Abstract
A synthetic 22-mer peptide (peptide 46) derived from the p53 C-terminal domain can restore the growth suppressor function of mutant p53 proteins in human tumor cells (G. Selivanova et al., Nat. Med. 3:632–638, 1997). Here we demonstrate that peptide 46 binds mutant p53. Peptide 46 binding sites were found within both the core and C-terminal domains of p53. Lys residues within the peptide were critical for both p53 activation and core domain binding. The sequence-specific DNA binding of isolated tumor-derived mutant p53 core domains was restored by a C-terminal polypeptide. Our results indicate that C-terminal peptide binding to the core domain activates p53 through displacement of the negative regulatory C-terminal domain. Furthermore, stabilization of the core domain structure and/or establishment of novel DNA contacts may contribute to the reactivation of mutant p53. These findings should facilitate the design of p53-reactivating drugs for cancer therapy.
The p53 tumor suppressor protein functions as a transcriptional activator that binds specifically to double-stranded DNA (for a review, see reference 15). p53 is normally expressed at low levels in a latent form that is unable to bind specifically to DNA. Under stress conditions, such as DNA damage, hypoxia, and oncogene activation, p53 is activated; it accumulates due to protein stabilization, leading to initiation of the p53-dependent biological response, i.e., cell cycle arrest and apoptosis. The N-terminal region of the p53 protein (residues 1 to 42) contains an acidic transactivation domain (24), and the hydrophobic core domain, corresponding to residues 102 to 292, has sequence-specific DNA binding activity (3, 8, 18, 25). The C-terminal region harbors a tetramerization domain (5, 21), plus a region that regulates specific DNA binding by the core domain (9), and binds single-stranded DNA ends (2).
The ability of p53 to activate transcription from promoters containing p53 binding sites is tightly linked to its tumor suppressor function. Most mutant p53 proteins found in human tumors have amino acid substitutions in the core domain, affecting either residues that are involved in direct contacts with DNA, e.g., Arg 248 and Arg 273 (DNA contact mutations), or residues that are important for maintaining the structural integrity of the core domain, e.g., Arg 175 and Arg 249 (structural mutations) (4). As a result, these mutant proteins are partially or completely deficient for specific DNA binding (reviewed in reference 15).
Activation of p53 in response to, for instance, DNA damage appears to involve a conformational shift that activates the specific DNA binding. Latent p53 can be activated for specific DNA binding in vitro by various manipulations of its C-terminal domain, including deletion of the last 30 residues, binding of monoclonal antibody PAb421, phosphorylation (9, 10, 11), and acetylation (6), suggesting a role for the C terminus as a negative regulator of p53 activity. This idea is further supported by the findings that short peptides derived from the C-terminal domain of p53 (residues 369 to 383) can activate specific DNA binding (1, 12, 20, 22). According to the allosteric model for regulation of p53 activity (11, 12), a putative interaction between the C-terminal domain and another region in p53 in a p53 tetramer locks the tetramer in a DNA binding-incompetent state. The addition of an excess of C-terminal peptide, as well as binding of antibody PAb421, C-terminal phosphorylation, or acetylation, would displace the C terminus from its binding site in the p53 molecule and thus allow specific DNA binding by the core domains in a tetramer.
Our previous observation that a synthetic peptide derived from the p53 C terminus (peptide 46) can restore the transactivation and growth suppression function of mutant p53 proteins in vivo raised the question of the mechanism of peptide 46-mediated p53 activation (20). While the current allosteric model can explain activation of wild-type p53 by the C-terminal peptide, the mechanism behind peptide-mediated reactivation of mutant p53 proteins carrying substitutions of residues that directly contact DNA or residues important for the structural stability of the core domain remains unclear. Understanding the molecular basis of peptide 46-mediated restoration of mutant p53 function is essential for the development of drugs that can reactivate mutant p53 protein in human tumors. Here we demonstrate that the ability of peptide 46 to activate p53 correlates with its ability to bind p53. Peptide 46-interacting regions were identified within both the core and C-terminal domains of p53. We present evidence that the direct binding of the C-terminal polypeptide, added in trans, to the isolated mutant core domains restores their specific DNA binding.
MATERIALS AND METHODS
Plasmid constructs.
The plasmids encoding the glutathione S-transferase (GST)–human wild-type p53 fusion protein and the deletion fusion proteins GST-p53(1-100), GST-p53(99-307), GST-p53(320-393), and GST-p53Δ30, have been described elsewhere (2, 19). GST-mutant p53 core domain constructs were generated by cloning PCR fragments amplified from mutant human p53 cDNAs (p53 His-273, p53 Trp-248, p53 Ala-143, p53 His-175, and p53 Ser-249) into the BamHI/EcoRI sites of the pGEX-2T vector (Pharmacia Biotech, Uppsala, Sweden). DNA sequencing confirmed the absence of additional mutations in the GST constructs. Plasmids encoding the GST–p53-I370/372/373 and GST–p53-I381/382/386 C-terminal mutant proteins were constructed by replacing the fragment encoding the C-terminal domain of the wild-type GST-p53 protein with the StuI/SalI fragment from pALTER plasmids encoding the p53-I370/372/373 and p53-I381/382/386 mutants, provided by Karen Vousden, Frederick Cancer Research and Development Center, Frederick, Md. The chloramphenicol acetyltransferase (CAT) reporter plasmid PG-CAT containing 13 repeats of a p53 consensus site (14), the wild-type p53 expression plasmid pC53-SN3, and the mutant p53 cDNA plasmids were gifts from Bert Vogelstein, Johns Hopkins Oncology Center, Baltimore, Md.
Synthetic peptides.
