2021;58:92–101. doi: 10.1016/j.cytogfr.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palliyaguru D.L., Chartoumpekis D.V., Wakabayashi N., Skoko J.J., Yagishita Y., Singh S.V., Kensler T.W. Withaferin A induces Nrf2-dependent protection against liver injury: role of Keap1-independent mechanisms. Free Radic. Biol. Med. 2016;101:116–128. doi: 10.1016/j.freeradbiomed.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh T., Lipton S.A. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci. 2007;30:37–45. doi: 10.1016/j.tins.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang C., Yan R., Luo D., Watabe K., Liao D.-F., Cao D. Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. J. Biol. Chem. 2009;284:26742–26748. doi: 10.1074/jbc.m109.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopachin R.M., Gavin T. Reactions of electrophiles with nucleophilic thiolate sites: relevance to pathophysiological mechanisms and remediation. Free Radic. Res. 2016;50:195–205. doi: 10.3109/10715762.2015.1094184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh T., McKercher S.R., Lipton S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013;65:645–657. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copple I.M., Shelton L.M., Walsh J., Kratschmar D.V., Lister A., Odermatt A., Goldring C.E., Dinkova-Kostova A.T., Honda T., Park B.K. Chemical tuning enhances both potency toward Nrf2 and in vitro therapeutic index of triterpenoids. Toxicol. Sci. 2014;140:462–469. doi: 10.1093/toxsci/kfu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Sun H., Xiao J., Wang F., Ding Y., Zhao J., Bai J., Cheng L., Gao K., Liu M., et al. Development of a liquid chromatography-tandem mass spectrometric (LC-MS/MS) method for simultaneous determination of epigallocatechin-3-gallate, silibinin, and curcumin in plasma and different tissues after oral dosing of Protandim in rats and its application in pharmacokinetic and tissue distribution studies. J. Pharmaceut. Biomed. Anal. 2019;170:54–62. doi: 10.1016/j.jpba.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y., Kumagai T., Yoshida C., Naito Y., Miyamoto M., Ohigashi H., Osawa T., Uchida K. Pivotal role of electrophilicity in GlutathioneS-transferase induction bytert-butylhydroquinone†. Biochemistry. 2003;42:4300–4309. doi: 10.1021/bi0340090. [DOI] [PubMed] [Google Scholar]
- 28.Gazaryan I., Thomas B. The status of Nrf2-based therapeutics: current perspectives and future prospects. Neural Regener. Res. 2016;11:1708,. doi: 10.4103/1673-5374.194706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attucks O.C., Jasmer K.J., Hannink M., Kassis J., Zhong Z., Gupta S., Victory S.F., Guzel M., Polisetti D.R., Andrews R., et al. Induction of heme oxygenase I (HMOX1) by HPP-4382: a novel modulator of Bach1 activity. PloS One. 2014;9:e101044,. doi: 10.1371/journal.pone.0101044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada S., Kanzaki H., Katsumata Y., Yamaguchi Y., Narimiya T., Attucks O.C., Nakamura Y., Tomonari H. Bach1 inhibition suppresses osteoclastogenesis via reduction of the signaling via reactive oxygen species by reinforced Antioxidation. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo J., Ibrahim L., Iaconelli J., Kwak J., Kumar M., Jung Y., Lairson L.L., Chatterjee A.K., Schultz P.G., Bollong M.J., et al. Discovery and SAR studies of 3-amino-4-(phenylsulfonyl)tetrahydrothiophene 1,1-dioxides as non-electrophilic antioxidant response element (ARE) activators. Bioorg. Chem. 2021;108:104614,. doi: 10.1016/j.bioorg.2020.104614. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T., Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Lastra D., Fernández-Ginés R., Manda G., Cuadrado A. In: Schmidt H.H.H.W., Ghezzi P., Cuadrado A., editors. Vol. 264. Springer, Cham; 2020. Perspectives on the clinical development of NRF2-targeting drugs. (Reactive Oxygen Species. Handbook of Experimental Pharmacology). [DOI] [PubMed] [Google Scholar]
- 34.Satoh T., Kosaka K., Itoh K., Kobayashi A., Yamamoto M., Shimojo Y., Kitajima C., Cui J., Kamins J., Okamoto S., et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. J. Neurochem. 2008;104:1116–1131. doi: 10.1111/j.1471-4159.2007.05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romo Vaquero M., Garcia Villalba R., Larrosa M., Yanez-Gascon M.J., Fromentin E., Flanagan J., Roller M., Tomas-Barberan F.A., Espin J.C., Garcia-Conesa M.T. Bioavailability of the major bioactive diterpenoids in a rosemary extract: metabolic profile in the intestine, liver, plasma, and brain of Zucker rats. Mol. Nutr. Food Res. 2013;57:1834–1846. doi: 10.1002/mnfr.201300052. [DOI] [PubMed] [Google Scholar]
- 36.Rezaie T., McKercher S.R., Kosaka K., Seki M., Wheeler L., Viswanath V., Chun T., Joshi R., Valencia M., Sasaki S., et al. Protective effect of carnosic acid, a pro-electrophilic compound, in models of oxidative stress and light-induced retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2012;53:7847–7854. doi: 10.1167/iovs.12-10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D., Lee B., Nutter A., Song P., Dolatabadi N., Parker J., Sanz-Blasco S., Newmeyer T., Ambasudhan R., McKercher S.R., et al. Protection from cyanide-induced brain injury by the Nrf2 transcriptional activator carnosic acid. J. Neurochem. 2015;133:898–908. doi: 10.1111/jnc.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira De. M.R. Carnosic acid as a promising agent in protecting mitochondria of brain cells. Mol. Neurobiol. 2018;55:6687–6699. doi: 10.1007/s12035-017-0842-6. [DOI] [PubMed] [Google Scholar]
- 39.Maynard M.E., Underwood E.L., Redell J.B., Zhao J., Kobori N., Hood K.N., Moore A.N., Dash P.K. Carnosic acid improves outcome after repetitive mild traumatic brain injury. J. Neurotrauma. 2019;36:2147–2152. doi: 10.1089/neu.2018.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komaravelli N., Tian B., Ivanciuc T., Mautemps N., Brasier A.R., Garofalo R.P., Casola A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic. Biol. Med. 2015;88:391–403. doi: 10.1016/j.freeradbiomed.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komaravelli N., Casola A. Respiratory viral infections and subversion of cellular antioxidant defenses. J. Pharmacogenomics Pharmacoproteomics. 2014;5 doi: 10.4172/2153-0645.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komaravelli N., Ansar M., Garofalo R.P., Casola A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway. Free Radic. Biol. Med. 2017;113:494–504. doi: 10.1016/j.freeradbiomed.2017.10.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olagnier D., Peri S., Steel C., van Montfoort N., Chiang C., Beljanski V., Slifker M., He Z., Nichols C.N., Lin R., et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014;10:e1004566,. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyler E., Franke V., Menegatti J., Kocks C., Boltengagen A., Praktiknjo S., Walch-Ruckheim B., Bosse J., Rajewsky N., Grasser F., et al. Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun. 2019;10:4878,. doi: 10.1038/s41467-019-12894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Kalamouni C., Frumence E., Bos S., Turpin J., Nativel B., Harrabi W., Wilkinson D., Meilhac O., Gadea G., Desprès P., et al. Subversion of the heme oxygenase-1 antiviral activity by zika virus. Viruses. 2018;11:2,. doi: 10.3390/v11010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., Hait A., Hernaez B., Knudsen A., Iversen M.B., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938,. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Que Y., Hu C., Wan K., Hu P., Wang R., Luo J., Li T., Ping R., Hu Q., Sun Y., et al. Cytokine release syndrome in COVID-19: a major mechanism of morbidity and mortality. Int. Rev. Immunol. 2021:1–14. doi: 10.1080/08830185.2021.1884248. 