Abstract
Background & Aims
Intrahepatic cholangiocarcinoma (iCCA) accounts for a fraction of primary liver cancers but has a 5-year survival rate of only 10%. Immune checkpoint inhibitors are effective in treating many solid cancers, but immune checkpoint inhibitor monotherapy has no clear benefit in iCCA. Mitogen-activated kinase (MEK) inhibitors, such as trametinib, have shown promising results in preclinical studies for iCCA by inhibiting cell proliferation and modifying the tumor microenvironment. This study aimed to show the potential benefit of combining trametinib with anti–programmed cell death protein 1 (PD-1) therapy in different iCCA mouse models.
Methods
Here, we assessed the in vitro cytotoxicity of trametinib in mouse (SB1 and LD-1) and human (EGI-1) cholangiocarcinoma cell lines. We examined the efficacy of single-agent trametinib, anti–PD-1, and a combination of both in subcutaneous, orthotopic, and plasmid-induced iCCA mouse models. Flow cytometry analysis was used to elucidate changes in the tumor immune microenvironment upon treatment. Whole-exome sequencing (WES) was performed on the SB1 tumor cell line to correlate this preclinical model with iCCAs in patients.
Results
Trametinib reduced tumor cell growth of SB1, LD-1, and EGI-1 tumor cells in vitro. Trametinib treatment led to up-regulation of major histocompatibility complex (MHC-I) and programmed cell death ligand 1 (PD-L-1) (programmed cell death ligand 1) on tumor cells in vitro. The combination of trametinib and anti–PD-1 reduced tumor burden in several iCCA tumor models and improved survival in SB1 tumor-bearing mice compared with either agent alone. Immunoprofiling of tumor-bearing mice showed an increase of hepatic effector memory CD8+ and CD4+ T cells, as well as an increased degranulation of CD8+ T cells, indicating enhanced cytotoxicity. WES and somatic mutational analysis showed no mutations of KRAS, BRAF, and ERK in SB1 tumor cells, and showed a similar genetic signature of SB1 found in a cohort of patients with iCCA.
Conclusions
Altogether, our study shows that trametinib improves the immunogenicity of tumor cells by up-regulating MHC-I surface expression. The combination with anti–PD-1 results in optimal treatment efficacy for iCCA. WES of SB1 cells suggests that KRAS wild-type iCCAs also respond to this combination therapy.
Keywords: Intrahepatic Cholangiocarcinoma, Anti–PD-1 Monoclonal Antibody, Trametinib
Abbreviations used in this paper: IC50, median inhibitory concentration; iCCA, intrahepatic cholangiocarcinoma; ICI, immune checkpoint inhibitor; IFNγ, interferon γ; KRASwt, KRAS wild-type; MHC-1, major histocompatibility complex; PBS, phosphate-buffered saline; PD-1, programmed cell death protein 1; PD-L1, programmed cell death ligand 1; WES, whole-exome sequencing
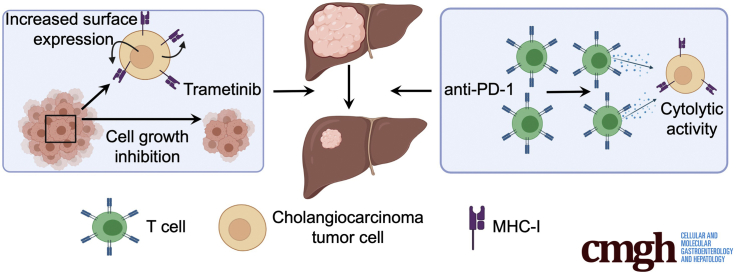
Graphical abstract
Summary.
Immune checkpoint inhibitors are not available for intrahepatic cholangiocarcinoma. Here, we show that trametinib improves the immunogenicity of tumor cells, leading to a robust response of the combination of trametinib and anti–PD-1 in different preclinical models.
See editorial on page 1153.
Intrahepatic cholangiocarcinoma (iCCA) accounts for 15% of primary liver cancer diagnoses and is associated with a dismal prognosis and a 5-year survival rate of only 10%.1,2 The worldwide incidence of iCCA is approximately 1.6 per 100,000 people.3 Although most cases of iCCA arise in individuals without risk factors, identifiable risk factors include hepatitis C virus infection, alcoholic liver disease, inflammatory bowel disease, and primary sclerosing cholangitis.4 Surgical resection is the primary curative therapy for iCCA, yet only 20%–40% of patients are eligible for surgical resection.5 Liver transplantation is a potential option for both patients with unresectable early disease and for those who respond to neoadjuvant strategies,6 but many patients do not meet criteria. If the disease is inoperable but remains intrahepatic, locoregional therapies, such as radiofrequency ablation or transarterial chemoembolization, can be used.7 Systemic therapies for unresectable disease are gemcitabine-based but have limited efficacy, with a median survival of just 12 months.7 Recently, immune checkpoint inhibitors (ICIs) have shown promising results for patients with hepatocellular carcinoma, but the role of ICIs in iCCA is largely unknown.8,9 As discussed in a review by Loeuillard et al,10 ICI monotherapy has limited efficacy for most patients with iCCA and studies are ongoing to explore their potential combination with locoregional therapies, immune microenvironment modulators, and molecular target therapies.
Mutations in the RAS–RAF–MEK–ERK pathway are common mechanisms of progression for several types of cancer. Although KRAS is one common mutated site in iCCA, KRAS mutations are found in only 18% of iCCAs.11 Mutant KRAS leads to the sustained downstream activity of MEK (mitogen-activated protein kinase/ERK kinase) proteins. MEK inhibitors, such as trametinib and cobimetinib, are approved to treat BRAF mutated melanoma and non–small-cell lung cancer,12,13 and are being investigated in many other cancer types, including biliary tract cancers. A recent study in mice suggested that KRAS wild-type (KRASwt) tumors also responded to MEK inhibition by modulating the tumor microenvironment.14 Taken together, MEK inhibitor and ICI monotherapies have not shown broad efficacy in iCCA.
Several clinical trials are ongoing to investigate the efficacy of MEK inhibitors and ICIs in gastrointestinal cancers, including pancreatic and colon cancer.15 Nevertheless, no data currently are available for the combination treatment of MEK inhibitors and ICIs in iCCA, but 2 clinical trials are ongoing that combine ICIs and MEK inhibitors for the unresectable, advanced biliary tract cancers (NCT02586987 and NCT03201458).
We investigated the effect of trametinib in combination with anti–PD-1 monoclonal antibody to treat iCCA in mice. We observed that trametinib led to up-regulation of major histocompatibility complex I (MHC-I) and programmed cell death ligand 1 PD-L1 on tumor cells, potentiating lymphocyte recognition when combined with anti–programmed cell death protein 1 (PD-1). The combination of trametinib and anti–PD-1 monoclonal antibodies reduced tumor size and increased survival time in subcutaneous and orthotopic models. Through whole-exome sequencing, we showed the KRASwt iCCA mouse tumor cell line, SB1, genetically resembles iCCAs found in a subset of patients.
Results
Trametinib Treatment Directly Inhibits Tumor Growth In Vitro and In Vivo
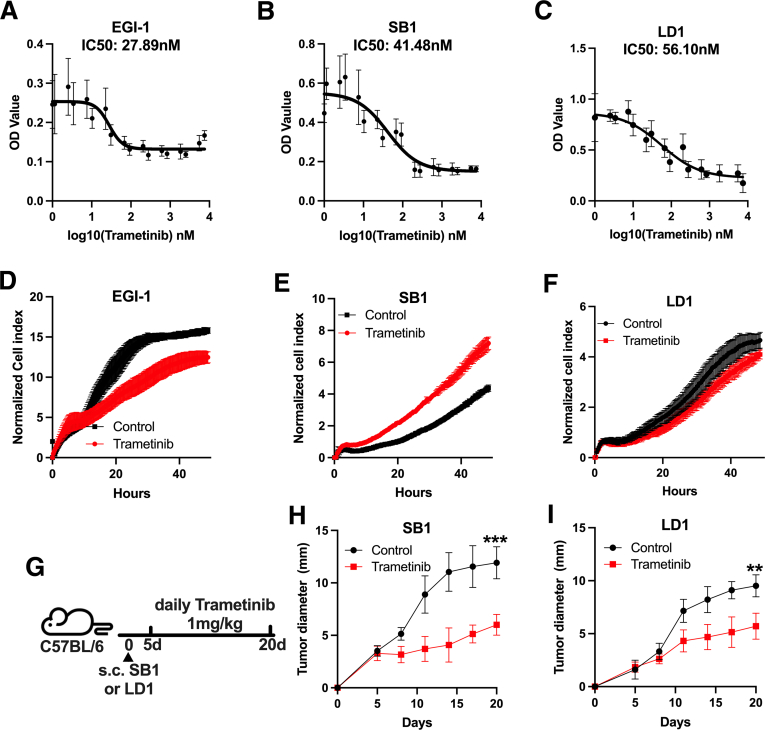
First, we tested the in vitro effect of trametinib on growth of the 2 available murine iCCA cell lines, SB1 and LD-1, and a human iCCA cell line (EGI-1). Dose-response studies showed a robust growth reduction in EGI-1 (Figure 1A, median inhibitory concentration [IC50], 27.89 nmol/L) cells. As expected, this growth inhibition was less pronounced in SB1 (Figure 1B, IC50, 41.50 nmol/L) and LD-1 (Figure 1C, IC50, 56.1 nmol/L) cells, which are KRASwt, in contrast to KRASG12D mutated EGI-1 cells. Similarly, we observed a decreased tumor cell proliferation of EGI-1, SB1, and LD-1 cells after 25 nmol/L trametinib treatment for 48 hours (Figure 1D–F). Other investigators have shown that trametinib also can lead to tumor growth inhibition in the absence of mutations of the mitogen-activated protein kinase/ERK pathway.14 Hence, the in vitro results prompted us to investigate whether trametinib treatment reduces iCCA tumor growth in vivo. SB1 or LD-1 tumor cells were injected subcutaneously and trametinib treatment was started after 5 days when tumors were established. Mice were treated with 1 mg/kg trametinib by daily gavage for 15 days and tumor growth was monitored (Figure 1G). Consistent with the in vitro results, trametinib treatment resulted in a significant reduction of tumor growth compared with control in SB1 (Figure 1H) and LD-1 (Figure 1I) flank tumors. Our results clearly show that trametinib can reduce iCCA tumor growth, even in KRASwt cell lines in vitro.
Figure 1.
Trametinib decreases tumor growth in vitro and in vivo. In vitro tumor cell growth of (A) EGI-1 (n = 8), (B) SB1 (n = 8), (C) LD-1 (n = 4), and with trametinib treatment for 48 hours. The 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromid (MTT) assay was used to measure cell viability. In vitro cell growth of (D) EGI-1, (E) SB1, and (F) LD-1 cells treated with or without 25 nmol/L trametinib treatment for 48 hours using the xCELLigence RTCA system (n = 3 per group). (G) Experimental set-up: C57BL/6 mice with subcutaneous injection of 106 SB1 or LD-1 cells treated with trametinib as indicated by daily gavage. (H and I) In vivo tumor growth of subcutaneous (A) SB1 and (B) LD-1 tumors over time. Two groups are shown: control (n = 5) vs trametinib treatment (n = 4). Tumor growth is shown as the largest tumor diameter in millimeters. Data represent means ± SD. ∗∗P < .01, ∗∗∗P < .001, ; Student t test. OD, optical density.
Trametinib Treatment Up-regulates MHC-I and PD-L1 on Tumor Cell Surface
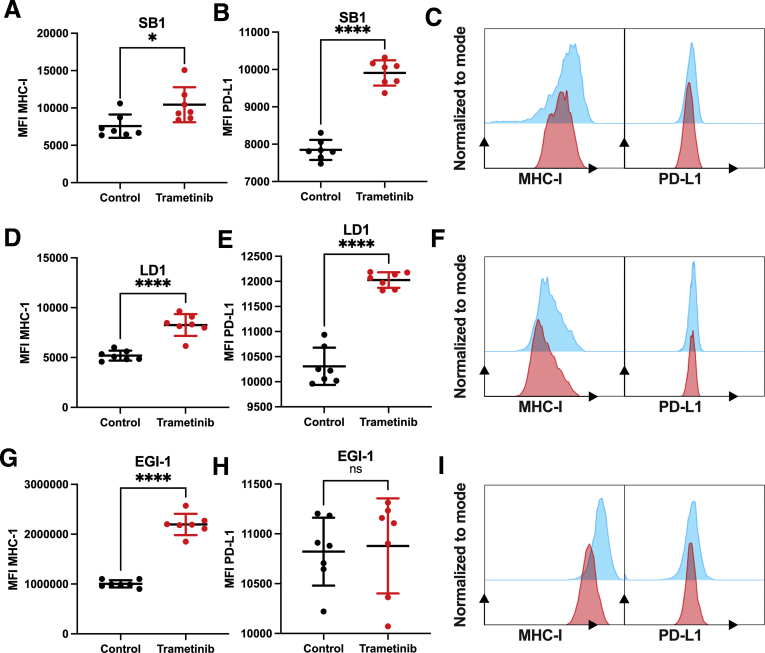
To further investigate the potential of trametinib treatment on antigen presentation of tumor cells, we treated SB1, LD-1, and EGI-1 with 25 nmol/L trametinib for 24 hours and measured PD-L1 and MHC-I surface expression by flow cytometry. Interestingly, trametinib increased the expression of PD-L1 and MHC-I in SB1 (Figure 2A–C) and LD-1 (Figure 2D–F). In EGI-1 cells, trametinib treatment increased MHC-I, but not PD-L1, expression (Figure 2G–I). Taken together, these results indicate that trametinib treatment has a potential effect on antitumor immunity. Therefore, we reasoned that the combination therapy using ICI might enhance the efficacy of trametinib.
Figure 2.