The 22-mer synthetic peptides 43 to 48 span residues 337 to 393 of the p53 C terminus. Each peptide is shifted by eight amino acid residues relative to the previous one. Peptides were synthesized by the Merrifield solid-phase method or purchased from Genosys (Cambridge, England). Purification by high-pressure liquid chromatography was performed on a Super Pac Pep-S column. The sequence of peptide 46, which corresponds to residues 361 to 382 in the p53 C terminus, is (in one-letter code) GSRAHSSHLKSKKGQSTSRHKK. Mutant derivatives of peptide 46 contained Ala substitutions for Lys 370 in peptide S21; Ser 371 in peptide S22; His 380 in peptide S30; Lys 370, 372, 373, 381, and 382 in peptide S33; and residues 363, 365, 368, 370, 372, and 373 in peptide S35 and Ile substitutions for Lys 370, 372, and 373 in peptide A3 and Lys 381 and 382 in peptide A4.
Transfections and CAT assays.
Transactivation assays using p53-responsive promoter constructs linked to the CAT reporter gene (PG-CAT) were performed as described previously (20). The wild-type p53 expression plasmid pC53-SN3 (0.5 μg) was cotransfected with the CAT reporter plasmid PG-CAT (2 μg) and β-galactosidase expression vector pRSV-β-gal (0.5 μg), with or without synthetic peptides (5 to 20 μg), using Lipofectamine (Gibco BRL). CAT activity was assayed 48 h after transfection. Transfection efficiency was quantified by measuring β-galactosidase activity in cell extracts.
DNA binding assays.
The GST-p53 proteins and baculovirus-produced p53 protein were prepared as described elsewhere (19, 20). Band shift assays were performed in binding buffer containing 100 mM HEPES (pH 7.5), 50 mM KCl, 1 mg of bovine serum albumin (BSA) per ml, 0.1% Triton X-100, 2 mM MgCl2, and 1 mM dithiothreitol essentially as described elsewhere (19). For the preparative band shift assay, 200 ng each of the GST-p53(99-307) and GST-p53(320-393) proteins and 5 ng of labeled BC oligonucleotide containing a consensus p53 binding site (7) were used. Bands corresponding to DNA-protein complexes were visualized by autoradiography and cut out from the gel. After boiling in sample buffer to denature proteins, the gel slices were put on top of sodium dodecyl sulfate (SDS)–8% polyacrylamide gels and subjected to a second electrophoresis. Western blot analysis was performed as outlined elsewhere (2).
Protein-protein and peptide-protein interaction assays.
Peptides were biotinylated by using biotinylation reagent (Amersham, Little Chalfont, England) as described by the manufacturer and immobilized on streptavidin-coated Dynabeads (Dynal AS, Oslo, Norway). To isolate proteins from SW480 colon carcinoma or Saos-2-His-273 cells (20) by using Dynabeads with immobilized peptides, cells were lysed in a buffer containing 1% Nonidet P-40, 100 mM HEPES (pH 7.5), 100 mM KCl, and 1 mM dithiothreitol and incubated with Dynabeads loaded with peptides for 1 h at room temperature. Dynabeads were blocked with 2% BSA in phosphate-buffered saline prior to the reaction. After three subsequent washings with binding buffer, proteins were eluted from the beads by boiling in Laemmli sample buffer and separated by SDS-polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose and probed with the p53-specific antibodies DO-1, PAb240, and PAb421 (Oncogene Research Products, Cambridge, Mass.) as described elsewhere (2). GST-p53 fusion proteins representing the N-terminal, core, and C-terminal p53 domains were assayed for peptide 46 binding by using the same conditions except that the reactions were carried out in the presence of 2% BSA.
For analysis of peptide-protein interactions in native gel mobility shift assays, peptides and proteins were incubated for 30 min at room temperature in DNA binding buffer (see above) containing 2% BSA. GST-p53 proteins and peptides were phosphorylated with [γ-32P]ATP and heart muscle kinase (Sigma, St. Louis, Mo.), using the conditions recommended by the manufacturer. Peptide-protein and protein-protein complexes were resolved on a 4% native polyacrylamide gel and visualized by autoradiography under the same conditions as used for detection of DNA-protein complexes in band shift assays (see above).
For the GST pull-down assays, 100 ng of 32P-labeled peptide 46 was incubated with the GST-p53 proteins (100 ng) immobilized on glutathione-Sepharose at 20°C for 30 min in DNA binding buffer containing 2% BSA. After washing with binding buffer, peptides retained on the beads were eluted by boiling in sample buffer and separated on an SDS–15% polyacrylamide gel. Quantitation was carried out by PhosphoImager (Molecular Dynamics, Sunnyvale, Calif.) analysis.
RESULTS
Peptide 46 interacts with the p53 protein in cell extracts.
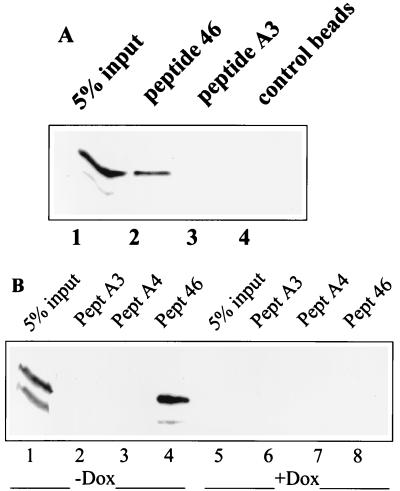
We have previously shown that peptide 46, corresponding to residues 361 to 382 of the p53 C terminus, can restore the growth suppression function of at least some mutant p53 proteins in human tumor cells (20). These findings raised the possibility that peptide 46-mediated activation of mutant p53 occurs through a direct interaction of the peptide with p53. To address this question, biotinylated peptide 46 immobilized on streptavidin-coated Dynabeads was incubated with lysates of SW480 colon carcinoma cells carrying His-273 mutant p53. After subsequent washing of the beads, bound proteins were analyzed by Western blotting. As shown in Fig. 1A, lane 2, peptide 46 efficiently precipitated the His-273 mutant p53 protein from the SW480 cell lysate. Control beads without peptide did not bring down p53 (Fig. 1A, lane 4). Furthermore, peptide A3, a mutant version of peptide 46 carrying Ile substitutions for Lys residues corresponding to positions 370, 372, and 373 in the p53 C-terminal domain that are essential for peptide 46-mediated activation of p53 in cells (20), failed to bind p53 (Fig. 1A, lane 3).