10.1080/08830185.2021.1884248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168. doi: 10.1016/j.cell.2020.11.025. e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryabkova V.A., Churilov L.P., Shoenfeld Y. Influenza infection, SARS, MERS and COVID-19: cytokine storm – the common denominator and the lessons to be learned. Clin. Immunol. 2021;223:108652,. doi: 10.1016/j.clim.2020.108652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato A., Nishida C., Sato-Kusubata K., Ishihara M., Tashiro Y., Gritli I., Shimazu H., Munakata S., Yagita H., Okumura K., et al. Inhibition of plasmin attenuates murine acute graft-versus-host disease mortality by suppressing the matrix metalloproteinase-9-dependent inflammatory cytokine storm and effector cell trafficking. Leukemia. 2015;29:145–156. doi: 10.1038/leu.2014.151. [DOI] [PubMed] [Google Scholar]
- 51.Shimazu H., Munakata S., Tashiro Y., Salama Y., Dhahri D., Eiamboonsert S., Ota Y., Onoda H., Tsuda Y., Okada Y., et al. Pharmacological targeting of plasmin prevents lethality in a murine model of macrophage activation syndrome. Blood. 2017;130:59–72. doi: 10.1182/blood-2016-09-738096. [DOI] [PubMed] [Google Scholar]
- 52.Olwal C.O., Nganyewo N.N., Tapela K. Djomkam zune, A.L.; owoicho, O.; bediako, Y.; Duodu, S. Parallels in sepsis and COVID-19 conditions: implications for managing severe COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.602848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bucova M., Majernikova B., Durmanova V., Cudrakova D., Gmitterova K., Lisa I., Klimova E., Kluckova K., Buc M. HMGB1 as a potential new marker of disease activity in patients with multiple sclerosis. Neurological sciences. Off. J. Italian Neurol. Soc. Italian Soc. Clin. Neurophysiol. 2019 doi: 10.1007/s10072-019-04136-3. 10.1007/s10072-019-04136-3. [DOI] [PubMed] [Google Scholar]
- 54.Wei J., Alfajaro M.M., Deweirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.-M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91. doi: 10.1016/j.cell.2020.10.028. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauter D., Kirchhoff F. Evolutionary conflicts and adverse effects of antiviral factors. eLife. 2021;10 doi: 10.7554/elife.65243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628,. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chow K.T., Gale M., Jr., Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 60.SFunabiki M., Kato H., Miyachi Y., Toki H., Motegi H., Inoue M., Minowa O., Yoshida A., Deguchi K., Sato H., et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S., Chu C., Wu Z., Liu F., Xie J., Yang Y., Qiu H. IFIH1 contributes to M1 macrophage polarization in ARDS. Front. Immunol. 2021;11 doi: 10.3389/fimmu.2020.580838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Downes K., Pekalski M., Angus K.L., Hardy M., Nutland S., Smyth D.J., Walker N.M., Wallace C., Todd J.A. Reduced expression of IFIH1 is protective for type 1 diabetes. PloS One. 2010;5:e12646,. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lincez P.J., Shanina I., Horwitz M.S. Reduced expression of the MDA5 gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 64.Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z., et al. New-onset diabetes in covid-19. N. Engl. J. Med. 2020;383:789–790. doi: 10.1056/nejmc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer H., Tschachler E., Eckhart L. Pangolins lack IFIH1/MDA5, a cytoplasmic RNA sensor that initiates innate immune defense upon coronavirus infection. Front. Immunol. 2020;11:939,. doi: 10.3389/fimmu.2020.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam T.T.-Y., Jia N., Zhang Y.-W., Shum M.H.-H., Jiang J.-F., Zhu H.-C., Tong Y.-G., Shi Y.-X., Ni X.-B., Liao Y.-S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 67.Carey C.M., Govande A.A., Cooper J.M., Hartley M.K., Kranzusch P.J., Elde N.C. Recurrent loss-of-function mutations reveal costs to OAS1 antiviral activity in primates. Cell Host Microbe. 2019;25:336–343. doi: 10.1016/j.chom.2019.01.001. e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davalos D., Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin. Immunopathol. 2012;34:43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 69.Butenas S., Orfeo T., Mann K.G. Tissue factor in coagulation. Arterioscler. Thromb. Vasc. Biol. 2009;29:1989–1996. doi: 10.1161/atvbaha.108.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pawlinski R., Wang J.-G., Owens A.P., Williams J., Antoniak S., Tencati M., Luther T., Rowley J.W., Low E.N., Weyrich A.S., et al. Hematopoietic and nonhematopoietic cell tissue factor activates the coagulation cascade in endotoxemic mice. Blood. 2010;116:806–814. doi: 10.1182/blood-2009-12-259267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu A., Nair K.S. Age effect on fibrinogen and albumin synthesis in humans. Am. J. Physiol. 1998;275:E1023–E1030. doi: 10.1152/ajpendo.1998.275.6.E1023. [DOI] [PubMed] [Google Scholar]
- 72.Gruenewald T.L., Seeman T.E., Ryff C.D., Karlamangla A.S., Singer B.H. Combinations of biomarkers predictive of later life mortality. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14158–14163. doi: 10.1073/pnas.0606215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji H.L., Zhao R., Matalon S., Matthay M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020 doi: 10.1152/physrev.00013.2020. 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maas C. Plasminflammation—an emerging pathway to bradykinin production. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baker S.K., Chen Z.L., Norris E.H., Revenko A.S., MacLeod A.R., Strickland S. Blood-derived plasminogen drives brain inflammation and plaque deposition in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E9687–E9696. doi: 10.1073/pnas.1811172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eslamifar Z., Behzadifard M., Soleimani M., Behzadifard S. Coagulation abnormalities in SARS-CoV-2 infection: overexpression tissue factor. Thromb. J. 2020;18 doi: 10.1186/s12959-020-00250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warr T.A., Rao L.V., Rapaport S.I. Disseminated intravascular coagulation in rabbits induced by administration of endotoxin or tissue factor: effect of anti-tissue factor antibodies and measurement of plasma extrinsic pathway inhibitor activity. Blood. 1990;75:1481–1489. [PubMed] [Google Scholar]
- 78.Colarusso C., Terlizzi M., Pinto A., Sorrentino R. A lesson from a saboteur: high-MW kininogen impact in coronavirus-induced disease 2019. Br. J. Pharmacol. 2020;177:4866–4872. doi: 10.1111/bph.15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van De Veerdonk F.L., Netea M.G., Van Deuren M., Van Der Meer J.W., De Mast Q., Brüggemann R.J., Van Der Hoeven H. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. eLife. 2020;9 doi: 10.7554/elife.57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the Airways of murine models after coronavirus infection. J. Virol. 2019;93 doi: 10.1128/jvi.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32:108175,. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Voinsky I., Gurwitz D. Smoking and COVID -19: similar bronchial ACE2 and TMPRSS2 expression and higher TMPRSS4 expression in current versus never smokers. Drug Dev. Res. 2020;81:1073–1080. doi: 10.1002/ddr.21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aitken R.J. COVID-19 and human spermatozoa—potential risks for infertility and sexual transmission? Andrology. 2021;9:48–52. doi: 10.1111/andr.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kam Y.-W., Okumura Y., Kido H., Ng L.F.P., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by Airway proteases enhances virus entry into human bronchial epithelial cells in. Vitro. PLoS ONE. 2009;4:e7870,. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abouhashem A.S., Singh K., Azzazy H.M., Sen C.K. Is low alveolar type II cell SOD3 in the lungs of elderly linked to the observed severity of COVID-19? Antioxidants Redox Signal. 2020 doi: 10.1089/ars.2020.8111. 10.1089/ars.2020.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hernandez-Saavedra D., Swain K., Tuder R., Petersen S.V., Nozik-Grayck E. Redox regulation of the superoxide dismutases SOD3 and SOD2 in the pulmonary circulation. Adv. Exp. Med. Biol. 2017;967:57–70. doi: 10.1007/978-3-319-63245-2_5. [DOI] [PubMed] [Google Scholar]