Trametinib increases surface expression of MHC-I on iCCA tumor cell lines. Median fluorescence intensity (MFI) of surface (A) MHC-I and (B) PD-L1 expression of SB1 cells treated with 25 nmol/L trametinib for 24 hours measured by flow cytometry (n = 7 per group). (C) Representative histogram plots of surface MHC-I (left section) and PD-L1 (right section) expression on SB1 cells after 25 nmol/L trametinib (red) treatment vs control (blue) for 24 hours. MFI of surface (D) MHC-I and (E) PD-L1 expression of LD-1 cells treated with 25 nmol/L trametinib for 24 hours measured by flow cytometry (n = 7 per group). (F) Representative histogram plots of surface MHC-I (left section) and PD-L1 (right section) expression on LD-1 cells after 25 nmol/L trametinib (red) treatment vs control (blue) for 24 hours. MFI of surface (G) MHC-I and (H) PDL-1 expression of EGI-1 cells treated with 25 nmol/L trametinib for 24 hours measured by flow cytometry (n = 7 per group). (F) Representative histogram plots of surface MHC-I (left section) and PD-L1 (right section) expression on EGI-1 cells after 25 nmol/L trametinib (red) treatment vs control (blue) for 24 hours. Data represent means ± SD. ∗P < .05, and ∗∗∗∗P < .0001; Student t test.
Anti–PD-1 Treatment Enhances Antitumor Immune Responses of Trametinib
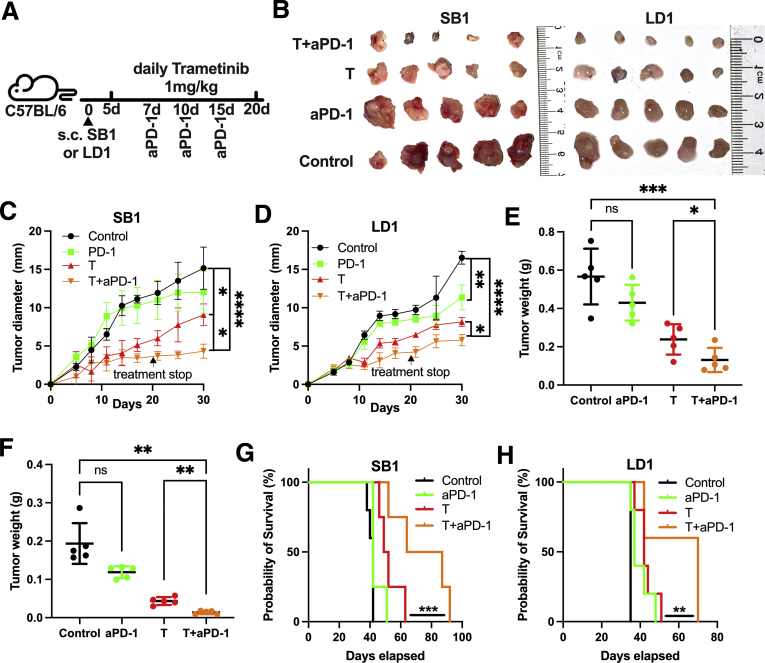
To investigate the potential synergic antitumor effect of ICI therapy and trametinib, we treated mice with SB1 and LD-1 flank tumors with trametinib and anti–PD-1 (Figure 3A). The tumor size of subcutaneous SB1 and LD-1 was reduced after trametinib, but not anti–PD-1 monotherapy. The combination treatment caused the greatest tumor growth reduction (Figure 3B and D) and end point measurement of SB1 and LD-1 tumor weights (Figure 3E and F). Combination treatment also improved the survival of SB1 tumor-bearing mice, whereas the single-agent treatment regimen failed to prolong survival in this tumor model (Figure 3G). Similar results were observed in LD-1 tumor-bearing mice after trametinib + anti–PD-1 treatment (Figure 3H).
Figure 3.
Trametinib in combination with anti–PD-1 improves survival of mice with subcutaneous SB1 tumors. (A) Experimental set-up: C57BL/6 mice with a subcutaneous injection of 106 SB1 or LD-1 cells treated with trametinib by daily gavage and received anti–PD-1 (200 μg/mouse at indicated time points). (B) Representative pictures of subcutaneous SB1 (left section, n = 5) and LD-1 (right section, n = 5) tumors. Experimental set-up is shown in Figure 3A. Tumor growth of subcutaneous (C) SB1 and (D) LD-1 flank tumors over time. Four groups are shown: control (n = 5) vs trametinib treatment (n = 4) vs anti–PD-1 (anti–PD-1, n = 5) treatment vs combination (Trametinib + anti–PD-1, n = 5). Tumor growth is shown as the largest tumor diameter in millimeters. Tumor weights of subcutaneous (E) SB1 and (F) LD-1 tumors. Experimental set-up is shown in Figure 3A. Survival of mice with subcutaneous (G) SB1 and (H) LD-1 tumors over time (n = 5 per group), log-rank test. Data represent means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001; Student t test. aPD-1, anti-Programmed cell death protein-1.
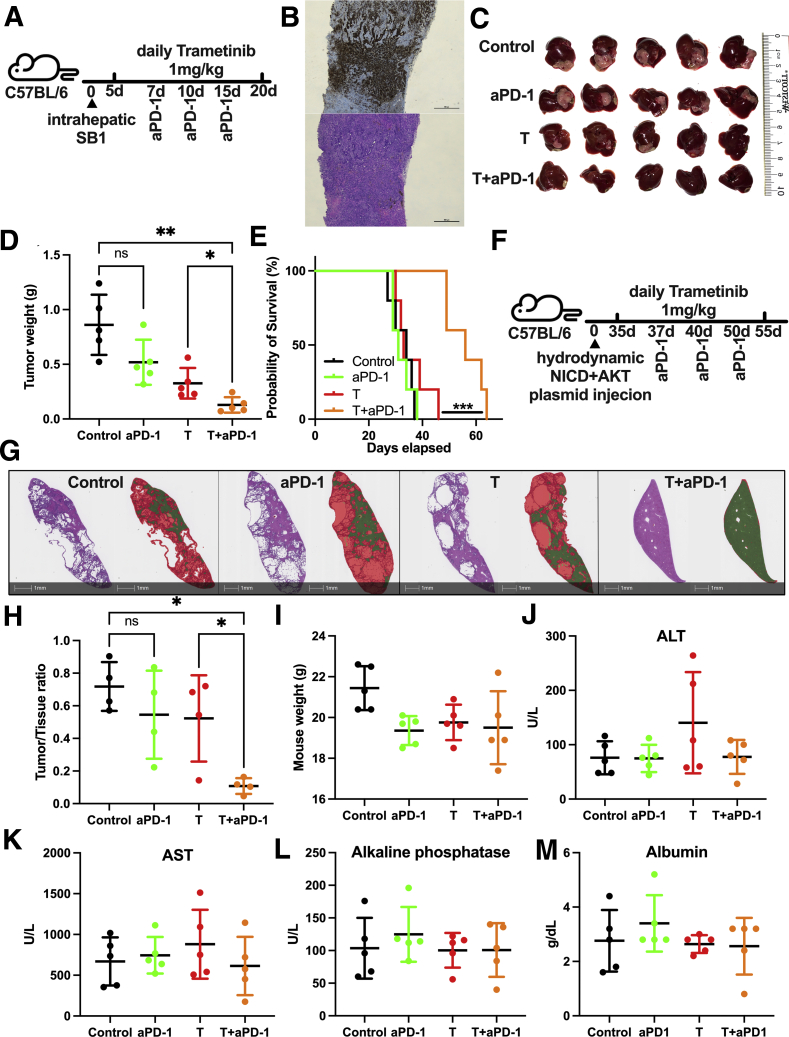
To better mimic the clinical setting of iCCA, we injected SB1 cells orthotopically into mouse livers (experimental outline shown in Figure 4A). The iCCA phenotype of intrahepatic SB1 tumors was confirmed by cytokeratine (CK) 19 staining (Figure 4B). Tumor burden after orthotopic SB1 injection was reduced in combination treatment compared with trametinib alone (Figure 4C and D). Survival of mice with orthotopic SB1 tumors showed a significant improvement of survival of mice treated with trametinib + anti–PD-1 (Figure 4E).
Figure 4.
Trametinib in combination with anti–PD-1 controls tumor growth in orthotopic iCCA models. (A) Experimental set-up: C57BL/6 mice with intrahepatic injection of 2 × 105 SB1 cells treated with trametinib by daily gavage and received anti–PD-1 (200 μg/mouse at the indicated time points). (B) Representative images of CK19 staining (upper section) and H&E staining (lower section) of intrahepatic SB1 tumors. Scale bar: 500 μm. (C) Representative pictures of intrahepatic SB1 tumors. Experimental set-up is shown in Figure 4A. (D) Tumor weights (g) of intrahepatic SB1 tumors. Experimental set-up is shown in Figure 4A. (E) Survival of mice with intrahepatic SB1 tumors over time (n = 5 per group), log-rank test. (F) Experimental set-up: C57BL/6 mice with hydrodynamic tail vein injection of NICD + AKT plasmid injection treated with trametinib by daily gavage and received anti–PD-1 (200 μg/mouse at the indicated time points). (G) Whole slide scan of a representative H&E stain (sections on the left) of a murine iCCA-bearing liver and tissue section after image analysis using HALO Random Forrest Classifier function (Indica Labs, Albuquerque, NM) and shown as a digital overlay indicating areas classified as tumor (red) and remaining normal liver (green). Scale bar: 1 mm. (H) Tumor to liver tissue ratio after NICD + AKT injection (n = 4 per group) corresponding to Figure 4F. Experimental set-up is shown in Figure 4A. (I) Weight of mice after treatment. Experimental set-up is shown in Figure 4E. (I) Alanine aminotransferase (ALT), (J) aspartate aminotransferase (AST), (K) alkaline phosphatase, and (L) albumin serum levels after treatment. Experimental set-up is shown in Figure 4A. Data represent means ± SD. ∗P < .05, ∗∗P < .01; Student t test. aPD-1, anti-Programmed cell death protein-1; T, trametinib.
Next, we extended our study to a primary iCCA model using AKT + Notch 1 hydrodynamic plasmid injections (experimental outline shown in Figure 4F). Again, the combination of anti–PD-1 and trametinib significantly reduced liver tumor burden compared with single therapy with anti–PD-1 or trametinib (Figure 4G and H).
Mouse weight; liver enzymes alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase; and albumin serum levels were equal among treatment groups, suggesting that the treatments were well tolerated (Figure 4I–M). Taken together, our results show that the combination of trametinib + anti–PD-1 is a potent therapy to reduce tumor burden in various iCCA tumor models in mice.
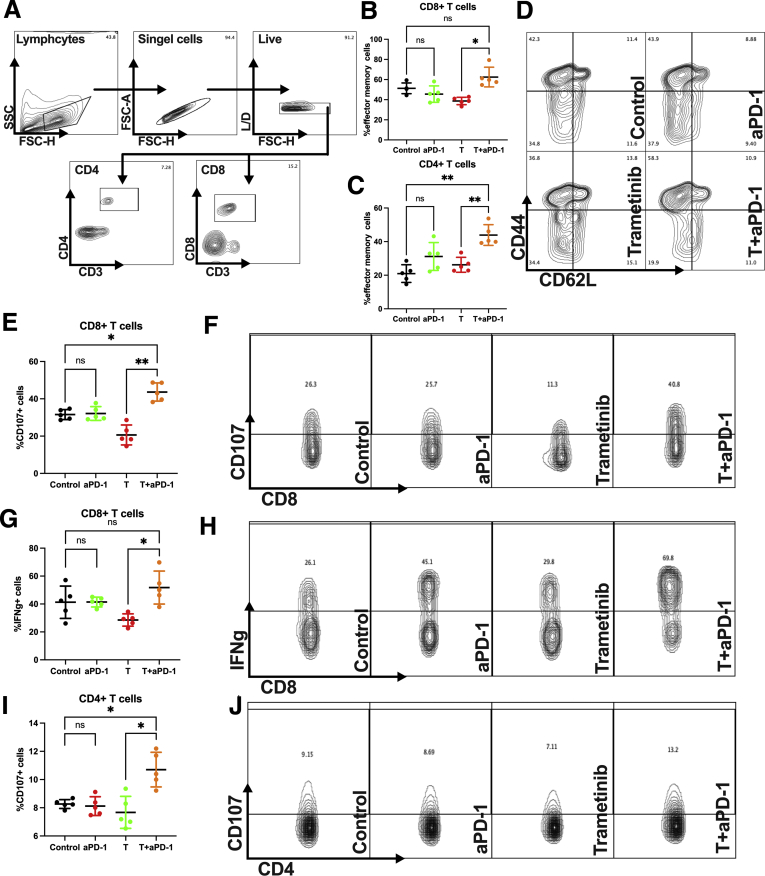
Combination of Anti–PD-1 + Trametinib Boosts Activation of T Cells
The immune profile of liver lymphocytes was analyzed to elucidate the underlying mechanism of the enhanced therapeutic efficacy of combination trametinib + anti–PD-1. In liver lymphocytes of orthotopic SB1 tumor-bearing mice, an up-regulation of CD44highCD62Llow effector memory CD8+ and CD4+ T cells (gating strategy shown in Figure 5A) was found in the combination treatment group (Figure 5B and C, gating strategy shown in Figure 5D). CD107 staining showed a significantly improved degranulation of CD8+ T cells upon anti–PD-1 + trametinib treatment (Figure 5E and F). In addition, more interferon γ (IFNγ)+CD8+ T cells were observed in the combination treatment group compared with the trametinib single-agent group (Figure 5G and H). To evaluate tumor-specific cytotoxicity, we stimulated splenocytes from SB1 flank-tumor-bearing mice with SB1 cells in vitro. The combination treatment increased the percentage of CD107+CD4+ T cells (Figure 5I and J). These results show that the combination treatment unleashes an immune response and creates a robust immune memory against iCCA in vivo.
Figure 5.
Trametinib + anti–PD-1 therapy unleashes the immune system against iCCA tumors. (A) Representative contour plot of CD4+ and CD8+ gating strategy of hepatic lymphocytes of orthotopic SB1 tumor-bearing mice after treatment. Experimental set-up is shown in Figure 4A. Percentage of hepatic (B) CD44highCD62Llow CD8+ and (C) CD4+ effector memory T cells after intrahepatic SB1 injection (n = 5 per group). Experimental set-up is shown in Figure 4A. (D) Representative contour plot and frequencies of CD44highCD62Llow CD8+ effector memory T cells of hepatic lymphocytes of orthotopic SB1 tumor-bearing mice after treatment (gating strategy CD3+CD8+). Experimental set-up is shown in Figure 4A. (E) Percentage and (F) representative contour plot and frequencies of CD107+CD8+ T cells of hepatic lymphocytes of orthotopic SB1 tumor-bearing mice after treatment (gating strategy CD3+CD8+) after ex vivo stimulation for 4 hours (n = 5 per group). Experimental set-up is shown in Figure 4A. (G) Percentage and (H) representative contour plot and frequencies of IFNγ+CD8+ T cells of hepatic lymphocytes of orthotopic SB1 tumor-bearing mice after treatment (gating strategy CD3+CD8+) after ex vivo stimulation for 4 hours (n = 5 per group). Experimental set-up is shown in Figure 4A. (I) Percentage and (J) representative contour plot and frequencies of CD107+CD4+ T splenocytes from SB1 flank-tumor-bearing mice after ex vivo stimulation with SB1 tumor cells for 5 hours (n = 5 per group). Group set-up is shown as in Figure 3A. Data represent means ± SD. ∗P < .05, ∗∗P < .01; Student t test. aPD-1, anti-Programmed cell death protein-1; FSC-A, forward scatter area; FSC-H, forward scatter high; L/D, live/dead; SSC,side scatter; T, trametinib.
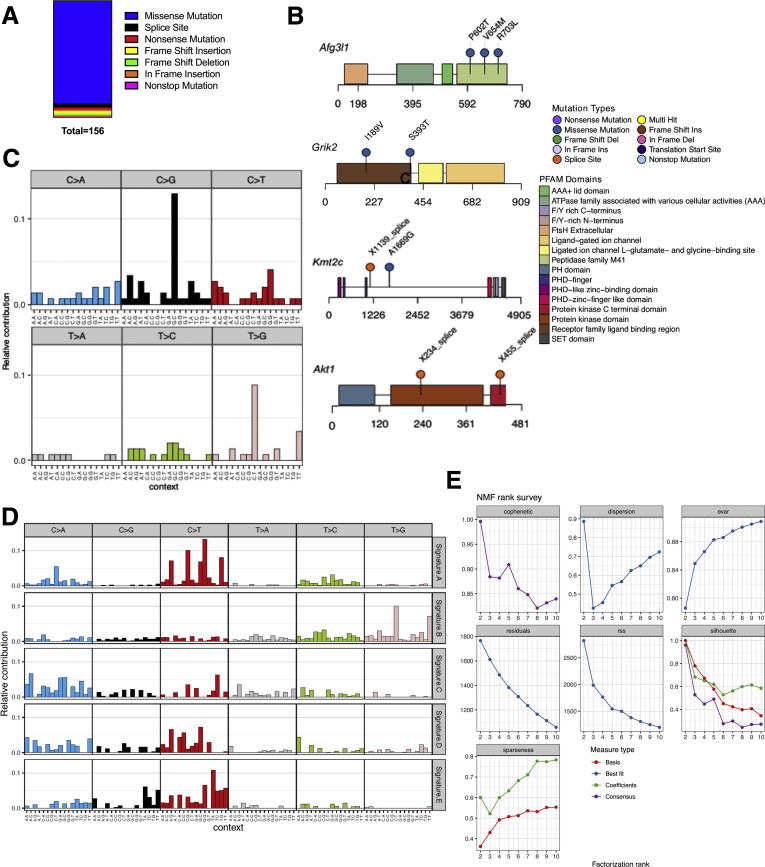
SB1 Is KRASwt and Shows a Similar Mutational Signature to a Subpopulation of iCCA Patients
To put our findings into clinical context and identify which subpopulation of iCCA patients might benefit from trametinib + anti–PD-1, we performed whole-exome sequencing (WES) of SB1 cells. SB1 cells were found to have 156 somatic mutations, which did not include KRAS, BRAF, and ERK (Figure 6A). The most frequently mutated gene loci were AFG5I1, AKT1, GRIK2, and KMT2C (Figure 6B). To compare the single-base substitution signature in SB1 (Figure 6C) with human data, 5 consensus signatures were derived from the human CCA (The cancer genome atlas (TCGA)-cholangiocarcinoma [CHOL]) data set. Importantly, the mutational signatures in SB1 matched most closely with consensus signature B. SB1 signature and signature B are both characterized primarily by T>G mutations (Figure 6D). We applied Cox regression modeling to evaluate the effect of each signature’s contribution to survival time reported in TCGA-CHOL. Signature B has a significant effect on survival, and that increased contribution from this signature confers an increased hazard for poor outcome (95% CI for odds ratio, 3.39–462.4). Therefore, trametinib + anti-PD-1 might be an attractive therapeutic option, especially for patients with a high risk for poor outcome and an unfavorable mutational signature.
Figure 6.
WES of SB1 tumor cells. (A) Summary of mutation number and type from WES of the SB1 cell line. (B) Lollipop plots for Afg3I1, Grik2, Kmt2C, and Akt1 showing a schematic of each protein with protein domains from PFAM (protein family database) highlighted in colored blocks. Location of mutations is annotated with pins along the length of the protein. (C) Single-base substitution (SBS) signature of SB1 cell line. Each of the 6 mutation classes, as indicated at the top of the panel, is separated into 16 categories based on the identity of the preceding and following nucleotide as shown below. (D) SBS signatures for the 5 consensus signatures (Signature A–E) derived from TCGA-CHOL, arranged in rows. (E) Diagnostic plots for NMF rank survey. Cophentic correlation (top-left) was used to determine the optimal rank (five). AAA, ATPases associated woth diverse cellular activities; ATPase, adenosine triphosphatase; Del, deletion; F/Y, phenylalanin/tyrosine; NMF, non-negative matrix factorisation; PHD, plant homeodomain; SET, suvar3-9, enhancer-of-zeste and trithorax.
Discussion
ICI is a promising treatment strategy that has shown benefits in patients with several malignancies. The use of ICI in patients with liver malignancies remained challenging because the promising results of ICI therapy were not reproducible in hepatocellular carcinoma or iCCA.10,16 Only recently, the first-line treatment of unresectable hepatocellular carcinoma changed from sorafenib to atezolizumab plus bevacizumab as a result of promising results in a phase 3 trial comparing this anti–PD-L1 + anti–vascular endothelial growth factor treatment regimen with sorafenib.17 Because of the tolerogenic landscape of the liver with a large amount of immune cells present, immunotherapy is particularly challenging.18 Thus, for patients with iCCA, there is still no ICI therapy available and selecting a potent combination partner might be a promising strategy to enhance the efficacy of ICI.
To investigate the effects of trametinib on iCCA tumor cells, we used a concentration of 25 nmol/L in vitro. This is a lower dose compared with recent publications that have used 100 nmol/L trametinib to treat Mia-PaCa2 cells.19 Monotherapy at this dosage showed a modest growth inhibition of tumor cells in vitro. Nevertheless, LD-1 and SB1 flank tumors in mice showed a robust response to trametinib treatment. Interestingly, when trametinib treatment was stopped, we observed an acceleration of tumor growth that resulted in no survival benefit of the group treated with trametinib alone. This is consistent with a recent clinical trial showing no improved survival resulting from trametinib treatment in biliary tract cancer.20 We hypothesized that trametinib has a direct effect on tumor cells that improves their immunogenicity. Indeed, trametinib treatment increased MHC-1 and PD-L1 surface expression on tumor cells in vitro. Thus, we reasoned that the addition of monoclonal antibodies targeting the PD-1/PD-L1 axis can enhance this therapy. Indeed, the combination treatment led to significantly improved survival of SB1 flank-tumor-bearing mice.
Another explanation of the insufficient long-term effect of trametinib might be the effect of trametinib on T cells. It has been shown by others that trametinib has a direct immune-suppressive effect on lymphocytes.21 In our study, we observed a similar trend in degranulation and IFNγ production of CD8 T cells of trametinib-treated mice, although this did not reach statistical significance. However, this immune-suppressive effect of trametinib was reversed in the combination treatment group. In addition, anti–PD-1 + trametinib increased the percentage of effector memory T cells and improved the re-recognition of SB1 tumor cells from CD4+ T cells of SB1 tumor-bearing mice in vitro. Our results show that the combination of anti–PD-1 + trametinib unleashes the immune system and represents a potential treatment for patients with iCCA.
Objective response rates for ICI therapy in solid tumors is approximately 15%–20%,22,23 but can provide long-term regression in carefully selected patients. We therefore performed WES of SB1 tumor cells to show mutational similarities of this cell line to subpopulations of iCCA patients. Indeed, we were able to identify a subpopulation with a similar mutational pattern compared with SB1 cells. This subpopulation is characterized by poor survival. In addition, WES showed that SB1 cells are not KRAS, BRAF, and ERK mutated. A limitation of this study was that we did not use a KRASmut iCCA cell line to show specific benefits of this therapy for KRASmut iCCA patients. To our knowledge, there is no KRASmut iCCA cell line available. However, we believe that our results show a promising treatment for patients with iCCA, especially with similar mutational pattern to SB1 cells.
Methods
Mouse Strains, Reagents, and Cell Lines
C57BL/6 mice were purchased from the Charles River Laboratory and were used at age 8–12 weeks for experiments. All experiments were conducted according to local institution guidelines and approved by the Animal Care and Use Committee of the National Institutes of Health (Bethesda, MD). Trametinib (Melkinist; Novartis, Basel, Swizerland) was obtained from the National Institutes of Health clinical center pharmacy. The murine CCA cell lines SB124 and LD-1, as well as the human CAA cell line EGI-1,25 were used in this study. LD-126 was established in our laboratory and is derived from a mouse after hydrodynamic injection with AKT and YAP plasmids. WES data from LD-1 as well as SB1 will be provided upon request.
Animal Studies
Subcutaneous injections were performed in 8-week-old C57BL/6 female mice. SB-1 (1 × 106) were resuspended in 100 μL phosphate-buffered saline (PBS) and the tumor cell suspension was injected in the lateral flank. All mice were randomized before initiation of treatment. Mice then were treated 5 days after injection with either vehicle control (saline), daily gavage of 1 mg/kg trametinib, or 200 μg anti–PD-1 (clone 29F.1A12; BioXCell, Lebanon, NH) on days 7, 10, and 15. One group received combined treatment with both trametinib and anti–PD-1. Tumor size was measured by caliper and mice were killed on day 20 after tumor cell injection. For subcutaneous survival experiments, mice were killed when tumors reached 20 mm in size or tumor necrosis exceeded 50% of the tumor surface. iCCA was established by injecting 2 × 105 SB1 cells in 20 μL of a 50:50 solution of PBS and Matrigel (Corning, Corning, NY) into the left liver lobe as described previously.27 CK19 staining of SB1 tumors confirmed a CCA phenotype (Figure 4B). Mice were treated as described earlier. For Notch intracellular domain 1 (NICD) + AKT iCCA, 20 μg NICD, 4 μg AKT, and 1 μg hyperactive sleeping beauty 2 transposase plasmids were diluted into 1.6 mL PBS and injected into the tail vein within 5–7 seconds.28 Treatment was initialized 5 weeks after injection as with either vehicle control (saline), daily gavage of 1 mg/kg trametinib or 200 μg anti–PD-1 (on days 37, 40, and 45), and the combination of anti–PD-1 and trametinib. Mice were killed on day 50 after injection. H&E-stained sections were scanned at 20× objective magnification using an Aperio AT2 digital whole slide scanner (Leica Biosystems, Wetzlar, Germany). Automated quantification using pattern recognition image analysis of tumor areas and normal liver tissue areas was performed using HALO image analysis software (Indica Labs, Albuquerque, NM).
SB1 WES
DNA was extracted from 106 SB1 tumor cells. Two exome samples were pooled and sequenced on NextSeq (Illumina, San Diego, CA) run using Agilent (Santa Clara, CA) SureSelect XT Mouse All Exon and paired-end sequencing mode. Reads were trimmed for adapters and low-quality bases using Trimmomatic software (Usadellab, Aachen, Germany, version 0.33) before alignment to the mouse mm10 reference genome using BWA mapping software (Illumina), version 0.7.15).29 Mapped reads then were de-duplicated using Picard Tools (Broad institute, Cambridge, MA)followed by re-alignment and base quality score recalibration using the Genome Analysis Toolkit (Broad institute) version 3.8.0.30 The sequencing run yielded 61.7 million total reads, >99% mapping to the reference genome, and mean exome coverage of 69×. The sequencing data of SB1 was uploaded as Sequence Read Archive (SRA) submission SRR13091994, BioProject accession PRJNA679801.
Somatic Variant Analysis
Somatic variant calling was performed using Mutect2 from Genome Analysis Toolkit version 3.8.0 in tumor only mode, following the best practices guidelines for exome-seq analysis provided by the Genome Analysis Toolkit authors.31 Variants were hard filtered for quality, annotated with functional and consequence prediction using Ensembl’s Variant Effect Predictor version 95 (Ensemble, Hinxton, UK),32 and converted to Mutation Annotation Format using the vcf2maf tool version 1.6.16 (Memorial Sloan Kettering, New York, NY).33
Variants were filtered further to exclude frequently mutated genes in exome data.34 Common single-nucleotide polymorphisms and indels—annotated with a frequency of greater than 0.01 in the ExAC (Broad institute), gnomAD (Broad institute), or 1000 Genomes (European Bioinformatics Institute, Hinxton, UK) databases—also were removed. In addition, variants with less than 20× depth and an alternate allele frequency of less than 5% in the SB1 cell line data were removed.
Mutational Signatures Analysis
Variants called by Mutect2 for the TCGA-CHOL data set were retrieved as Mutation Annotation Format files using the R (Vienna, Austria) package TCGABiolinks35 (version 2.12.6). For each variant site, adjacent 5’ and 3’ bases were extracted from the genomic sequence to generate trinucleotide substitution frequencies using maftools36 (version 2.0.16). This matrix was used to compute consensus signatures from all 50 TCGA-CHOL samples using the NMF package37 (version 0.23.0). The number of signatures was optimized by comparing cophenetic correlation from 3 to 10 signatures as described by Brunet et al,38 and 5 signatures was chosen as the optimal number of signatures (Figure 6E). The contribution of these 5 signatures (signatures A–E) in individual signatures from each sample were computed using MutationalPatterns (Bioconductor, Fred Hutchinson Cancer Research Cener, Seattle, WA)39 (version 1.10.0).
Survival Analysis
Clinical data provided with TCGA-CHOL were retrieved using TCGABiolinks(Bioconductor, Fred Hutchinson Cancer Research Cener). Cox proportional hazards regression modeling was performed to model the effect of each signature contribution independently, controlling for patient age and sex as covariates. Modeling was performed using the survival R package40 (version 3.2-7), and visualized using survminer41 (version 0.4.8).
In vitro Tumor Cell Viability Assay, Proliferation Assay, and Antigen Profiling
A total of 5000–10,000 SB1, LD-1, and EGI-1 cells per well were treated with trametinib in 200 μL media (RPMI, 10% fetal bovine serum, 1% penicillin/streptomycin) for 48 hours. Cell viability was measured with the MTT Cell Proliferation kit (ab211091; Abcam, Cambridge, UK). Cell proliferation also was analyzed using the xCELLigence RTCA system (Agilent) according to the manufacturer’s instructions. A total of 5000–10,000 SB1, LD-1, and EGI-1 cells were seeded in a 16X E-Pate (00 300 600 890; Agilent) and treated with 200 nmol/L trametinib for 48 hours. For MHC-I and PD-L1 surface staining, 3 × 105 cells/well were treated with 200 nmol/L trametinib for 24 hours and cells were analyzed using flow cytometry.
CD107 Degranulation Assay
SB1 cells were seeded overnight (2 × 104 cells/well) in a 96-well, flat-bottom plate. The next day, SB1 flank-tumor-bearing mice were killed and splenocytes were isolated. A total of 106 splenocytes/well were used for co-culture with SB1 cells and incubated for 5 hours, and then washed and stained. CD107 expression on lymphocytes was analyzed to determine tumor-specific degranulation.
Flow Cytometry
For surface marker staining, cells were stained with antibodies for 15 minutes at room temperature. For ex vivo activation, cells were stimulated with a stimulation cocktail (51-2042E; BD Biosciences, Franklin Lakes, NJ) and simultaneously stained with fluorochrome-coupled anti-CD107a antibody. Dead cells were excluded by using a live/dead cell staining kit (ThermoFisher scientific, Waltham, MA). The following antibodies were used for flow cytometry analysis: anti–CD3-Brilliant violet (BV) 605 (clone 17A2; Biolegend, San Diego, CA), CD8-Alexa Fluor 700 (clone 53-6.7; Biolegend), anti-CD8-Pacific Blue (PB) (clone 53-6.7; Biolegend), anti–CD44-Phycoerythrin (PE)/Cy7 (clone 1M7; Biolegend), anti-CD62L PerCP/Cy5 (clone MEL-14; Biolegend), anti–CD4-Alexa Fluor 700 (clone GK1.5; Biolegend), anti–IFNγ- Allophycocyanin (APC) (clone XMG1.2; Biolegend), anti–CD107a–PE (clone 1D4B; BD Biosciences), anti–MHC-I–APC (clone 28-8-6; Biolegend), anti–MHC-I–APC (clone W6/32; Invitrogen, Carlsbad, CA), and anti–PD-L1–PerCP/Cy5.5 (clone 10F.9G2; Biolegend).
Statistical Analysis
For all models, randomization of animals and blinding of examiners were performed. Statistical analysis was performed with GraphPad Prism 8 (GraphPad, San Diego, CA). The significance of the difference between groups was calculated by the Student unpaired t test. P < .05 was considered statistically significant.
CRediT Authorship Contributions
Simon Wabitsch, MD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead)
Mayank Tandon (Formal analysis: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Benjamin Ruf, MD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Qianfei Zhang, PhD (Investigation: Supporting; Writing – review & editing: Supporting)
Justin D McCallen, BS (Investigation: Supporting; Writing – review & editing: Supporting)
John C McVey, BA (Investigation: Supporting; Writing – review & editing: Supporting)
Chi Ma, MD PhD (Conceptualization: Supporting; Writing – original draft: Equal)
Benjamin Green, MD (Validation: Supporting; Writing – review & editing: Supporting)
Laurence P Diggs, MD (Conceptualization: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Bernd Heinrich, MD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Tim F Greten, MD (Conceptualization: Equal; Supervision: Lead; Writing – review & editing: Equal)
Acknowledgements
The authors would like to thank Dr Jonathan Hernandez for providing the EGI-1 cell line. The graphical abstract was created with BioRender.com.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by Deutsche Forschungsgemeinschaft grant WA-4610/1-1 (S.W.), and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute (ZIA BC 011345, ZO1 BC010870) (T.F.G.).
References
- 1.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertuccio P., Malvezzi M., Carioli G., Hashim D., Boffetta P., El-Serag H.B., La Vecchia C., Negri E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J Hepatol. 2019;71:104–114. doi: 10.1016/j.jhep.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Mosadeghi S., Liu B., Bhuket T., Wong R.J. Sex-specific and race/ethnicity-specific disparities in cholangiocarcinoma incidence and prevalence in the USA: an updated analysis of the 2000-2011 Surveillance, Epidemiology and End Results registry. Hepatol Res. 2016;46:669–677. doi: 10.1111/hepr.12605. [DOI] [PubMed] [Google Scholar]
- 4.de Groen P.C., Gores G.J., LaRusso N.F., Gunderson L.L., Nagorney D.M. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 5.Tan J.C., Coburn N.G., Baxter N.N., Kiss A., Law C.H. Surgical management of intrahepatic cholangiocarcinoma--a population-based study. Ann Surg Oncol. 2008;15:600–608. doi: 10.1245/s10434-007-9627-x. [DOI] [PubMed] [Google Scholar]
- 6.Mazzaferro V., Gorgen A., Roayaie S., Droz Dit Busset M., Sapisochin G. Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:364–377. doi: 10.1016/j.jhep.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Bridgewater J., Galle P.R., Khan S.A., Llovet J.M., Park J.W., Patel T., Pawlik T.M., Gores G.J. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 8.Duffy A.G., Makarova-Rusher O.V., Greten T.F. The case for immune-based approaches in biliary tract carcinoma. Hepatology. 2016;64:1785–1791. doi: 10.1002/hep.28635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim R.D., Chung V., Alese O.B., El-Rayes B.F., Li D., Al-Toubah T.E., Schell M.J., Zhou J.M., Mahipal A., Kim B.H., Kim D.W. A phase 2 multi-institutional study of nivolumab for patients with advanced refractory biliary tract cancer. JAMA Oncol. 2020;6:888–894. doi: 10.1001/jamaoncol.2020.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeuillard E., Conboy C.B., Gores G.J., Rizvi S. Immunobiology of cholangiocarcinoma. JHEP Rep. 2019;1:297–311. doi: 10.1016/j.jhepr.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura H., Arai Y., Totoki Y., Shirota T., Elzawahry A., Kato M., Hama N., Hosoda F., Urushidate T., Ohashi S., Hiraoka N., Ojima H., Shimada K., Okusaka T., Kosuge T., Miyagawa S., Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 12.Kim C., Giaccone G. MEK inhibitors under development for treatment of non-small-cell lung cancer. Expert Opin Investig Drugs. 2018;27:17–30. doi: 10.1080/13543784.2018.1415324. [DOI] [PubMed] [Google Scholar]
- 13.Heppt M.V., Dietrich C., Graf S.A., Ruzicka T., Tietze J.K., Berking C. The systemic management of advanced melanoma in 2016. Oncol Res Treat. 2016;39:635–642. doi: 10.1159/000448904. [DOI] [PubMed] [Google Scholar]
- 14.Wang P., Song X., Utpatel K., Shang R., Yang Y.M., Xu M., Zhang J., Che L., Gordan J., Cigliano A., Seki E., Evert M., Calvisi D.F., Hu X., Chen X. MEK inhibition suppresses K-Ras wild-type cholangiocarcinoma in vitro and in vivo via inhibiting cell proliferation and modulating tumor microenvironment. Cell Death Dis. 2019;10:120. doi: 10.1038/s41419-019-1389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin M.H., Kim J., Lim S.A., Kim J., Lee K.M. Current insights into combination therapies with MAPK inhibitors and immune checkpoint blockade. Int J Mol Sci. 2020;21:2531. doi: 10.3390/ijms21072531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteside T.L., Demaria S., Rodriguez-Ruiz M.E., Zarour H.M., Melero I. Emerging opportunities and challenges in cancer immunotherapy. Clin Cancer Res. 2016;22:1845–1855. doi: 10.1158/1078-0432.CCR-16-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y., Kudo M., Breder V., Merle P., Kaseb A.O., Li D., Verret W., Xu D.Z., Hernandez S., Liu J., Huang C., Mulla S., Wang Y., Lim H.Y., Zhu A.X., Cheng A.L. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 18.Ruf B., Heinrich B., Greten T.F. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18:112–127. doi: 10.1038/s41423-020-00572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinsey C.G., Camolotto S.A., Boespflug A.M., Guillen K.P., Foth M., Truong A., Schuman S.S., Shea J.E., Seipp M.T., Yap J.T., Burrell L.D., Lum D.H., Whisenant J.R., Gilcrease G.W., 3rd, Cavalieri C.C., Rehbein K.M., Cutler S.L., Affolter K.E., Welm A.L., Welm B.E., Scaife C.L., Snyder E.L., McMahon M. Publisher correction: protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:861. doi: 10.1038/s41591-019-0433-3. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda M., Ioka T., Fukutomi A., Morizane C., Kasuga A., Takahashi H., Todaka A., Okusaka T., Creasy C.L., Gorman S., Felitsky D.J., Kobayashi M., Zhang F., Furuse J. Efficacy and safety of trametinib in Japanese patients with advanced biliary tract cancers refractory to gemcitabine. Cancer Sci. 2018;109:215–224. doi: 10.1111/cas.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vella L.J., Pasam A., Dimopoulos N., Andrews M., Knights A., Puaux A.L., Louahed J., Chen W., Woods K., Cebon J.S. MEK inhibition, alone or in combination with BRAF inhibition, affects multiple functions of isolated normal human lymphocytes and dendritic cells. Cancer Immunol Res. 2014;2:351–360. doi: 10.1158/2326-6066.CIR-13-0181. [DOI] [PubMed] [Google Scholar]
- 22.Xu-Monette Z.Y., Zhang M., Li J., Young K.H. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response? Front Immunol. 2017;8:1597. doi: 10.3389/fimmu.2017.01597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Qin S. Immune checkpoint inhibitors in hepatocellular carcinoma: opportunities and challenges. Oncologist. 2019;24(Suppl 1):S3–S10. doi: 10.1634/theoncologist.2019-IO-S1-s01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizvi S., Fischbach S.R., Bronk S.F., Hirsova P., Krishnan A., Dhanasekaran R., Smadbeck J.B., Smoot R.L., Vasmatzis G., Gores G.J. YAP-associated chromosomal instability and cholangiocarcinoma in mice. Oncotarget. 2018;9:5892–5905. doi: 10.18632/oncotarget.23638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf L.M., Leitzel K.E., Pegg A.E., Harvey H.A., Lipton A. The in vitro interaction of alpha-difluoromethyl ornithine (DFMO) and several interferons on human cell lines. J Biol Response Mod. 1985;4:391–395. [PubMed] [Google Scholar]
- 26.Zhang Q., Ma C., Duan Y., Heinrich B., Rosato U., Diggs L.P., Ma L., Roy S., Fu Q., Brown Z.J., Wabitsch S., Thovarai V., Fu J., Feng D., Ruf B., Cui L.L., Subramanyam V., Frank K.M., Wang S., Kleiner D.E., Ritz T., Rupp C., Gao B., Longerich T., Kroemer A., Wang X.W., Ruchirawat M., Korangy F., Schnabl B., Trinchieri G., Greten T.F. Gut microbiome directs hepatocytes to recruit MDSC and promote cholangiocarcinoma. Cancer Discov. 2021;11:1248–1267. doi: 10.1158/2159-8290.CD-20-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown Z.J., Heinrich B., Greten T.F. Establishment of orthotopic liver tumors by surgical intrahepatic tumor injection in mice with underlying non-alcoholic fatty liver disease. Methods Protoc. 2018;1:21. doi: 10.3390/mps1020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan B., Malato Y., Calvisi D.F., Naqvi S., Razumilava N., Ribback S., Gores G.J., Dombrowski F., Evert M., Chen X., Willenbring H. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122:2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Memorial Sloan Kettering Cancer Center vcf2maf. 2013. https://github.com/mskcc/vcf2maf Available from: Accessed August 22, 2020.
- 34.Shyr C., Tarailo-Graovac M., Gottlieb M., Lee J.J., van Karnebeek C., Wasserman W.W. FLAGS, frequently mutated genes in public exomes. BMC Med Genomics. 2014;7:64. doi: 10.1186/s12920-014-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaujoux R., Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blokzijl F., Janssen R., van Boxtel R., Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 2018;10:33. doi: 10.1186/s13073-018-0539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Therneau T.M. A package for survival analysis in R. https://CRAN.R-project.org/package=survival Available from: Retrieved March 20 2020.
- 41.Kassambara A., Kosinski M., Biecek P. 2020. survminer: drawing survival curves using 'ggplot2'. [Google Scholar]