FIG. 1.
Peptide 46 binds to the His-273 mutant p53 protein in a cellular extract. (A) Biotinylated peptide 46 and its mutant derivative peptide A3, carrying Ile substitutions for Lys residues 370, 372, and 373, were immobilized on streptavidin-coated Dynabeads and used for precipitation of p53 from an SW480 cell extract. Bound proteins were analyzed by Western blotting using the p53-specific monoclonal antibody DO-1. Peptide 46 precipitated mutant p53 protein from the SW480 cell extract (lane 2), whereas no p53 was precipitated by control peptide A3 (lane 3) or Dynabeads without peptide (lane 4). Lane 1, 5% of input of cell lysate. (B) Western blot analysis of p53 precipitated from Saos-2-His-273 cells grown in the absence (p53 on; lanes 1 to 4) or presence (p53 off; lanes 5 to 8) of doxycycline. Peptide 46 (lane 4), but neither the mutant control peptide A3 (lane 2), peptide A4 that carries Ile substitutions for Lys residues 381 and 382 (lane 3), nor unloaded Dynabeads (not shown), precipitated p53. Lanes 1 and 5, 5% of input of lysate from cells grown in the absence and presence of doxycycline, respectively. Experimental conditions were as for panel A.
Similar results were obtained for a subline of Saos-2 cells expressing His-273 mutant p53 under the control of a tetracycline-repressible promoter (20). Peptide 46, but not mutant peptide A3 or A4 or control beads without peptide, bound the His-273 mutant p53 protein from Saos-2-His-273 cells grown in the absence of doxycycline (Fig. 1B). As expected, no p53 was precipitated in the presence of doxycycline, which downregulates His-273 p53 expression. Taken together, these results demonstrate that peptide 46 is able to bind the p53 protein even in the presence of a multitude of cellular proteins.
Peptide 46 binds to both the core domain and the C-terminal domain of p53.
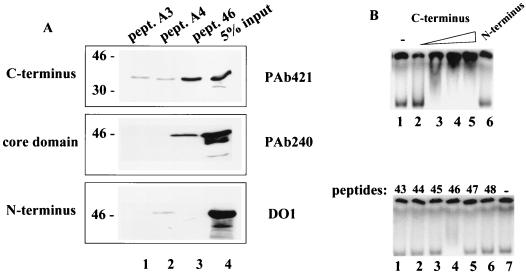
To map the region of peptide 46 binding in the p53 protein, we tested peptide 46 binding to GST-p53 fusion proteins representing the N-terminal [GST-p53(1-100)], core [GST-p53(99-307)], and C-terminal [GST-p53(320-393)] domains of p53. The core and C-terminal domains were efficiently precipitated by the biotinylated peptide 46 bound to streptavidin-coated beads (Fig. 2A). The N-terminal domain showed only weak binding (less than 1% of input, visible after longer exposure), whereas the GST protein alone did not bind at all (data not shown). The A3 and A4 mutant control peptides carrying Ile substitutions of Lys residues did not bring down the core domain and were less efficient in binding the C-terminal domain.
FIG. 2.
Peptide 46 interacts with the core and C-terminal domains of p53. (A) Biotinylated peptide 46 or its mutant derivative peptides A3 and A4, carrying Ile substitutions for Lys 370, 372, and 373 and of Lys 381 and 382, respectively, were immobilized on streptavidin-coated Dynabeads and incubated with p53 deletion mutant proteins representing the N-terminal [GST-p53(1-100)], core [GST-p53(99-307)], and C-terminal [GST-p53(320-393)] domains. Bound proteins were eluted and analyzed by Western blotting using the p53-specific monoclonal antibodies PAb421 (top), PAb240 (middle), and DO-1 (bottom) to detect the C-terminal, core, and N-terminal domain polypeptides, respectively. (B) The 32P-labeled GST-p53(99-307) core domain protein was incubated with increasing amounts (100 and 200 ng) of the GST-p53(320-393) C-terminal domain protein (lanes 2 to 5, upper panel), 200 ng of the GST-p53(1-100) N-terminal domain protein (lane 6, upper panel), or a series of overlapping synthetic peptides spanning the p53 C terminus (lower panel). The protein-protein and peptide-protein complexes were separated by native polyacrylamide gel electrophoresis and visualized by autoradiography.
The interaction between peptide 46 and the p53 domains was further examined in a native gel mobility shift assay. This assay takes advantage of the fact that protein-protein complexing causes a shift in the mobility of a protein in a native polyacrylamide gel, analogous to the mobility shift of a DNA probe bound to protein. Incubation of 32P-labeled GST-p53(99-307), corresponding to the core domain, with increasing concentrations of GST-p53(320-393), corresponding to the C-terminal domain, prior to loading shifted the mobility of the core domain (Fig. 2B, upper panel, lanes 3 to 5), indicating complexing between the core domain and the C-terminal domain.
A series of overlapping synthetic peptides spanning the p53 C terminus was also tested for binding to the core domain in the native gel mobility shift assay. Peptide 46, the only C-terminal peptide that can activate the specific DNA binding of p53 (data not shown), changed the mobility of the core domain, whereas the other C-terminal peptides did not (Fig. 2B, lower panel). Conversely, the mobility of the C-terminal domain protein was shifted upon incubation with the core domain protein (data not shown).
The ability of peptide 46 to bind to the core and C-terminal domains of p53 was further confirmed in pull-down experiments using the GST-p53(99-307) and GST-p53(320-393) proteins and 32P-labeled peptide 46 (data not shown).
C-terminal Lys residues at positions 370, 372, 373, 381, and 382 are critical for the interaction with the core domain.
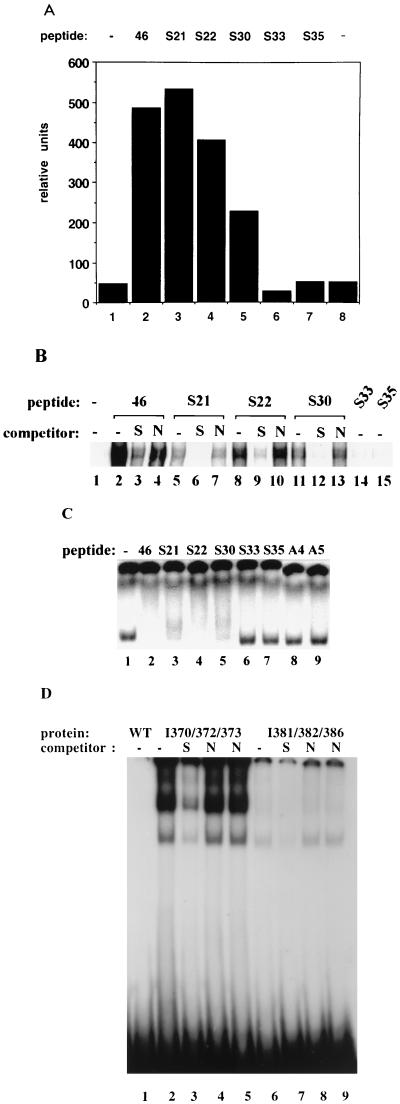
We next analyzed a series of mutant peptides to identify residues that are critical for core domain binding, activation of specific DNA binding, and stimulation of p53-mediated transactivation of a reporter gene. Ala substitution for Ser 371 did not significantly affect the activation properties of peptide S22 or its ability to bind to the core domain (Fig. 3A to C, lanes 3). A single Ala substitution for Lys 370 or His 380 (peptide S21 or S30, respectively) resulted in a reduced ability of the peptide to activate p53 in DNA binding (Fig. 3B) and/or in CAT reporter assays (Fig. 3A) but did not markedly affect core domain binding (Fig. 3C, lanes 3 and 5). However, simultaneous replacement of more than one Lys residue completely abolished the ability of the peptide to stimulate both specific DNA binding and p53-mediated transactivation (Fig. 3A and B). Peptide S33, carrying Ala substitutions for Lys 370, 372, 373, 381, and 382, and peptide S35, carrying Ala substitutions for Lys 370, 372, and 373, Arg 363, His 365, and His 368, activated neither specific DNA binding of p53 (Fig. 3B) nor p53-mediated transcription (Fig. 3A), in correlation with their inability to bind the core domain (Fig. 3C). Moreover, peptides A3 and A4, carrying Ile substitutions for Lys residues at positions 370, 372, and 373 and at positions 381 and 382, respectively, failed to bind the core domain and to activate p53 for specific DNA binding (Fig. 3C and 2A and data not shown). Thus, Lys residues in peptide 46 are critical both for the activation of p53 and for binding to the core domain. These results demonstrate that the ability of peptides to interact with the core domain correlates with their ability to activate specific DNA binding and transcriptional transactivation by p53.
FIG. 3.
Lysine residues at positions 370, 372, 373, 381, and 382 are critical for both p53 activation and core domain binding by peptide 46. (A) Plasmid PG-CAT was cotransfected with a wild-type p53 expression plasmid and a series of mutant derivatives of peptide 46. Ala substitution for Lys residues at positions 370, 372, 381, and 382 in peptide S33 or for residues at positions 363, 365, 368, 370, 372, and 373 in peptide S35 abolished the ability of peptide 46 to stimulate p53-mediated CAT reporter gene activation (columns 6 and 7), whereas the single Ala substitutions in peptides S21 (Lys 370), S22 (Ser 371), and S30 (His 380) had little or no effect (columns 3 to 5). (B) Mutant derivatives of peptide 46 were tested for the ability to activate the specific DNA binding of baculovirus- produced p53 in a band shift assay as described in Materials and Methods. Peptides S21, S22, and S30, carrying single Ala substitutions, were able to stimulate specific DNA binding of p53, although with variable efficiency, whereas peptides S33 and S35, carrying Ala substitutions for several Lys residues, were inactive. The sequence specificity of p53 DNA binding induced by the peptides was demonstrated by competition of p53-DNA complexing with specific (S) oligonucleotide BC (lanes 3, 6, 9, and 12) but not with nonspecific (N) control oligonucleotide MN (lanes 4, 7, 10, and 13). (C) The 32P-labeled GST-p53(99-307) core domain protein was incubated with synthetic peptides and analyzed in a native gel mobility shift assay as described for Fig. 2B. Peptides 46, S21, S22, and S30, but not peptides S33, S35, A3, and A4, caused a shift in the migration of the core domain protein on the native gel. (D) GST–full-length p53 proteins carrying Ile substitutions for Lys residues at positions 370, 372, and 373 (I370/372/373; lanes 2 to 5) and 381, 382, and 386 (I381/382/386; lanes 6 to 9) were tested for specific DNA binding in a band shift assay as described for panel B. Lane 1, GST–wild-type (WT) p53 protein. The specific oligonucleotide BC (S) competed out p53-DNA complexes (lanes 3 and 7), whereas the nonspecific (N) control oligonucleotide MN with either blunt (lanes 4 and 8) or protruding (lanes 5 and 9) ends did not compete.
The allosteric model for regulation of p53 activity predicts that disruption of the interaction between the C-terminal domain and the core domain will activate specific DNA binding by the core domain. Since replacement of Lys residues at positions 370, 372, and 373 or at positions 381 and 382 abolished peptide binding to the core domain, we asked whether the same substitutions in the context of the full-length p53 protein would disrupt the C terminus-core domain interaction and cause activation of specific DNA binding. GST-p53 proteins carrying Ile substitutions corresponding to the substitutions in peptides A3 and A4 were tested for specific DNA binding in the absence of the activating antibody PAb421. The p53-I370/372/373 mutant protein, carrying Ile substitutions at positions 370, 372, and 373, showed very potent specific DNA binding (Fig. 3D, lane 2). The p53-I381/382/386 mutant protein, carrying Ile substitutions at positions 381, 382, and 386, also exhibited increased specific DNA binding compared to wild-type p53 but was significantly less active than p53-I370/372/373 (lane 6). These results indicate that the Lys residues at positions 370, 372, and 373 are the residues most important for the interaction between the C terminus and the core domain, and they support the idea that the C terminus negatively regulates specific DNA binding by interacting with the core domain.
Interaction of the isolated core domain with the C-terminal domain in trans facilitates specific DNA binding.
We next explored whether the interaction of the C-terminal peptide added in trans with the core domain is transient and serves only to displace the C-terminal domain from its binding site on the core domain or whether this interaction is rather stable and able to directly affect the DNA binding of the core domain. To discriminate between these two possibilities, we examined the specific DNA binding of the GST-p53(99-307) core domain protein in the presence of the GST-p53(320-393) C-terminal domain protein. A protein corresponding to the C-terminal p53 domain (residues 311 to 393) was shown to stimulate specific DNA binding of full-length wild-type p53 and of the p53 core domain (residues 96 to 312) when added in trans (13).
Incubation with the GST-p53(320-393) C-terminal polypeptide stimulated GST-p53(99-307) protein-DNA complexing as well (Fig. 4A, lanes 1 and 2). A new, faster-migrating complex, complex II, appeared in addition to the GST-p53(99-307)-DNA complex (complex I). To examine the specificity of a newly induced DNA binding, we used a competition assay. Protein-DNA complexes I and II were abolished by excess unlabeled specific BC oligonucleotide (lanes 3 and 4), while similar quantities of the unrelated MN control oligonucleotide failed to compete (lanes 5 and 6). Thus, both protein-DNA complexes I and II could be attributed to a specific interaction between the core domain and DNA. The GST-p53(320-393) protein did not bind DNA under the conditions used (lane 7). When increasing amounts of GST-p53(320-393) protein were added, complex I was gradually replaced with complex II in a dose-dependent manner (lanes 8 to 10).
FIG. 4.
The isolated wild-type core domain binds DNA more efficiently upon complexing with the C-terminal polypeptide. (A) Interaction of the p53 C-terminal domain protein (residues 320 to 393) with the core domain protein (residues 99 to 307) stimulated the sequence-specific DNA binding of the core domain in a band shift assay (lanes 1 and 2). Addition of the GST-p53(320-393) protein induced the formation of a new protein-DNA complex, complex II (lane 2), in a dose-dependent manner (lanes 8 to 10). Complex II was efficiently competed by a two- or fivefold excess of the specific (S) oligonucleotide BC (lanes 3 and 4) but not by the nonspecific control (contr) oligonucleotide MN (lanes 5 and 6). The GST-p53(320-393) protein alone did not bind DNA under the conditions used (lane 7). (B) The C-terminal domain protein is present in a core domain-DNA complex, as determined by Western blot analysis using the monoclonal antibodies PAb240 (upper panel) and PAb421 (lower panel) to detect proteins eluted from the region of a preparative gel corresponding to complex II (lanes 1, 3, and 5) and complex I (lanes 2, 4, and 6) after incubation of DNA with the C-terminal domain protein only (lanes 1 and 2), both the C-terminal and core domain proteins (lanes 3 and 4), and the core domain protein only (lanes 5 and 6). Lane 7, control GST-p53(320-393) protein alone; lane 8, control GST-p53(99-307) protein alone. (C) The GST-p53Δ30 protein, which lacks the 30 C-terminal amino acid residues and is constitutively activated for specific DNA binding, was further activated by peptide 46 in a band shift assay (lanes 1 and 3). Lane 1, GST-p53Δ30 in the absence of peptide 46; lane 2, GST-p53Δ30 in the presence of the activating antibody PAb421; lane 3, GST-p53Δ30 in the presence of peptide 46. The GST-p53Δ30-DNA complex induced by peptide 46 was competed by the specific (S) oligonucleotide BC (lane 4) but not by the nonspecific (N) control oligonucleotide MN (lane 5).
To analyze the protein content of complexes I and II, the regions of a preparative gel corresponding to complex I and II were cut out, and proteins were eluted and analyzed by Western blotting. The core domain protein was detected by monoclonal antibody PAb240 (Fig. 4B, upper panel), and the C-terminal domain protein was detected by monoclonal antibody PAb421 (Fig. 4B, lower panel). After incubation of both the C-terminal and core domain proteins with the DNA probe (producing mainly complex II), both proteins were detected in complex II (lane 3, upper and lower panels) but not in the region of the gel corresponding to complex I (lane 4, upper and lower panels). When only the core domain was incubated with DNA, it was detected in complex I (lane 6, upper panel) but not in the region of the gel corresponding to complex II (lane 5, upper panel). As expected, the C-terminal domain protein was not detected (lanes 5 and 6, lower panel). When only the C-terminal domain protein was incubated with DNA, neither protein was detected in the regions of the gel corresponding to complexes I and II (lanes 1 and 2, upper and lower panels). Hence, the interaction between the C-terminal domain polypeptide added in trans and the isolated core domain facilitated specific DNA binding by the core domain; moreover, the C-terminal polypeptide remained in a complex with the core domain upon binding of the latter to DNA.
This conclusion was further supported by the observation that peptide 46 was able to increase the level of specific DNA binding by the already constitutively activated p53Δ30 truncation mutant that lacks the C-terminal regulatory domain. Incubation of peptide 46 with the GST-p53Δ30 protein resulted in a robust stimulation of DNA binding as well as the appearance of a more slowly migrating complex, suggesting that the peptide remained bound to the GST-p53Δ30-DNA complexes (Fig. 4C, lane 3). Thus, p53 activation by the C-terminal peptide occurs not only as a result of displacement of the C terminus from its binding site on the core domain but also due to increased DNA binding of the core domain–C-terminal peptide complex.
The C-terminal polypeptide restores specific DNA binding of tumor-derived mutant core domains.
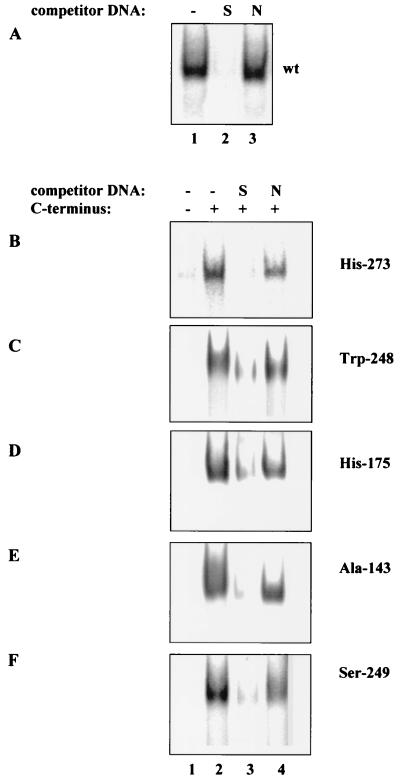
We have previously suggested that restoration of the specific DNA binding of mutant p53 proteins by the C-terminal peptide occurs through the establishment of novel core domain-DNA contacts due to the binding of the basic C-terminal peptide to the core domain (20). To test this idea, we examined the DNA binding of isolated core domains representing the most common tumor-derived mutant p53 proteins in the presence of the C-terminal domain polypeptide. GST-p53 core domain proteins carrying the tumor-derived mutations His-273 and Trp-248 (DNA contact mutants) and His-175, Ala-143, and Ser-249 (structural mutants) all failed to bind DNA in a band shift assay (Fig. 5B to F, lanes 1), whereas the wild-type core domain bound efficiently to the labeled probe (Fig. 5A, lane 1). The only exception was the His-273 mutant core domain, which showed a weak residual DNA binding ability seen only after longer exposure of the gel (not shown). Incubation of the mutant core domain proteins with the GST-p53(320-393) C-terminal polypeptide resulted in the appearance of a DNA-protein complex (Fig. 5B to F, lanes 2). Whereas 0.2 pmol of wild-type core domain was sufficient to detect complexing with DNA, 1 to 2 pmol of mutant core domain proteins and 5 pmol of the C-terminal polypeptide were required to restore DNA binding (Fig. 5B, C, E, and F). An even higher amount, 4.5 pmol, of the His-175 mutant core domain was required to detect DNA complexing (Fig. 5D). This probably reflects the fact that the His-175 mutant has the most extensive structural defects (4).
FIG. 5.
The sequence-specific DNA binding of isolated mutant core domains is restored by the C-terminal polypeptide. (A) The wild-type (wt) GST-p53(99-307) protein bound to the labeled specific BC oligonucleotide in a band shift assay (lane 1). The core domain-DNA complex was competed by a 10-fold excess of the specific (S) oligonucleotide BC (lane 2) but not by a 10-fold excess of the nonspecific (N) MN oligonucleotide (lane 3). GST-p53(99-307) proteins carrying tumor-derived mutations His-273 (B), Trp-248 (D), His-175 (D), Ala-143 (E), and Ser-249 (F) did not bind the labeled BC oligonucleotide in the absence of the C-terminal domain polypeptide (lanes 1) but formed a complex with BC in the presence of the C-terminal polypeptide (lanes 2). The DNA-mutant core domain complexes were competed out by a 10-fold excess of the unlabeled specific (S) oligonucleotide BC (lane 3) but not by the nonspecific (N) oligonucleotide MN (lane 4).
The mutant core domain-DNA complexes induced by the C-terminal polypeptide were efficiently competed by the unlabeled specific oligonucleotide BC but not by the nonspecific oligonucleotide MN, in the same way as wild-type core domain DNA binding was competed, either in the absence or in the presence of the C-terminal polypeptide (Fig. 5A, lanes 2 and 3; Fig. 5B to F, lanes 3 and 4; Fig. 4A, lanes 3 to 6). Therefore, the C-terminal polypeptide was able to restore the specific DNA binding of mutant p53 core domains.
DISCUSSION
Activation of p53 in response to genomic injury or cellular stress involves both increased protein stability and the transition from a latent to an active conformation. According to the allosteric model, the C-terminal domain binds to a site in another p53 molecule within a p53 tetramer, locking the DNA binding core domains in a latent conformation (12). C-terminal modification in vivo—for instance, acetylation (6)—would disrupt this interaction, resulting in a conformational shift that allows DNA binding by the core domains. In vitro activation of p53 by synthetic C-terminal peptides may occur as a result of competitive displacement of the p53 C terminus from its binding site. So far, however, direct evidence supporting this model is lacking. The crystal structure of the full-length p53 protein has not yet been defined, and an interaction between the C-terminal negative regulatory domain and the core or N-terminal domains of p53 has not been demonstrated.
Our results demonstrate a direct interaction between the C terminus of p53 and the p53 core domain. First, peptide 46, derived from the C terminus of p53, was able to bind p53 in cellular extracts (Fig. 1). Second, peptide 46 bound to the GST-p53(99-307) protein representing the core domain in pull-down experiments, using either the biotinylated peptide or the GST-p53(99-307) protein as a bait, and in a native gel mobility shift assay (Fig. 2 and data not shown). Third, a DNA band shift assay showed that the isolated C-terminal domain interacted with the core domain in complex with DNA (Fig. 4).
The Lys residues at positions 370, 372, 373, 381, and 382 in the p53 C terminus appeared critical for the interaction with the core domain, as shown by the observation that the two mutant peptides carrying substitutions for Lys 370, 372, and 373 or for Lys 381 and 382 failed to bind the core domain (Fig. 2A and 3C). Notably, substitutions for the same residues in the context of the full-length p53 protein (and Lys 386 in the p53-I381/382/386 mutant) resulted in the activation of specific DNA binding, analogous to the removal of the C-terminal domain by truncation. The fact that p53 mutants with substitutions for C-terminal Lys residues critical for the binding to the core domain were constitutively active for specific DNA binding provides direct evidence for the idea that the interaction of the C-terminal and core domains locks p53 in a conformation that is latent for specific DNA binding.
Binding of the GST-p53(320-393) protein representing the C-terminal domain to the GST-p53(99-307) protein representing the core domain did not inhibit specific DNA binding by the core domain. Rather, a new protein-DNA complex containing both the core and the C-terminal domain proteins appeared on the gel. This demonstrates that the complexing of the C-terminal and core domains per se does not inhibit specific DNA binding. Hence, the negative regulation of the specific DNA binding of the full-length p53 protein by the C terminus does not occur simply by steric hindrance, i.e., occupation of the DNA binding site in the core domain by the C-terminal domain. Instead, the interaction of the C-terminal domain with the core domain of either the same or the adjacent subunit within a p53 tetramer may prevent the proper orientation of the core domains required for the formation of the DNA binding interface. The existence of a flexible linker of approximately 35 residues between the core domain and the tetramerization domain (4) and a stretch of at least 10 flexible residues between the tetramerization domain and the basic C-terminal domain (16) should permit an essential degree of freedom of orientation for these domains. It is conceivable that the positioning of the C terminus on the core domain in latent p53 leads to tight packing of the linker between the core and teramerization domains. This should prevent alignment of the DNA binding core domains along the DNA. In the active state when the C terminus is not bound to the core, the linker is extended, allowing cooperative binding of the core domains to a specific DNA binding site. The shift between active and latent states is probably mediated by the tetramerization domain (8, 26).
Whereas activation of wild-type p53 by C-terminal p53 peptides could be due to displacement of the C-terminal domain from the core domain through competitive inhibition of this interaction, the restoration of the specific DNA binding and growth suppression function of mutant p53 proteins by the C-terminal peptide 46 (20) could hardly be explained by this mechanism alone. An understanding of the molecular mechanism of reactivation of mutant p53 proteins by peptide 46 is obviously of great importance for the development of p53-activating anticancer drugs. Our observation that peptide 46 binds to mutant p53 proteins within the context of multiple cellular proteins indicates that peptide-mediated activation of p53 in living cells occurs by a direct interaction between the peptide and p53. This notion is further supported by the findings that the same amino acid residues are critical for peptide-mediated activation of p53 in vitro (12; this study) (Fig. 3B) and in living cells (Fig. 3A) (20) and for binding to the core domain (Fig. 2A and 3C). Thus, amino acid substitutions in the peptide that abrogate peptide-mediated activation of p53 abolish peptide binding to the core domain, and vice versa.
We have demonstrated that the isolated wild-type core domain in complex with the C-terminal polypeptide binds DNA more efficiently than the core domain alone (Fig. 4A). Moreover, the C-terminal domain polypeptide was able to restore the specific DNA binding of isolated mutant core domains (Fig. 5B to F). This provides an important clue as to the mechanism of restoration of mutant p53 function by the C-terminal peptide. We believe that the interaction of the core domain with C-terminal peptides can facilitate the establishment of novel core domain-DNA contacts due to the net basic charge of the C-terminal peptide. This mechanism may be particularly important for the restoration of the DNA binding of DNA contact mutant p53 proteins like p53 His-273 and p53 Trp-248, as introduction of positively charged residues in the vicinity of a mutant residue can rescue specific DNA binding of mutant p53 (27). As for structural mutants such as p53 Ala-143, p53 Ser-249, and p53 His-175, it is possible that the interaction of the C-terminal peptide with the partially unfolded core domain will increase the stability of the core domain structure, resulting in proper positioning of the residues contacting DNA.
A recent study has demonstrated a positive cooperativity between the C-terminal (residues 363 to 393) and N-terminal (residues 80 to 95) domains in C-terminal peptide binding (17). Consistent with these results, we found that the binding of peptide 46 to the full-length p53 protein is much more effective than the binding of separate domains in pull-down assays (20a). Thus, it is conceivable that all three domains are involved in the formation of a peptide binding pocket in the full-length p53 protein. Under the conditions used, the interaction of peptide 46 with the separate N-terminal domain is apparently too weak to be detected (Fig. 2A). The fact that the interaction between the C-terminal peptide and the core domain, but not the interaction between the C-terminal peptide and N-terminal or C-terminal domains, is inhibited at high salt concentrations (>150 mM) (20a) may explain why this interaction was not observed by Muller-Tiemann et al. (17).
The interaction of peptide 46 with the C-terminal domain could reflect the involvement of the C-terminal basic region in the formation of p53 oligomers, as previously suggested (23). Moreover, the effective displacement of the C terminus of the full-length p53 protein from its binding pocket may require the simultaneous binding of the activating peptide to the C-terminal domain and the putative N-terminal/core domain pocket.
Based on the results described here, we suggest a dual mechanism for p53 activation by C-terminal peptides. First, the disruption of the interaction between the C terminus and a pocket formed by the core and N-terminal domains by C-terminal peptides will result in a conformational shift that allows correct positioning of the core domains for specific DNA binding. Second, the complexing of the peptide with the core domain will restore the specific DNA binding by the mutant core domain through stabilization of the core domain structure and/or establishment of additional protein-DNA contacts.
Our demonstration that the specific DNA binding of the isolated core domains representing the five most common tumor-derived mutant p53 proteins, including both DNA contact and structural mutants, was reactivated upon interaction with the C-terminal polypeptide in trans points to the feasibility of this approach for restoring the function of a broad subset of mutant p53 proteins in human tumors. Of note, even the DNA binding of the His-175 mutant core domain, which represents a significantly unfolded mutant p53 protein (4), could be restored by the C-terminal polypeptide. Of all mutant core domain proteins tested, only the His-273 mutant showed residual DNA binding. Our data thus suggest that a p53-reactivating strategy based on C-terminal peptides does not require that the mutant p53 protein have any significant residual DNA binding activity. Studies evaluating the effect of the p53 C-terminal peptide on a wide range of different mutant p53 proteins in living cells are now under way.
ACKNOWLEDGMENTS
We thank Bert Vogelstein, Johns Hopkins Oncology Center, for plasmids pC53-SN3 and PG-CAT; Moshe Oren, The Weizmann Institute, for the recombinant p53 expression baculovirus; Thierry Soussi, CNRS, Paris, France, for the p53 purification protocol; Karen Vousden, Frederick Cancer Research and Development Center, for the p53-I370/372/373 and p53-I381/382/286 mutant plasmids; and Michael Fritsche, Institute for Experimental Cancer Research, Freiburg, Germany, for the Saos-2-His-273 cells. We also thank Alexej Protopopov and Robert Skraban for help with preparation of the figures.
This work was supported by the Swedish Cancer Society, Åke Wibergs Stiftelse, and Concern Foundation for Cancer Research.
REFERENCES
- 1.Abarzua P, LoSardo J E, Subler M L, Spathis R, Lu Y-A, Felix A, Neri A. Restoration of the transcription activation function to mutant p53 in human cancer cells. Oncogene. 1996;13:2477–2482. [PubMed] [Google Scholar]
- 2.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K P, Szekely L, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 5.Clore G M, Omichinski J G, Sakagushi K, Zambrano N, Sakamoto H, Apella E, Groneborn A M. High resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994;265:386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- 6.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 7.Halazonetis T D, Davies L J, Kandil A N. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 1993;12:1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halazonetis T D, Kandil A N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 10.Hupp T R, Meek D W, Midgley C A. Activation of the cryptic DNA binding function of mutant forms of p53. Nucleic Acids Res. 1993;21:3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 12.Hupp T R, Sparks A, Lane D P. Small peptides activate the latent sequence-specific binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman L, Prives C. Activation of p53 sequence specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 14.Kern S E, Pietenpol J A, Thagaligam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–832. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 15.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 16.Lee W, Harvey T S, Yin Y, Yau P, Litchfield D, Arrowsmith C H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 17.Muller-Tiemann B F, Halazonetis T D, Elting J J. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavletich N P, Chambers K A, Pabo C O. The DNA binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 19.Selivanova G, Iotsova V, Kiseleva E, Ström M, Bakalkin G, Grafström R C, Wiman K G. The single stranded DNA end binding site of p53 coincides with the C-terminal regulatory region. Nucleic Acids Res. 1996;24:3560–3567. doi: 10.1093/nar/24.18.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selivanova G, Iotsova V, Okan I, Fritsche M, Ström M, Groner B, Grafström R C, Wiman K G. Restoration of the growth suppressor function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med. 1997;3:632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 20a.Selivanova, G., L. Ryabchenko, and K. G. Wiman. Unpublished data.
- 21.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 23.Stürzbecher H-W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 24.Unger T, Nau M M, Segal S, Minna J D. p53: a transdominant regulator of transcription whose function is ablated by mutations occurring in human cancer. EMBO J. 1992;11:1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 26.Waterman J L F, Shenk J L, Halazonetis T D. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieczorek A M, Waterman J L F, Waterman M J F, Halazonetis T D. Structure-based rescue of common tumor-derived p53 mutants. Nat Med. 1996;2:1143–1146. doi: 10.1038/nm1096-1143. [DOI] [PubMed] [Google Scholar]