- 89.Kropski J.A., Blackwell T.S. Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. J. Clin. Invest. 2018;128:64–73. doi: 10.1172/jci93560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka K.I., Ishihara T., Azuma A., Kudoh S., Ebina M., Nukiwa T., Sugiyama Y., Tasaka Y., Namba T., Ishihara T., et al. Therapeutic effect of lecithinized superoxide dismutase (PC-SOD) on bleomycin-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2009 doi: 10.1152/ajplung.00289.2009. [DOI] [PubMed] [Google Scholar]
- 91.Tanaka K.-I., Tanaka Y., Miyazaki Y., Namba T., Sato K., Aoshiba K., Azuma A., Mizushima T. Therapeutic effect of lecithinized superoxide dismutase on pulmonary emphysema. J. Pharmacol. Exp. Therapeut. 2011;338:810–818. doi: 10.1124/jpet.111.179051. [DOI] [PubMed] [Google Scholar]
- 92.Guan S.P., Seet R.C.S., Kennedy B.K. Does eNOS derived nitric oxide protect the young from severe COVID-19 complications? Ageing Res. Rev. 2020;64:101201,. doi: 10.1016/j.arr.2020.101201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lotz C., Muellenbach R.M., Meybohm P., Mutlak H., Lepper P.M., Rolfes C.B., Peivandi A., Stumpner J., Kredel M., Kranke P., et al. Effects of inhaled nitric oxide in COVID-19–induced ARDS – is it worthwhile? Acta Anaesthesiol. Scand. 2020 doi: 10.1111/aas.13757. 10.1111/aas.13757. [DOI] [PubMed] [Google Scholar]
- 94.Cernadas M.A.R., De Miguel L.S.N., GarcíA-DuráN M., GonzáLez-FernáNdez F., MilláS I., MontóN M., Rodrigo J., Rico L., FernáNdez P., De Frutos T., et al. Expression of constitutive and inducible nitric oxide synthases in the vascular wall of young and aging rats. Circ. Res. 1998;83:279–286. doi: 10.1161/01.res.83.3.279. [DOI] [PubMed] [Google Scholar]
- 95.Fang W., Jiang J., Su L., Shu T., Liu H., Lai S., Ghiladi R.A., Wang J. The role of NO in COVID-19 and potential therapeutic strategies. Free Radic. Biol. Med. 2021;163:153–162. doi: 10.1016/j.freeradbiomed.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bougaki M., Searles R.J., Kida K., Yu J., Buys E.S., Ichinose F. Nos3 protects against systemic inflammation and myocardial dysfunction in murine polymicrobial sepsis. Shock. 2010;34:281–290. doi: 10.1097/SHK.0b013e3181cdc327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korhonen R., Lahti A., Kankaanranta H., Moilanen E. Nitric oxide production and signaling in inflammation. Curr. Drug Targets - Inflamm. Allergy. 2005;4:471–479. doi: 10.2174/156801005452635f9. [DOI] [PubMed] [Google Scholar]
- 98.Kumar V., Dhanjal J.K., Bhargava P., Kaul A., Wang J., Zhang H., Kaul S.C., Wadhwa R., Sundar D. Withanone and withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J. Biomol. Struct. Dyn. 2020:1–27. doi: 10.1080/07391102.2020.1775704. 10.1080/07391102.2020.1775704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kumar V., Dhanjal J.K., Kaul S.C., Wadhwa R., Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1772108. 10.1080/07391102.2020.1772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shawan M.M.A.K., Halder S.K., Hasan M.A. Luteolin and abyssinone II as potential inhibitors of SARS-CoV-2: an in silico molecular modeling approach in battling the COVID-19 outbreak. Bull. Natl. Res. Cent. 2021;45 doi: 10.1186/s42269-020-00479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rai H., Barik A., Singh Y.P., Suresh A., Singh L., Singh G., Nayak U.Y., Dubey V.K., Modi G. Molecular docking, binding mode analysis, molecular dynamics, and prediction of ADMET/toxicity properties of selective potential antiviral agents against SARS-CoV-2 main protease: an effort toward drug repurposing to combat COVID-19. Mol. Divers. 2021 doi: 10.1007/s11030-021-10188-5. 10.1007/s11030-021-10188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]