Abstract
Antigen identification is an important step in the vaccine development process. Computational approaches including deep learning systems can play an important role in the identification of vaccine targets using genomic and proteomic information. Here, we present a new computational system to discover and analyse novel vaccine targets leading to the design of a multi-epitope subunit vaccine candidate. The system incorporates reverse vaccinology and immuno-informatics tools to screen genomic and proteomic datasets of several pathogens such as Trypanosoma cruzi, Plasmodium falciparum, and Vibrio cholerae to identify potential vaccine candidates (PVC). Further, as a case study, we performed a detailed analysis of the genomic and proteomic dataset of T. cruzi (CL Brenner and Y strain) to shortlist eight proteins as possible vaccine antigen candidates using properties such as secretory/surface-exposed nature, low transmembrane helix (< 2), essentiality, virulence, antigenic, and non-homology with host/gut flora proteins. Subsequently, highly antigenic and immunogenic MHC class I, MHC class II and B cell epitopes were extracted from top-ranking vaccine targets. The designed vaccine construct containing 24 epitopes, 3 adjuvants, and 4 linkers was analysed for its physicochemical properties using different tools, including docking analysis. Immunological simulation studies suggested significant levels of T-helper, T-cytotoxic cells, and IgG1 will be elicited upon administration of such a putative multi-epitope vaccine construct. The vaccine construct is predicted to be soluble, stable, non-allergenic, non-toxic, and to offer cross-protection against related Trypanosoma species and strains. Further, studies are required to validate safety and immunogenicity of the vaccine.
Subject terms: Vaccines, Computational biology and bioinformatics
Introduction
New data-driven approaches, such as reverse vaccinology1,2, systems vaccinology3, and machine learning4, have started to capitalize on the vast amount of omics data available for vaccine design. Several computational studies have analysed genomes or proteomes of individual pathogenic strains or species to predict vaccine candidates5–10. In one of these studies, researchers have used the protein–protein interaction dataset and a network biology approach to prioritize vaccine targets for Borrelia burgdorferi11. Moreover, Goodswen et al.12 used a machine learning approach to distinguish between true and false vaccine candidates for eukaryotes including Caenorhabditis elegans, Toxoplasma gondii and Plasmodium sp.12.
There are several tools, resources, and databases available in the immuno-informatics domain that have contributed to the development of vaccines in the recent past13–15. In 2019, Dalsass et al. compared six open-source standalone Reverse Vaccinology (RV) programs designed for bacterial pathogens: NERVE, VaxiJen, Vaxign, Bowman-Heinson, Jenner-predict, and VacSol and tested them on eleven different bacterial proteomes16. Several advantages, as well as limitations, have been reported in the existing pipelines or tools. For instance, most of the programs and algorithms have been built around bacterial and prokaryotic systems with only a little work with eukaryotic pathogens, including Trypanosoma cruzi. Furthermore, the issue of false-positive predictions remains a challenge. (See “Supplementary Website”).
Despite significant advancements in vaccinology, computational proteomics, machine learning, and reverse vaccinology, finding vaccine candidates, producing them in the laboratory, and confirming their efficacy in animal models remains a complicated undertaking. Thus, there is an urgent need for building pipelines or computational frameworks, to integrate diverse algorithms and databases using a single input and provide meaningful results for researchers working on vaccine development.
In this work, we are introducing an integrated framework that combines immuno-informatics approaches, bioinformatics tools, and supervised machine learning-based tools for vaccine discovery. Here, we rank or classify pathogen proteins based on their propensity to be good vaccine candidates and to design safe and effective multiple epitope vaccine candidates using a set of tools such as PsortB, WoLF PSORT, BLAST, HMMTop, ProtParam, FungalRV, NetCTL, VaxiJen 2.0, or IEDB tools. As a proof of concept, we applied our system to different pathogens including Mycobacterium tuberculosis, Plasmodium vivax, Candida albicans, and Influenza A virus and identified several key vaccine candidates.
Since we have a long-term interest in the development of vaccines against neglected tropical diseases, we performed a detailed analysis of genomic and proteomic datasets of T. cruzi, the causative agent of Chagas disease (CD). CD affects an estimated 6.5 million people (healthdata.org), particularly those living in extreme poverty in Latin-America and certain areas of the USA, such as South Texas. An estimated 10,000–20,000 patients succumb to CD annually17 and previous studies have reported several issues in the development of a vaccine against CD18. Since monovalent vaccines had only partial success, the idea of combining vaccine candidates was proposed19,20. Recently, Sanchez Alberti et al. designed Traspain, a chimeric antigen including the N-terminal domain of Cruzipain (Cz), the central region of the Amastigote surface protein 2 and a subdominant region of an inactive trans-sialidase21 as a potential vaccine candidate.
Clinical studies, like the BENEFIT trial, have shown limited benefits of therapeutic drugs (i.e., benznidazole) in halting the progression of CD-associated cardiovascular disease22,23. Even with available antiparasitic drugs, patients with chronic Chagasic cardiomyopathy (CCC) experience cardiac inflammation and fibrosis leading to heart failure, conduction abnormalities, or sudden death. Several studies have demonstrated that CCC from chronic T. cruzi infection in the heart can be controlled by therapeutic vaccines in animal models, but so far, no vaccine has entered human clinical trials24,25. Using computational techniques, we not only identified vaccine targets against T. cruzi but also designed a putative multi-epitope vaccine along with an in-silico model of immune stimulation that predicts responses associated with protective immunity.
Methodology
The Vax-ELAN pipeline (https://vac.kamalrawal.in/vaxelan/) was developed using computational tools to screen the pathogen proteomes. Vax-ELAN evaluates and shortlists proteins that show the relevant characteristics (features) to qualify them as potential vaccine candidates (Supplementary Fig. 1, Fig. 1).
Figure 1.
Methodology for developing a multi-epitope subunit vaccine construct (Draw.io—https://www.diagrams.net/-14.6.10).
Features and thresholds
The features used in Vax-ELAN include subcellular localization26, secretory/non-secretory protein27, stability28, cleavage sites29, adhesion property30, CTL epitope prediction, MHC class-I binding31, transmembrane helix prediction32, essentiality33, virulence34, molecular weight28, non-homology with host proteins, etc.
In Supplementary Table 1, we summarize various research studies to provide the rationale for the selection of particular features and thresholds. For example, Pizza et al. reported that the main cause of failed cloning and expression of 250 out of 600 vaccine candidates from Neisseria meningitidis B was due to the presence of more than one transmembrane spanning region (TM)6. Thus, we decided to have no more than two predicted TMs as an a priori requirement. Further, to avoid autoimmunity, the vaccine targets should not be similar to human proteins, therefore BLASTp was utilized to filter those proteins having > 30% identity with human proteins [E-value < 0.005]35.
Because the immune system readily recognizes surface-exposed proteins on the pathogen, predicting the subcellular localization of the proteins serves as one of the major criteria for designing a vaccine candidate. Therefore, we used tools such as PSORTb2.0, WoLF PSORT, TargetP and CELLO36,37 to identify the localization of proteins as extracellular, outer membrane, cytoplasmic, periplasmic, and inner membrane.
Tools
To compute these features, we used different bioinformatics and immunoinformatics tools/databases such as TargetP26, SignalP27, ProtParam28, PSORTb38, WoLF PSORT39, TMPred40, NetMHC41, NetChop29, BLAST42, Virulence Factor Database [VFdb]34 and microbial virulence database [MvirDB]43 (Table 1).
Table 1.
Tools used for extraction of features along with their cut-off values.
| S. no. | Features | Tool | Cut-off | References |
|---|---|---|---|---|
| 1 | Proteins with less number of trans-membrane helices |
TmPred TMHMM HMMtop |
≤ 1 |
Monterrubio-López et al. (2015)5 Naz et al. (2019)44 Solanki et al. (2018)45 |
| 2 | Non-homology with human | BLAST with human proteome | e-value:10e − 5, identity > 30%, query coverage ≥ 70% | Pearson et al. (2013)35 |
| 3 | Stability (instability index value) | ProtParam | < 40 | Solanki et al. (2018)45 |
| 4 | Non-allergen | Blastp with AllerBase | e-value:10e − 5, identity > 30% | Pearson et al. (2013)35 |
| 5 | Adhesion prediction | FungalRv | ≥ − 1.2 | Monterrubio-López et al. (2015)5 |
| 6 | Essential genes prediction | DEG Database | e-value:10e − 5, identity > 30%, query coverage ≥ 70% | Solanki et al. (2018)45 |
| 7 | Virulence factor | Blastp with VFDB | e-value:10e − 5, identity > 30% | Solanki et al. (2018)45 |
| 8 | Molecular weight | ProtParam | < 110 kDa | Naz et al. (2019)44 |
| 9 | Secretory/non-secretory protein | Signalp (dvalue) | ≥ 0.5 | Liebenberg et al. (2012)46 |
| 10 | Non-bacterial pathogen/BLAST with gut flora | Blastp with GutfloraDB | e-value:10e − 5, identity > 30%, query coverage ≥ 70% | Naz et al. (2019)44 |
| 11 | Sub-cellular localization | Targetp | ≥ 0.8 | Goodswen et al. (2014)47 |
| 12 | MHC Class-1 binding (number of high binders) | NetMHC | ≥ 4.9 | Schroeder and Aebischer (2011)48 |
| 13 | MHC Class-1 binding (number of weak binders) | NetMHC | ≥ 5.05 | Schroeder and Aebischer (2011)48 |
| 14 | Number of cleavage sites | NetChop | ≥ 110 | Dhanda et al. (2017)49 |
| 15 | Number of peptides | NetMHC | < 500 | Schroeder and Aebischer (2011)48 |
| 16 | Number of amino acids | NetChop | < 500 | Dhanda et al. (2017)49 |
| 17 | Cytotoxic T lymphocytes (CTL epitope prediction) (number of MHC ligands) | NetCTL | < 7.5 | Solanki et al. (2018)45 |
| 18 | Antigenicity | Vaxijen | > 0.4 | Monterrubio-López et al. (2015)5 |
| 19 | Subcellular localization | Psortb | > 9.5 | Muruato et al. (2017)50 |
| 20 | MHC Class-1 binding prediction | IEDB (HLA02*01) | > 50 nM | Schroeder and Aebischer (2011)48 |
| 21 | Subcellular localization | Psortb |
Cell wall Extracellular |
Naz et al. (2019)44 Muruato et al. (2017)50 Solanki et al. (2018)45 |
| 22 | Subcellular localization | Psortb | Outer membrane, extracellular and periplasmic | Naz et al. (2019)44 |
| 23 | Subcellular localization | Wolf Psort | Extracellular or plasma membrane | Watanabe et al. (2021)51 |
Strategies
Vax-ELAN has the provision to scan protein sequences (or proteomes) using multiple strategies (See Supplementary Fig. 1). For instance, in strategy 1, we used subcellular localization prediction programs to identify outer membrane and periplasmic proteins. Since, there are no specific algorithms available for protozoa or parasites, we used tools such as PSORTB (Strategy 1A) as well as WoLF PSORTB (Strategy 1B) for the prediction of subcellular localization (See Supplementary Fig. 1).
Subsequently, we employed various filters to prioritize proteins based on features that are associated with antigenicity, including adhesion, allergenicity, and non-homology with the host proteome. The filtering strategy has been reported to find vaccine targets in Shigella sonnei52, Brucella sp.53 and Helicobacter pylori54.
Pearce et al.55 had reported the induction of protective immunity against Schistosoma mansoni by vaccination with schistosome paramyosin (Sm97), a nonsurface parasite antigen in a mouse model55. Therefore, we designed strategy 2 (without sub-cellular localisation filter) in Vax-ELAN, so that there is minimal risk of filtering important (non-surface) antigens.
In another alternative approach based upon inclusion (strategy 4), we use all possible tools (without elimination/filtering) to perform a comprehensive evaluation of a given protein sequence.
In this approach, we also convert the outputs from different tools (N) into binaries (1/0) using threshold values (Supplementary Tables 2, 3). Second, a row-wise sum corresponding to all the properties [i.e., Si] was computed. This is followed by the computation of probability value (Pi = Si/N). Higher Pi indicates the propensity of a given protein molecule to possess desirable properties in order to be a good vaccine candidate (Supplementary Table 4).
For instance, trans-sialidases (TS) were found to be among the top-ranking hits (with a comparatively higher Pi value of 0.75). TS have been reported to be important vaccine candidates in numerous preclinical immunological studies in TC-CLB (Supplementary Table 5). Likewise, important vaccine targets were reported as top-scoring hits from other pathogens as well. For example, ferric enterobactin receptor protein (Si = 9; Pi 0.75) [present in N. gonorrhoeae] was shortlisted as a vaccine target56 (Supplementary File 1).
In the next section, we describe the approach for building a machine learning-based tool using components of the Vax-ELAN framework.
Optimisation of thresholds
Though threshold values (listed in Table 1) are supported by literature evidence there is no guarantee of optimality when they are used in machine learning systems. Therefore, we decided to optimise these cut-offs using a quantitative approach. For this reason, we collected protein sequences (antigenic) with experimental evidence from different organisms and labelled them as examples of a positive dataset (see, VaxiDL supplementary). Similarly, another dataset consisting of non-immunogenic proteins (negative dataset) was also compiled. Next, we compared the distributions of each property in the positive and negative datasets. Subsequently, we harnessed Receiver Operating Curves (ROC) to find thresholds at which positive and negative examples could be discriminated against (See Supplementary Fig. 2). With the help of optimized thresholds generated for each property (Supplementary Table 6), we converted the numerical/categorical values of each property into a binary score (0 or 1).
Machine learning approach
A dataset containing positive and negative protein sequences (PVCs) was compiled using text data mining and manual curation. A total of 11 biological and 1436 physicochemical features were computed for the dataset using several bioinformatics tools. Further, this dataset was subdivided into training, testing, and validation datasets, followed by scaling and normalization of data. Next, a DL model with Fully Connected Layers (FCLs) was constructed, hyper-tuned and trained. The Vaxi-DL model was benchmarked against known PVC prediction tools such as VaxiJen and Vaxign-ML. The preliminary results have shown that the Vaxi-DL model surpassed other PVC-prediction servers in terms of accuracy and efficiency (See, https://vac.kamalrawal.in/vaxidl/). Areas under the receiver operating characteristics curves (AUC) were primarily used to assess the algorithm. On an independent dataset, the algorithm achieved an AUC of 0.90 (95% CI 0.91–0.93) for detecting potential vaccine candidates (Manuscript in Preparation).
Screening of proteomes of pathogens to shortlist vaccine candidates
Using Vax-ELAN (strategy 4), we screened proteomes of 21 pathogens [seven bacterial, four fungal, five protozoan, and five viral proteomes] to shortlist and rank proteins as potential vaccine targets (Supplementary Table 7). We found that the highest scoring results were enriched in vaccine targets (with experimental evidence reported in the literature) (Supplementary File 1). To illustrate, GPI anchored protein was predicted as one of the top vaccine targets (Pi score 0.75) while screening the Aspergillus fumigatus proteome57. VAX-Elan also predicted Glycerol-3-phosphate acyltransferase (GPAT3) (having Pi score 0.71) in M. pneumoniae58. In addition, Histone 2B59 was shortlisted as one of the vaccine targets (Pi = 0.67) in Plasmodium vivax, and CyRPA60 (Pi = 0.67) was shortlisted as one of the candidates in Plasmodium falciparum. Cysteine protease61 (Pi = 0.67), 24-c-methyltransferase62 (Pi = 0.58) and iron superoxide dismutase63 (Pi = 0.58) in Leishmania donovani were found as potential vaccine targets.
Evaluation of experimentally known antigenic and non-antigenic proteins
Protective antigens are proteins that can evoke an adaptive immune response against infectious and non-infectious diseases64. To begin with, we collected four datasets of protective antigens belonging to bacteria, protozoa, fungi, and viruses. Each set is composed of antigenic and non-antigenic sequences collected from previously reported resources such as Protegen65. For example, we collected 1237 bacterial antigen sequences as a positive dataset (Supplementary File 2). To create a negative/control dataset, we randomly selected those proteins (from the same species) which had less than 10% sequence similarity with sequences belonging to the positive dataset. We also removed redundancies in each dataset by filtering protein sequences that had sequence similarities of more than 30%36. Thus, the filtered positive dataset had 670 unique bacterial antigens whereas 677 sequences were obtained for the negative dataset66. Similarly, we created independent datasets for protozoan, fungal, and viral pathogens (Supplementary Table 8). Subsequently, we applied the Vax-ELAN tool on sequences of positive and negative datasets (Supplementary Fig. 3a–d, Fig. 2). We found that known antigens had comparatively higher Pi values when compared to non-antigens (Mann–Whitney U test, p-value < 0.005).
Figure 2.
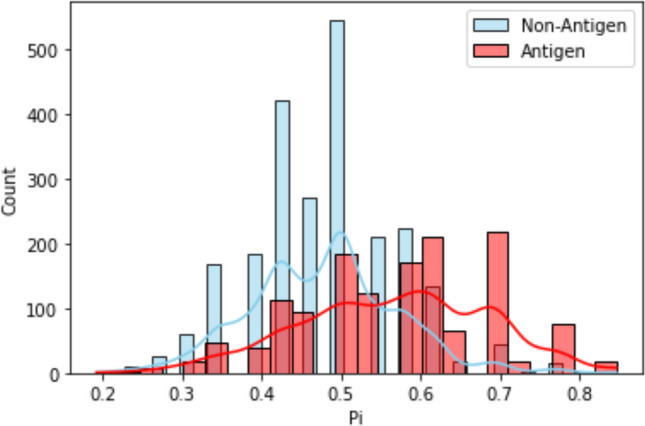
Frequency distribution of Pi values based on the results from positive and negative datasets of bacteria, protozoa, fungi, and viruses. The Y-axis represents sequence count. The X-axis represents the *Pi score values for each sequence. Blue depicts non-antigen and red antigen sequences. *Pi stands for probability value where Pi = Si/N (where, Si refers to the row-wise sum values and N refers to the total number of the tools).
Application of Vax-ELAN on T. cruzi
Retrieval of genome and proteome sequences for vaccine designing
We applied Vax-ELAN on two different strains of T. cruzi, CL Brenner and Y. The whole-genome sequences of T. cruzi (strains CL Brenner and Y) were obtained from NCBI (Accession ID: NZ_AAHK00000000 and Accession ID: NMZO00000000) along with protein sequences in FASTA format. The results of TC-CLB are shown in the subsequent sections of the manuscript whereas the results of Y strain are shown in the Supplementary File 5.
Vax-ELAN pipeline for prediction of vaccine candidates
T. cruzi protein sequences were screened based on several parameters such as cellular localization26, transmembrane helices27, instability index value28, allergenicity67, antigenicity66, the probability of having adhesion-like characteristics30, and non-homology with human proteins. Additionally, the T. cruzi proteins were also screened against the Database of Essential Genes [DEG]33, using the BLAST tool [bit score of 100, cut-off (E-value) of 1E − 5, and BLOSUM 62 matrix]. Further, virulent proteins were extracted using the Virulence Factor Database [VFdb]34 and microbial virulence database [MvirDB]43. Ideally, the vaccine targets should not be similar to the human proteins, therefore BLASTp was utilized to filter those T. cruzi proteins having > 35% identity with human proteins [E-value < 0.005] (See Supplementary Table 9, Supplementary Fig. 4).
Alternate strategies adopted for protein filtering
Apart from the methods mentioned in the previous section, we also used alternate strategies to identify potential vaccine targets from T. cruzi CL Brenner (TC-CLB). For example, in one of the experiments on proteome screening, we filtered TC-CLB proteins using a set of bioinformatics tools. First, we used the PSORTb tool, to check subcellular localization, followed by the BLASTp tool to evaluate non-homology with human proteins. Subsequently, we used ProtParam to compute the stability of proteins, succeeded by a BLASTp search against the allergen database to filter non-allergen proteins. Furthermore, we used the VaxiJen2.0 server66 to check the antigenicity of the filtered set of proteins and then used FungalRV30 to predict adhesion molecule-like properties. This strategy generated a set of potential vaccine candidates. As an alternative strategy (1B), we used the WoLF PSORT tool for screening in the first step instead of PSORTb. Additionally, we repeated this analysis after the randomizing order of the application of filters (See Supplementary File-A).
Conversion of proteins’ feature/property values into binary values
A row-wise sum corresponding to all the properties [i.e., total score] was computed for TC-CLB. Thereafter, all the proteins of TC-CLB were ranked according to the total score (Si or Pi). Finally, the top 100 unique proteins were selected for further analysis [See, Supplementary File-A (Strategy-4)].
Strategy—ORF-based screening of TC-CLB
To perform comprehensive screening for all possible vaccine candidates, we downloaded the T. cruzi CL Brenner and TC-Y genomes from NCBI to find out all possible ORFs. We used Prodigal68 to predict 121,349 in the genome. Next, the predicted ORFs were subjected to evaluation with tools such as WoLF PSORT/PSORTB, BLAST, ProtParam, Vaxijen, and Fungal RV to filter proteins.
Comparison of different strategies to find top ranking proteins
We collected the top-ranking hits from different strategies and used python-based programs to find common and unique proteins (See, Supplementary File-B). Shortlisted proteins reported from multiple strategies were used in subsequent steps such as epitope prediction and vaccine construction.
Interspecies and inter-strain comparison of trypanosoma
We retrieved proteomes from thirteen strains of T. cruzi (See Table 2) and four related species of Trypanosoma (See Supplementary File 3). Subsequently, we applied the Vax-ELAN server to obtain top-ranking hits using strategy 4.
Table 2.
Different strains and species of Trypanosoma used for the identification of key vaccine candidates.
| Trypanosoma cruzi (different strains) | Trypanosoma species with its strain |
|---|---|
| Trypanosoma cruzi Berenice | Trypanosoma brucei brucei (927/4 GUTat10.1) |
| Trypanosoma cruzi BrazilcloneA4 | Trypanosoma brucei equiperdum(IVM-t1) |
| Trypanosoma cruzi Dm28c Dm28c | Trypanosoma brucei gambiense (MHOM/CI/86/DAL972) |
| Trypanosoma cruzi G | Trypanosoma congolense (strain IL3000) |
| Trypanosoma cruzi Sylvio_X10_1 | |
| Trypanosoma cruzi Marinkellei B7 | |
| Trypanosoma cruzi YcloneC6 | |
| Trypanosoma cruzi CL | |
| Trypanosoma cruzi CL Brenner | |
| Trypanosoma cruzi CruziDm28c | |
| Trypanosoma cruzi Dm28c | |
| Trypanosoma cruzi TCC | |
| Trypanosoma cruzi Y |
Design of multi-antigenic and multi-epitope vaccines against TC-CLB
Identification of epitopes
Numerous studies have suggested that epitope-based antigens can induce protective immunity against different infectious agents69–71. Various methods have been described in the literature to determine the B and T-cell epitopes which include; functional assays wherein the antigen is sometimes mutated and antibody-antigen interaction is evaluated, 3D structure analysis of antigen–antibody complexes or screening the peptide library of antibody binding, utilization of MHC multimers, and lymphoproliferation by ELISPOT assays72. Apart from these time-consuming and expensive experimental techniques, scores of computational methods have also been developed in the past few years. In the subsequent section, we shall describe different approaches for the prediction of B-cell, MHC-I, and MHC-II epitopes in potential vaccine candidates.
Selection of linear B-cell epitopes
Linear B-cell epitopes are effective antigenic peptide sequences for stimulating B-cell immune responses. There are different methods for B-cell epitope predictions which can be classified into sequence-based and machine learning-based methods. We used multiple tools for predictions which include BCEPRED73, ABCPred74, and BepiPred75 servers (See, Supplementary File-C). We selected the top-scoring epitopes simultaneously predicted by different servers for the final vaccine peptide. Besides, we also used VaxiJen 2.0 along with the IEDB server conservancy analysis to rank and shortlist epitopes. To illustrate, only those epitopes which had shown 100% conservation were selected (Fig. 3).
Figure 3.
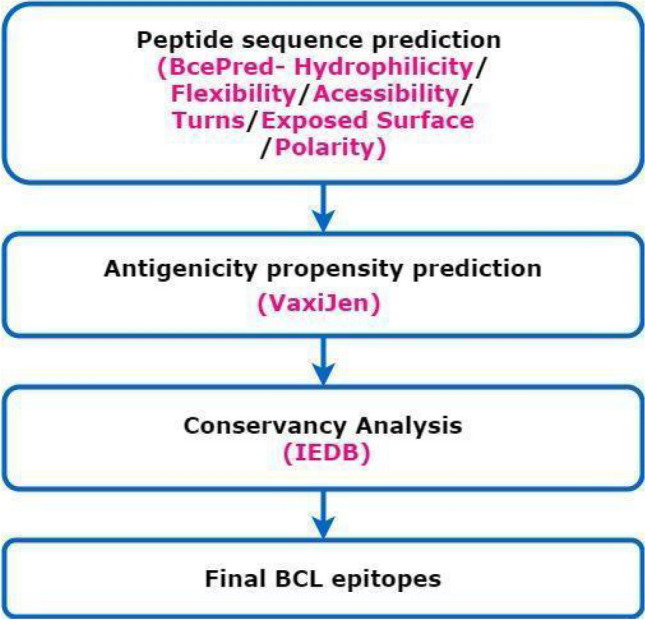
Workflow for the selection of B-cell epitope sequences (Draw.io—https://www.diagrams.net/-14.6.10).
T-cell epitope prediction
The objective of T-cell epitope prediction is to identify short peptide sequence within an antigen that can act as a stimulant of CD4+ or CD8+ T-cells. There are several methods available to predict MHC binding peptides which can be divided into data-driven approaches or structure-based methods. Structure-based methods are not used commonly because of their poor accuracy and requirement of intensive computational infrastructure. Data-driven methods are based on peptides [i.e., anchor residues, PSSM] known to bind with MHC molecules which are stored in databases such as IEDB, EPIMHC76, and AntiJen77. Further, there are machine learning-based methods that have been trained on data sets consisting of peptides that either bind or do not bind to MHC molecules. The presence of hundreds of allelic variants of human leukocyte antigens [HLAs] encoding MHCs presents another set of challenges for epitope prediction78. We used different methods such as NetCTL79, Propred80, EpiJen78 and NetMHC81 tools. Different tools for predictions were used during the study but for brevity, we shall describe results from one of the best-known tools (i.e., NetCTL) in subsequent sections.
Selection of cytotoxic T lymphocytes [CTL] epitopes
NetCTL1.2 server has demonstrated comparatively high-level accuracy for CTL epitope predictions therefore a docker image of this tool was created for its execution on local systems (See, Supplementary File-D). It predicts the MHC-class I binding peptide sequences, with proteasome C-terminal cleavage and transporter associated with TAP efficiency (Transporter associated with Antigen Processing). Using this server, the CTL epitopes were predicted based on default parameters and cut-offs [MHC supertype A1, the threshold as 0.75, and weight on C-terminal cleavage as 0.15, and weight on TAP transport efficiency as 0.05]. Further, these epitopes were subjected to antigenic propensity analysis using the VaxiJen 2.0 and immunogenicity analysis (by IEDB class-1 Immunogenicity servers). The epitopes showing poor scores, or overlaps were discarded (Fig. 4).
Figure 4.
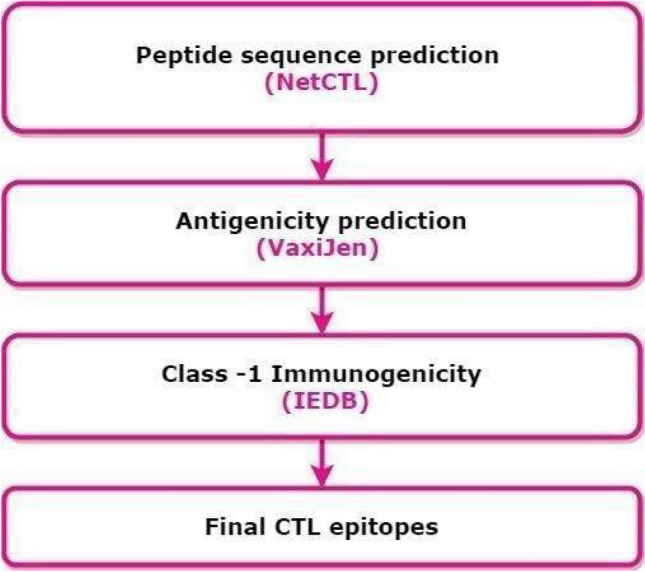
Workflow for selecting cytotoxic-T-lymphocyte epitope sequences (Draw.io—https://www.diagrams.net/-14.6.10).
Selection of helper T cells [HTL] epitopes
Prediction of HTL epitopes was performed using the IEDB-MHC-II binding tool (http://tools.iedb.org/mhcii/). This tool utilizes different methods to predict the epitopes, including a consensus method combining NN-align, SMM-align, and other combinatorial approaches. Epitopes obtained through the MHC-II Binding server were subjected to allergenicity prediction using the AlgPred82 and AllerTop83 servers. Next, using the VaxiJen 2.0 server, non-allergenic epitopes were tested for their antigenic propensity. To predict the toxicity status of epitopes, the antigenic epitopes were subjected to the ToxinPred server84. Finally, by employing the IFNepitope server15, IFN gamma induction analysis was performed on the non-toxic epitopes. Epitopes that possess the potential to induce the release of IFN gamma were selected as potential epitope candidates for vaccine construction (Fig. 5) (See, Supplementary File-E).
Figure 5.
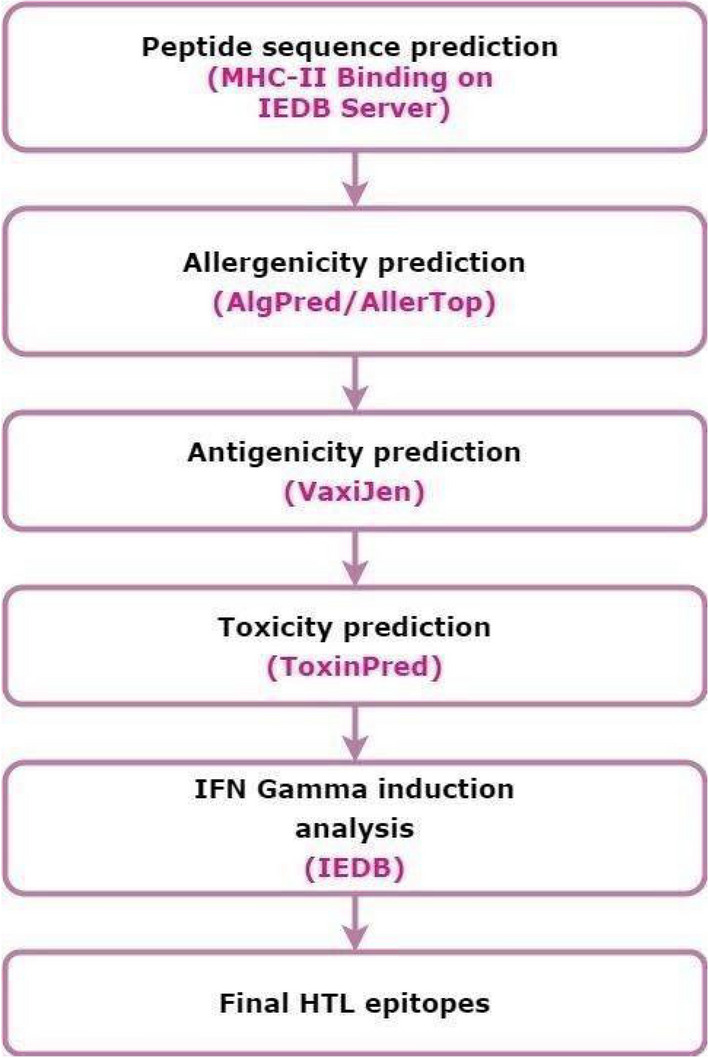
Workflow for selecting Helper-T-lymphocyte epitope sequences (Draw.io—https://www.diagrams.net/-14.6.10).
The assemblage of multi-epitope vaccine candidate sequence
Three potential vaccine candidates were constructed from top-ranking B-Cell, CTL, and HTL epitopes predicted using various bioinformatics tools. Immunogenicity of the constructs was enhanced by adding adjuvants such as β-defensin [Accession ID: AGV15514.1], L7/L12 50s ribosomal protein [Accession ID: WP_088359560.1, Flavobacteria JJC], and HABA protein [Accession ID: AGV15514.1; Mycobacterium. tuberculosis]. The adjuvant was attached to the first top CTL epitope [Protein ID: XP_804513.1] using an EAAAK linker. The other top CTL epitopes, belonging to the eight proteins filtered using the RV pipeline, were joined with each other through the GGGS linker. Next, the AAY linker was used to connect the CTL epitope to the HTL epitope sequence as well as all the HTL epitopes with each other. The KK linker was used to bridge the HTL epitope to the BCL epitopes as well as the BCL epitopes with each other. Finally, an EAAAK linker was added at the end to improve the stability of the constructs.
Evaluation of antigenicity and allergenicity of vaccine construct
The antigenic propensity prediction for the vaccine construct was performed through VaxiJen 2.0 and ANTIGENpro (http://scratch.proteomics.ics.uci.edu/) servers. The VaxiJen tool is based on the principle of auto cross-covariance [ACC] transformation of protein sequences into vectors using the physicochemical properties of amino acids.
The AlgPred and AllerTOP (http://www.ddg-pharmfac.net/AllerTOP) servers were used to predict the allergenicity of vaccine constructs. AlgPred is a web-based tool for predictions of allergens that combines bioinformatics and machine learning approaches such as IgE epitope scanning, MEME/ MAST motif-based search, amino acid composition, or dipeptide composition-based SVM methods, hybrid method, and BLAST on ARPs. The authors have reported an accuracy of 93.5% for their tool. On the other hand, AllerTOP v2.0 is based on auto and cross-variance transformation, amino acid E-descriptors, and machine learning methods such as k-nearest neighbours [KNN], algorithm. AllerTOP v2.0 was reported with 85.3% accuracy at fivefold cross-validation.
Analysis of solubility and physicochemical properties
To evaluate the solubility of the designed vaccine sequence, Protein-Sol85 [https://protein-sol.manchester.ac.uk/] server was used. Furthermore, it was assessed for several physicochemical parameters by using the ProtParam server. The properties evaluated include molecular weight, theoretical isoelectric point [pI], half-life, instability index [II], aliphatic index, and hydropathicity or GRAVY value.
Prediction of the secondary structure of the construct
PSIPRED86 and CFSSP87 tools were employed for secondary structure analysis. The consensus of both tools was taken into consideration. PSIPRED 3.2 is a freely accessible online server that utilizes a position-specific iterated BLAST for the identification and selection of specific sequences that show significant similarity with the designed vaccine construct. Further, it is reported to show a Q3 score of 81.6% and is available at http://bioinf.cs.ucl.ac.uk/psipred/.
CFSSP (Chou and Fasman Secondary Structure Prediction Server) is an online protein secondary structure prediction server. This server predicts regions of the secondary structure of the protein sequence such as alpha-helix, beta-sheet, and turns from the amino acid sequence in a linear sequential graphical view. CFSSP implements the Chou-Fasman algorithm, which is based on an analysis of the relative frequencies of each amino acid in alpha helices, beta sheets, and turns based on the known protein structures solved by X-ray crystallography.
Tertiary structure assessment of the vaccine construct
For homology modelling, the final multi-epitope vaccine construct was subjected to the Iterative Threading ASSEmbly Refinement (I-TASSER)88 server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). It is used for generating automated protein structures and performing predictions. It is reported to design a 3D atomic model by utilizing the multiple threading alignments and iterative structural assembly simulations of the submitted amino acid sequence.
Refinement of the tertiary structure
Using the I-TASSER server, a three-dimensional model of the chimeric protein was obtained. Next, we refined the 3D model using two-step refinement process consisting of 3Drefine89 (http://sysbio.rnet.missouri.edu/3Drefine/) and GalaxyRefine90 (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) online protein structure refinement servers. The 3Drefine refinement protocol utilizes iterative optimization of hydrogen bonding network combined with atomic-level energy minimization on the optimized model using a composite physics and knowledge-based force field for efficient protein structure refinement. Whereas GalaxyRefine rebuilds side chains and performs side-chain repacking and subsequent overall structure relaxation by molecular dynamics simulation.
Validation of the model stability
Validation is essential for the evaluation of stability and to find inherent errors that might be present in the predicted 3D protein models. For validation of the 3D model, the ProSA-web server (https://prosa.services.came.sbg.ac.at/prosa.php) was used to calculate the overall quality score in context with all the known protein structures. For generating the Ramachandran plot, MolProbity and RAMPAGE servers were used. MolProbity (http://molprobity.biochem.duke.edu/) is an all-atom structure validation online server that offers Ramachandran analysis. Ramachandran plots are used to visualize the energetically allowed and disallowed dihedral angles, psi [ψ], and phi [ϕ], of amino acids. RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) is another freely accessible server that integrates the PROCHECK91 principle for validation of the protein model by applying a Ramachandran plot and segregating the Glycine and Proline residues plot.
Prediction of discontinuous B-cell epitopes for the vaccine construct
Antibodies must interact with antigen epitopes to remove the infectious agent. Therefore, the prediction of conformational epitopes such as discontinuous B-cell epitopes is important. It has been found that discontinuous B-cell epitopes comprise residues remotely located in the primary structure that are brought into proximity due to the folding of the protein and 90% of B-cell epitopes are discontinuous92. There are several tools for discontinuous B-cell epitopes prediction such as BEPro93, Ellipro94, and Epitopia95. Ellipro is based on the notion that residues that protrude from the protein surface are more accessible for antibody binding and that these protruding residues can be identified by treating the protein as an ellipsoid. