Abstract
The prevalence of SARS-CoV-2 is a great threat to global public health. However, the relationship between the viral pathogen SARS-CoV-2 and host innate immunity has not yet been well studied. The genome of SARS-CoV-2 encodes a viral protease called 3C-like protease. This protease is responsible for cleaving viral polyproteins during replication. In this investigation, 293T cells were transfected with SARS-CoV-2 3CL and then infected with Sendai virus (SeV) to induce the RIG-I like receptor (RLR)-based immune pathway. q-PCR, luciferase reporter assays, and western blotting were used for experimental analyses. We found that SARS-CoV-2 3CL significantly downregulated IFN-β mRNA levels. Upon SeV infection, SARS-CoV-2 3CL inhibited the nuclear translocation of IRF3 and p65 and promoted the degradation of IRF3. This effect of SARS-CoV-2 3CL on type I IFN in the RLR immune pathway opens up novel ideas for future research on SARS-CoV-2.
Keywords: SARS-CoV-2 3CL, RLR signalling pathway, IFNs, Innate immune, IRF3
1. Introduction
Late in 2019, a novel coronavirus strain causing a fatal respiratory disease was reported that subsequently spread worldwide [1]. The World Health Organization (WHO) gave the pathogen an interim name, novel coronavirus 2019 (2019-nCoV) [2]. Following a large number of reports describing the complete gene sequencing of this pathogen, the virus could then be quickly identified in patients by reverse transcription-polymerase chain reaction (RT–PCR) [3]. The WHO subsequently renamed the 2019-nCoV pathogen SARS-CoV-2 and the disease it caused coronavirus disease 2019 (COVID-19) before 12 February 2020 [4]. This was followed by the WHO officially recognizing COVID-19 as a pandemic [5]. It is worth noting that SARS-CoV-2 is an emerging virus, and a large number of studies are still underway. The spread of this pathogen involves several mechanisms, many of which are yet to be well characterized [6].
While the primary target of SARS-CoV-2 is the human immune system, the associated mechanisms remain to be characterized in detail [7]. SARS-CoV-2 is an enveloped virus with a single-stranded positive-sense RNA genome that is approximately 30 kb in length [8]. As an aetiological RNA viral pathogen, SARS-CoV-2 targets the innate immune pathway [9]. This aspect is an emergent avenue of current and future research [10], [11]. SARS-CoV-2 3CL is the 3-chymotrypsin-like protease (also known as nonstructural protein 5, NSP5) that cleaves the polyprotein at 11 different sites to produce various nonstructural proteins [12]. This makes SARS-CoV-2 3CL a vital protein in the replication of virus particles, unlike the structural or accessory protein-coding genes. Thus, SARS-CoV-2 3CL is a potential target for screening molecules that inhibit viral replication [13].
The RLR pathway, a vital part of the innate immune system, regulates the production of type I IFNs to directly or indirectly control viral replication [14], [15]. Given the essential role of SARS-CoV-2 3CL in viral replication and transcription, the protease emerges as a crucial protein [16]. Once a cell is infected by the virus, the activation of the immune system results in the expression of relevant genes in the system to mediate viral replication [17], [18]. To escape the immune system, the virus changes its unfavourable environment into a favourable environment that facilitates viral replication and its subsequent release. This involves unique mechanisms to destroy the immune defence system of the host [19]. Thus, SARS-CoV-2 3CL is a vital target of study, as it may serve as an important foundation for specific treatments [20].
In this study, SARS-CoV-2 3CL was transfected into 293T cells, followed by activation of the RLR antiviral immune pathway by Sendai virus (SeV) infection to examine the effect of the viral protease on this antiviral immune pathway. We aimed to understand the effect of SARS-CoV-2 3CL on the production of type I IFN and the possible mechanism of action.
2. Materials and methods
2.1. Cell culture, plasmids, and reagents
Human 293T cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO). Cells lines in our study have been tested and found free of Mycoplasma. 3CL of SARS-CoV-2 SARS-CoV-2/Wuhan-Hu-1 strain) was synthesized and cloned into expression vector pCGGS with the HA-tag at N-terminus. The IFN-β promoter, ISRE promoter or NF-κB promoter, Myc-MAVS, Flag-RIG-I-N, Myc-TBK1, Flag-p65, Flag-IRF3/5D plasmids have been previously described [21]. For immunoblot analysis, the following antibodies were used: Anti-IRF3 (sc-9082, Santa Cruz), anti-p65 (8242, CST), anti-PCNA (sc-56, Santa Cruz), anti-β-Actin (4970S, CST).
2.2. SeV infection
Once the cells were infected by SeV, the culture medium was removed from the plate and washed twice with phosphate buffered saline (PBS). Serum-free medium containing SeV ([MOI] = 1) was added at the specified time followed by replacement with the medium containing 2% fetal bovine serum.
2.3. RNA Real-time PCR
Total RNA was extracted using TRIzol (Invitrogen), and cDNA was synthesized using M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. The analysis of the relative gene expression levels was performed using a StepOnePlus PCR System (Applied Biosystems) and the following PCR primers: hIFN-β (IFNB1) forward, 5′-AACTGCAACCTTTCGAAGCC-3′; hIFN-β (IFNB1) reverse, 5′-TGTCGCCTACTACCTGTTGTGC-3′; hISG56 (IFIT1) forward, 5′-TTCGGAG-AAAGGCATTAGA-3′; hISG56 (IFIT1) reverse, 5′-TCCAGGGCTTCATTCATAT-3′; hRantes (CCL5) forward, 5′-TGCCTGTTTCTGCTTGCTCTTGTC-3′; hRantes (CCL5) reverse, 5′-TGTGGTAGAATCTGGGCCCTTCAA-3′; SeV M forward, 5′-GTGATTTGGGCGGCATCT-3′; SeV M reverse, 5′-GATGGCCGGTTGGAACAC. GAPDH served as an internal control using the PCR primers, GAPDH forward, 5′-TTGTCTCCTGCGACTTCAAC-AG-3′; GAPDH reverse, 5′-GGTCTGGGATGGAAATTGTGAG-3′.
2.4. Luciferase assay
Following transfection of IFN-β, ISRE, or NF-κB promoters, a β-Gal plasmid, and the relevant plasmids required for the various experiments in 24-well plates, cells were collected after infection with SeV. Twenty-four hours later, cells were lysed using lysis buffer. After centrifugation, luciferase assays were performed with a luciferase assay kit (Beyotime).
2.5. Subcellular fractionation
Transfected cells (5 × 107) that were infected with SeV or uninfected, were washed with PBS and lysed using a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology) according to the manufacturer’s instructions.
2.6. Codon optimization
Due to the availability of a large amount of genomics data, along with increased knowledge of protein expression and structure relationships, codon optimization can significantly improve the level of gene expression [22]. A variety of vital factors involved in different stages of protein expression, such as GC content, mRNA secondary structure, and codon adaptability, can be considered during this process [23]. In this study, the 3CL gene of SARS-CoV-2 (SARS-CoV-2/Wuhan-Hu-1 strain) was optimized accordingly for overexpression in Escherichia coli using a commercial proprietary algorithm, NG® Codon Optimization Technology (Synbio Technologies, NJ, USA).
2.7. Construction of the recombinant expression vector pET-30a-SARS-CoV-2 3CL
The codon-optimized 3CL gene of SARS-CoV-2 (SARS-CoV-2/Wuhan-Hu-1 strain) sequence was synthesized from Make Research Easy. The plasmid pET-30a (+) was linearized using the enzymes BamHI and EcoRI, and the linearized vector was purified using a DNA purification kit (Zoman Biotechnology). The SARS-CoV-2 3CL sequence was cloned into the prokaryotic expression vector pET-30a (+) with the His-tag at the N-terminus using T4 DNA ligase and incubated at 16 °C overnight. DH5α competent cells were provided by Shanghai Weidi Biotechnology. The recombinant plasmid was identified by restriction enzyme digestion and DNA sequencing and was named pET-30a-SARS-CoV-2 3CL.
2.8. Expression and enrichment of the SARS-CoV-2 3CL fusion protein
Escherichia coli (E. coli) BL21 (DE3) competent cells were transformed with the plasmid pET-30a-SARS-CoV-2 3CL to produce the fusion protein BL-pET-30a-SARS-CoV-2 3CL. Cells from a single colony of E. coli BL-pET-30a-SARS-CoV-2 3CL were inoculated into 5 mL of Luria-Bertani (LB) medium containing kanamycin and incubated at 37 °C overnight. Cells were then inoculated into 500 mL of fresh LB medium containing kanamycin until the optical density (OD) 600 of the culture medium reached a value of 0.6–0.8. A final concentration of 0.5 mM isopropyl β-D-thiogalactoside (IPTG) was used to induce protein expression for 5 h at 37 °C. The BL-pET-30a fusion protein was induced using the same conditions by the same method. This was followed by the harvest of noninduced BL-pET-30a, BL-pET-30a-SARS-CoV-2 3CL bacteria and induced BL-pET-30a and pET-30a-SARS-CoV-2 3CL bacteria. Then, PBST buffer solution (containing a protease inhibitor) was added to the harvested bacteria, and the bacterial cells were disrupted under ultrasonic conditions. Finally, the lysed bacterial suspension was centrifuged at 12,000 rpm at 4 °C, and the supernatant and inclusion bodies were separated. Furthermore, the recombinant protein was purified by Ni2+-NTA agarose affinity chromatography using a His-tagged protein purification kit. The supernatant was loaded on a Ni2+-NTA agarose column and washed with washing buffer (20 mM Tris, 0.5 M NaCl, 10 mM imidazole). Then, the bound protein was eluted with elution buffer (20 mM Tris, 0.5 M NaCl, 50–500 mM imidazole).
2.9. Preparation of the anti-SARS-CoV-2 3CL polyclonal antibody
New Zealand rabbits were given a subcutaneous injection of 3CL protein at a concentration of 1 mg/mL emulsified with the same amount of Freund's complete adjuvant. For the second and third immunizations at Day 22 and Day 36, respectively, 1 mL of antigen and an equal volume of Freund's incomplete adjuvant were emulsified to prepare the mixture. Ten days after the last immunization, the rabbits’ carotid arteries were bled to collect approximately 10 mL of whole blood, and the samples were placed in a refrigerator at 4 °C overnight. The serum was collected by centrifugation and divided into 4-mL aliquots, which were stored at –80 °C for later use.
2.10. Immunoblot analysis and SDS–PAGE
Following the recovery of 293T cells showing 3CL eukaryotic expression plasmid transfection for 24 h, the transfected cell proteins were resolved by SDS–PAGE. The protein was transferred to a PVDF membrane, with 5% skimmed milk used for blocking, followed by incubation of the membrane with diluted 3CL antibodies (1:1000) at 4 °C overnight. Then, a secondary antibody labelled with horseradish peroxidase was added, and the final reaction product was visualized using an enhanced chemiluminescence (ECL) kit (Beyotime Technology).
2.11. Statistical analysis
Statistical significance was determined by applying an unpaired Student’s t-test using GraphPad Prism. Values of P < 0.05 were considered to be statistically significant.
3. Results
3.1. SARS-CoV-2 3CL inhibits interferon-β and downstream ISG mRNA expression following SeV infection
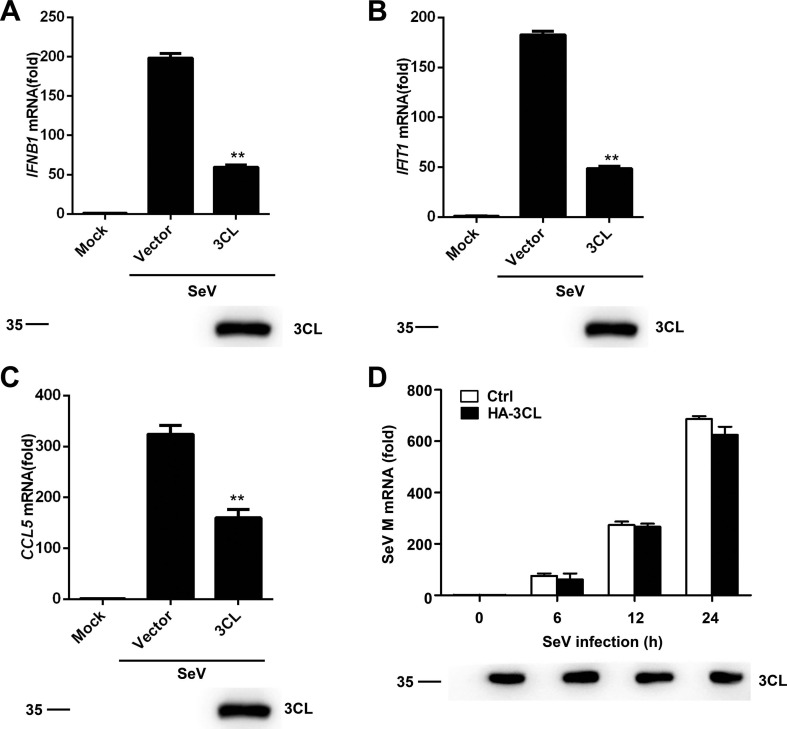
To study whether SARS-CoV-2 3CL inhibits the production of interferon (IFN)-β, 293T cells were transfected with SARS-CoV-2 3CL. At 24 h after transfection, the cells were infected or mock-infected with Sendai virus (SeV) for a period of 8 h. As shown in Fig. 1 A, SARS-CoV-2 3CL significantly inhibited the level of IFN-β mRNA. The mRNA levels of the downstream genes of type I interferon IFIT1 and CCL5 (encoding ISG56 and Rantes, respectively) were also significantly suppressed (Fig. 1B and C). To further investigate whether the expression of 3CL impacts SeV replication, the mRNA expression level of the SeV M gene was monitored in SARS-CoV-2 3CL-overexpressing 293T cells and control (Ctrl) 293T cells. No statistically significant difference in the mRNA expression level of the SeV M gene was observed between SARS-CoV-2 3CL-overexpressing 293T and Ctrl 293T cells (Fig. 1D). These results indicate that SARS-CoV-2 3CL can inhibit the activation of IFN-β when infected with SeV.
Fig. 1.
SARS-CoV-2 3CL inhibits IFN-β and ISG mRNA expression. (A-C) Quantitative PCR analysis of IFNB1 (A), IFIT1 (B), and CCL5 (C) mRNA levels in 293 T cells transfected with empty vector or the HA-3CL-expressing plasmid for 24 h followed by infection with SeV for 8 h (top). Expression of the indicated transfected plasmids was detected by immunoblotting with an anti-3CL antibody (1:1000) (below). (D) Quantitative PCR analysis of SeV M mRNA levels in 293T cells transfected with HA-3CL-expressing plasmid or an empty vector and then treated with SeV for the indicated time periods (top). The expression of the indicated transfected plasmids was detected by immunoblotting (below). *P < 0.05; **P < 0.001; ***P < 0.001 (unpaired, two-tailed Student’s t-test). Data are representative of at least three independent experiments.
To detect the successful expression of 3CL, we prepared a polyclonal antibody against SARS-CoV-2 3CL. The full-length SARS-CoV-2 3CL gene sequence was synthesized by Make Research Easy and cloned into the pET-30a vector using the EcoR I and BamH I sites (supplementary Fig. S1A). The presence of the inserted DNA sequence in the recombinant plasmid was confirmed by restriction enzyme digestion (supplementary Fig. S1B) and then further ascertained by DNA sequencing (supplementary Fig. S1C). Following transformation with pET-30a-SARS-CoV-2 3CL, E. coli Rosetta cells were induced to express the fusion protein. Both the unpurified and purified 3CL products were found to be approximately 38 kDa (supplementary Fig. S2A and B). For the analysis of the titer of the 3CL antibody in serum, rabbit serum injected with 3CL antigen was incubated at ratios of 1:100, 1:500, 1:1000, 1:5000 or 1:10,000. The results showed that the target band was observed at a titer ratio range of 1:5000 to 1:100 (supplementary Fig. S3A). In the following studies, an anti-3CL antibody diluted 1:1000 was utilized. We next detected the expression of 3CL by western blotting analysis (Fig. 1A-D).
3.2. SARS-CoV-2 3CL inhibits the promoter activities of IFN-β, ISRE and NF-κB
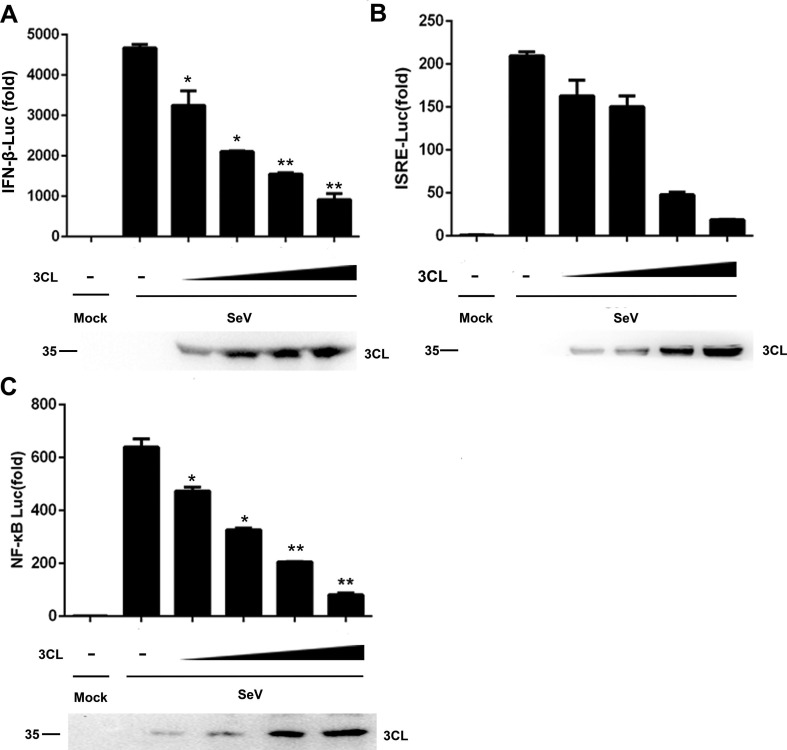
Reporter assays revealed that SARS-CoV-2 3CL downregulated the activation of the IFN-β promoter induced by SeV infection in 293T cells in a dose-dependent manner (Fig. 2 A). Genes encoding type I IFNs are mainly regulated by the IRF3 and NF-κB signalling pathways [24]. Therefore, we next determined the pathways involved in IFN-β activation that were regulated by SARS-CoV-2 3CL. The SeV-induced activation of the ISRE and NF-κB promoters hypothesized to stimulate the RIG-I signalling pathway was inhibited by SARS-CoV-2 3CL in a dose-dependent manner (Fig. 2B and C). These results demonstrated that SARS-CoV-2 3CL inhibited IRF3- and NF-κB-mediated innate immune responses in a dose-dependent manner.
Fig. 2.
SARS-CoV-2 3CL inhibits IFN-β, ISRE, and NF-κB promoter activity. (A-C) Luciferase activity of lysates from 293T cells transfected with IFN-β-Luc for 24 h (A), ISRE-Luc (B), or NF-κB-Luc (C), along with 100 ng to 400 ng of the HA-3CL-expressing plasmid followed by SeV exposure for 6 h. The results are presented relative to the luciferase activity in control cells with the luciferase reporter and empty vector (top). Expression of the indicated transfected plasmids was detected by immunoblotting with an anti-3CL antibody (1:1000) (below). *P < 0.05; **P < 0.001; ***P < 0.001 (unpaired, two-tailed Student’s t-test). Data are representative of at least three independent experiments.
3.3. SARS-CoV-2 3CL inhibits the RIG-I-like signalling pathway
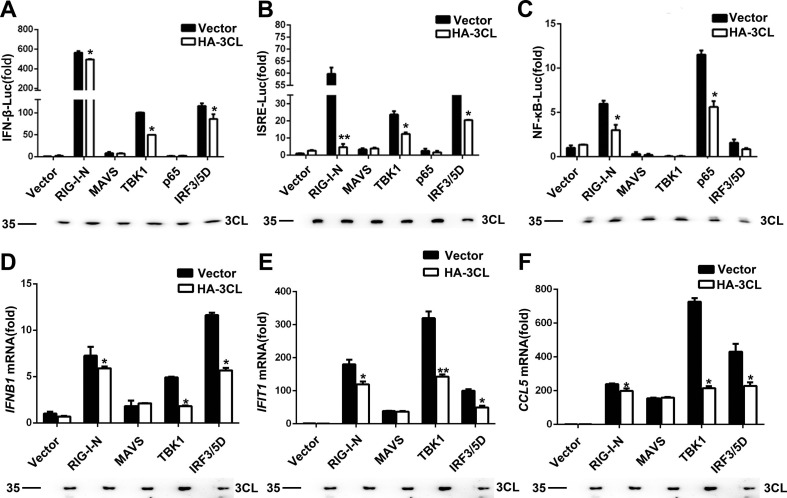
We next examined the signalling cascade at which SARS-CoV-2 3CL blocks the antiviral innate immune response. We cotransfected the plasmid expressing SARS-CoV-2 3CL with those encoding key signalling proteins of the RIG-I-like signalling pathway. This was followed by the determination of the activation of the IFN-β, ISRE, and NF-κB promoters. As shown in Fig. 3 A-C, the overexpression of SARS-CoV-2 3CL inhibited RIG-I-N-, MAVS-, TBK1-, p65-, and IRF3/5D (a constitutively active IRF3 mutant)-triggered IFN promoter activation. Additionally, the results were similar for the mRNA expression of endogenous IFNB1, IFIT1, and CCL5 induced by RIG-I-N, MAVS, TBK1, and IRF3/5D (Fig. 3D-F). Taken together, the results suggest that SARS-CoV-2 3CL suppressed the RIG-I-like signalling pathway.
Fig. 3.
SARS-CoV-2 3CL inhibits RIG-I-like signalling pathways. (A–C) Luciferase activity of lysates from 293T cells transfected with luciferase reporter constructs IFN-b-Luc for 24 h (A), ISRE-Luc (B) or NF-κB-Luc (C), plus Flag-RIG-I-N, Flag-MAVS, Myc-TBK1, Flag-p65, or Flag-IRF3/5D, along with HA-3CL or an empty vector. The results are presented relative to the luciferase activity in control cells expressing the luciferase reporter and empty vector (top). Expression of the indicated transfected plasmids was detected by immunoblotting with an anti-3CL antibody (1:1000) (below). (D–F) Quantitative PCR analysis of IFNB1 (D), IFIT1 (E), and CCL5 (F) mRNA levels in 293T cells transfected for 24 h with the indicated plasmids and HA-3CL or an empty vector (top). The expression of the indicated transfected plasmids was detected by immunoblotting (below). *P < 0.05; **P < 0.001 (unpaired, two-tailed Student’s t-test). Data are representative of at least three independent experiments.
3.4. SARS-CoV-2 3CL targets IRF3 to regulate the RIG-I signalling pathway
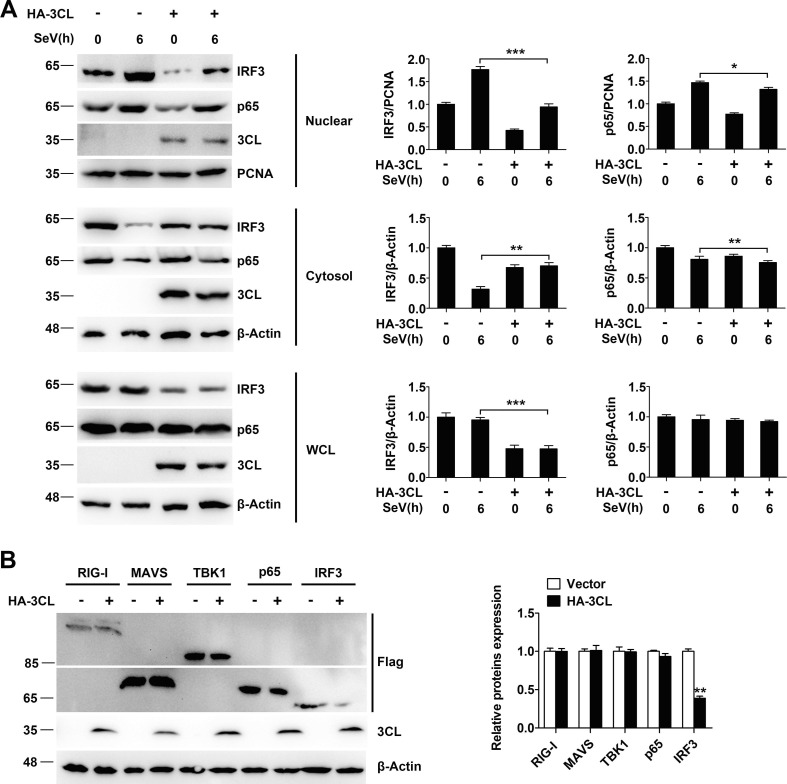
IRF3 and p65 proteins translocate from the cytoplasm to the nucleus to activate the transcription of type I IFNs. We next assessed the effect of SARS-CoV-2 3CL on the nuclear translocation of IRF3 and p65. Subcellular fractionation and relative quantification analysis demonstrated that IRF3 and p65 rapidly translocated from the cytoplasm to the nuclear fraction in 293T cells compared with SARS-CoV-2 3CL-overexpressing 293T cells after SeV infection (Fig. 4 A). In an attempt to identify candidates that are direct targets for the proteolysis of SARS-CoV-2 3CL, we initially tested the effect of SARS-CoV-2 3CL on different components of the RLR pathway in terms of their stability. We found that SARS-CoV-2 3CL promoted the degradation of IRF3 (Fig. 4B). The quantifications of the western blots are shown on the right sides of the blots. Together, these data suggest that IRF3 is the target protein of SARS-CoV-2 3CL in the suppression of RIG-I-mediated type I IFN production.
Fig. 4.
. SARS-CoV-2 3CL targets IRF3 to regulate the RIG-I signalling pathway. (A) Immunoblot analysis of the indicated proteins in 293T cells transfected with HA-3CL or an empty vector and then infected with SeV for 6 h, followed by nuclear-cytoplasm extraction. The expression levels of the indicated proteins were analysed with the indicated antibodies (left). Relative quantification analysis of IRF3 and p65 (right). (B) Immunoblot analysis of lysates of 293T cells transfected with Flag-RIG-I, Flag-MAVS, Flag-TBK1, Flag-p65, or Flag-IRF3, along with HA-3CL or an empty vector for 24 h. The expression levels of the indicated proteins were analysed with the indicated antibodies (left). Relative quantification analysis of RIG-I, MAVS, TBK1, p65 and IRF3 (right). *P < 0.05; **P < 0.001; ***P < 0.001 (unpaired, two-tailed Student’s t-test). Data are representative of at least three independent experiments.
4. Discussion
The production of interferons (IFNs) is essential to eliminate invading viruses and maintain immune homeostasis [25], [26]. Moreover, type I interferon inhibits viral replication while also regulating the expression of downstream genes [27]. The production of type I interferon in the antiviral immune pathway involving RIG-I-like receptors is influenced by multiple target proteins [28], [29]. SARS-CoV-2 is a novel strain of coronavirus that cause an infectious respiratory disease and was discovered in humans in late 2019 [30]. The virus is a serious threat to life [31]. The SARS-CoV-2 3CL protease plays an important role in the replication of the virus [32]. In this study, the role of SARS-CoV-2 3CL in the innate immune antiviral pathway was examined. We found that SARS-CoV-2 3CL can inhibit the production of interferon-β and its downstream ISGs (ISG56 and Rantes) in SeV-stimulated 293T cells.
Coronavirus 3C-like protease is an important protease in viral replication [33], [34]. To study the mechanisms of coronavirus infection, several extensive studies have been conducted [35]. The genetic sequence of the virus was also released [36]. It is known that the first line of defence of the immune system plays an important role in the process of resisting viral invasion [37]. Infection by an RNA virus activates the RIG-I-like receptor signalling pathway and promotes the expression of IFN-I along with several downstream genes, including ISGs [38]. In our work, 293 T cells were transfected with plasmids expressing the SARS-CoV-2 3CL protein followed by infection with SeV. This was followed by detecting the expression of IFN-β and ISGs using q-PCR. The results showed that the SARS-CoV-2 3CL protein had suppressive effects on the mRNA levels of IFNs and the ISGs studied. However, a recent report showed that no significant difference in SeV-mediated IFN-β activation was detected in the expression of 3CL (also known as NSP5) [39]. There are some possible reasons for this discrepancy. The different experimental details lead to different results, such as the plasmid transfection dose or time points in response to SeV infection. Very recently, Fung et al. reported the suppressive effect of 3CL on IFN-β gene transcription. They revealed that 3CL proteins encoded by SARS-CoV and SARS-CoV-2 antagonized IFN production by retaining phosphorylated IRF3 in the cytoplasm [35].
Host proteins possibly influence important target proteins in the RLR pathway, such as MAVS, RIG-I-N, TBK1, p65, and IRF3, to affect the production of type I interferon. In our study, we found that SARS-CoV-2 3CL can downregulate the production of type I interferon in the RLR pathway. Subcellular fractionation demonstrated that SARS-CoV-2 3CL inhibited the nuclear translocation of IRF3 and p65. In an attempt to identify candidates that are direct targets for proteolysis of SARS-CoV-2 3CL, we tested the effects of SARS-CoV-2 3CL on different components of the RLR pathway in terms of their stability. We found that SARS-CoV-2 3CL significantly promoted the degradation of IRF3 upon SeV infection. Taken together, these data suggested that IRF3 was the target protein of SARS-CoV-2 3CL to suppress RIG-I-mediated type I IFN production. The target of the protease to inhibit the production of type I interferon and the associated mechanisms will be the subject of future research.
The construction and expression of the prokaryotic expression vector SARS-CoV-2 3CL with the synthesized SARS-CoV-2 3CL protein was injected into rabbits along with an antigen adjuvant to produce a SARS-CoV-2 3CL polyclonal antibody. In this study, our results showed that the target band was observed at a titer ratio range of 1:5000 to 1:100, and we incubated the PVDF membrane with diluted 3CL antibodies (1:1000) at 4 °C overnight in immunoblot analysis. The preparation of this polyclonal antibody has also laid a foundation for future research.
In our study, the production of type I interferon in the RLR antiviral immune pathway was significantly affected by SARS-CoV-2 3CL. This finding provides a foundation for future detailed studies on SARS-CoV-2, thereby making a definite contribution to research that can affect human life and health.
CRediT authorship contribution statement
Wenwen Zhang: Methodology, Formal analysis, Writing – original draft. Zhenling Ma: Conceptualization, Methodology, Funding acquisition. Yaru Wu: Methodology, Validation. Xixi Shi: Methodology, Validation. Yanyan Zhang: Validation, Data curation. Min Zhang: Software. Menghao Zhang: Investigation. Lei Wang: Methodology, Software. Wei Liu: Conceptualization, Methodology, Resources, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by grants from the National Natural Science Foundation of China (No. 31802164), the research start-up fund to topnotch talents of Henan Agricultural University (No. 30500618 and 30500424), and the Scientific and Technological Research Project of Henan Province (No. 212102310898).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cyto.2021.155697.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Pius T., Nabaasa S., Kusiima N., Eze E.D., Robinson S. Combating the Spread of COVID-19, the Challenges Faced and Way forward for the International Community: A Review. Open Access Library J. 2020;7(11):1–7. [Google Scholar]
- 2.Guo Y., Cao Q., Hong Z., Tan Y., Chen S. The origin, transmission and clinical therapies on coronavirus disease 2019(COVID-19) outbreak-an update on the status. Mil. Med. Res. 2020;7(01):99–109. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., Veer B.V.D., Brink S.V.D., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhama K., Khan S., Tiwari R., Sircar S., Rodriguez-Morales A.J. Coronavirus Disease 2019 - COVID-19. Clin. Microbiol. Rev. 2020;33(4):e00028–e120. doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naqvi A., Fatima K., Mohammad T., Fatima U., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta. Mol. Basis. Dis. 2020:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shomuradova A., Vagida M., Sheetikov S., Zornikova K., Kiryukhin D., Titov A., Peshkova I., Khmelevskaya A., Dianov D., Malasheva M., Shmelev A., Serdyuk Y., Bagaev D., Pivnyuk A., Shcherbinin D., Maleeva A., Shakirova N., Pilunov A., Malko D., Khamaganova E., Biderman B., Ivanov A., Shugay M., Efimov G. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity. 2020;53(6):1245–1257. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esmaeilzadeh A., Jafari D., Tahmasebi S., Elahi R., Khosh E. Immune-Based Therapy for COVID-19. Adv. Exp. Med. Biol. 2021;1318 doi: 10.1007/978-3-030-63761-3_26. [DOI] [PubMed] [Google Scholar]
- 8.Wu F., Zhao S., Yu B., Chen Y.M., Zhang Y.Z. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature. 2020;580(7803):E7. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H., Yingying L., Hou F. SARS-CoV-2 infection and the antiviral innate immune response. J. Mol. Biol. 2020;12(12):963–967. doi: 10.1093/jmcb/mjaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., Hui H., Bray B., Gonzalez G.M., Zeller M., Anderson K.G., Knight R., Smith D., Wang Y., Carlin A.F. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection - ScienceDirect. 2021;35(6):109091. doi: 10.1016/j.celrep.2021.109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallenave J.M., Guillot L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in Covid-19: Key Therapeutic Targets? Front. Immunol. 2020;11:1229. doi: 10.3389/fimmu.2020.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C., Yu X., Kuo C., Min J., Chen S., Wu S., Ma L., Liu K., Guo R. Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases. FEBS J. 2021 doi: 10.1111/febs.15696. [DOI] [PubMed] [Google Scholar]
- 13.Roe M.K., Junod N.A., Young A.R., Beachboard D.C., Stobart C.C. Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19. J. Gen. Virol. 2021;102(3) doi: 10.1099/jgv.0.001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Sastre A., Biron C. Type 1 Interferons and the Virus-Host Relationship: A Lesson in Détente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 15.Cao Z., Xia Z., Zhou Y., Yang X., Hao H., Peng N., Liu S., Zhu Y. Methylcrotonoyl-CoA carboxylase 1 potentiates RLR-induced NF-κB signaling by targeting MAVS complex. Sci. Rep. 2016;6(1):33557. doi: 10.1038/srep33557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand K., Ziebuhr J., Wadhwani P., Mesters J., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 17.Matta B., Barnes B.J. Coordination between innate immune cells, type I IFNs and IRF5 drives SLE pathogenesis. Cytokine. 2019;132(5):154731. doi: 10.1016/j.cyto.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scutigliani E.M., Kikkert M. Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Factor Rev. 2017;37:17–27. doi: 10.1016/j.cytogfr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco R., Aguayo F. Role of BamHI-A Rightward Frame 1 in Epstein-Barr Virus-Associated Epithelial Malignancies. Biology (Basel) 2020;9(12):461. doi: 10.3390/biology9120461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tahir Ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Z., Zhang W., Fan W., Wu Y., Zhang M., Xu J., Li W., Sun L., Liu W., Liu W. Forkhead box O1-mediated ubiquitination suppresses RIG-I-mediated antiviral immune responses. Int. Immunopharmacol. 2020;90:107152. doi: 10.1016/j.intimp.2020.107152. [DOI] [PubMed] [Google Scholar]
- 22.Gao C.Y., Xu T.T., Zhao Q.J., Li C.L. Codon optimization enhances the expression of porcine β-defensin-2 in Escherichia coli. Genet. Mol. Res : GMR. 2015;14(2):4978–4988. doi: 10.4238/2015.May.12.1. [DOI] [PubMed] [Google Scholar]
- 23.Liu B., Kong Q., Zhang D., Yan L. Codon optimization significantly enhanced the expression of human 37-kDa iLRP in Escherichia coli, 3. Biotech. 2018;8(4):210. doi: 10.1007/s13205-018-1234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiscott J. Convergence of the NF-κB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18(5–6):483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Porritt R.A., Hertzog P.J. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36(3):150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Braciale T.J., Sun J., Kim T.S. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 2012;12(4):295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W., Li J., Zheng W., Shang Y., Zhao Z. Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. eLife. 2017;6:e24425. doi: 10.7554/eLife.24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao C., An R., Yu Y., Dai H., Qu Z., Gao M., Wang J. BICP0 Negatively Regulates TRAF6-Mediated NF-κB and Interferon Activation by Promoting K48-Linked Polyubiquitination of TRAF6. Front. Microbiol. 2019;10:3040. doi: 10.3389/fmicb.2019.03040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y., Oanh N., Heo J., Kim S., Lee H., Lee H., Lee J., Kang H., Lim W., Yoo Y., Cho H. Dual targeting of RIG-I and MAVS by MARCH5 mitochondria ubiquitin ligase in innate immunity. Cell Signal. 2020;67:109520. doi: 10.1016/j.cellsig.2019.109520. [DOI] [PubMed] [Google Scholar]
- 30.Mbaye M.M., Khalfi B.E., Hafiane S.E., Soukri A. Characteristics of the New Coronavirus Named SARS-CoV-2 Responsible for an Infection Called Coronavirus Disease 2019 (COVID-19) Annual Res. Rev. Biol. 2020:68–77. [Google Scholar]
- 31.Mai A., Yun X.B., Fei X.B., Ys A. Diagnosis of COVID-19, vitality of emerging technologies and preventive measures. Chem. Eng. J. 2021;423:130189. doi: 10.1016/j.cej.2021.130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Biomed. 2020;40(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Cao H., Cheng Y., Zhang X., Han H. Inhibition of Porcine Epidemic Diarrhea Virus Replication and Viral 3C-Like Protease by Quercetin. Int. J. Mol. Sci. 2020;21(21):8095. doi: 10.3390/ijms21218095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo C.J., Liang P.H. Characterization and inhibition of the main protease of severe acute respiratory syndrome coronavirus. ChemBioEng Rev. 2015;2(2):118–132. [Google Scholar]
- 35.Fung S.Y., Siu K.L., Lin H., Man L.Y., Jin D.Y. SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int. J. Dev. Biol. 2021;17(6):1547–1554. doi: 10.7150/ijbs.59943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan F.W., Kok K.H., Zhu Z., Chu H., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jun E.J., Kim Y.K. Activation of innate immune system during viral infection: role of pattern-recognition receptors (PRRs) in viral infection. J. Bacteriol. Virol. 2009;39(3):145–157. [Google Scholar]
- 38.Vazquez C., Horner S.M., Sullivan C.S. MAVS coordination of antiviral innate immunity. J. Virol. 2015;89(14):6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei X., Dong X., Ma R., Wang W., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):1015–1024. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.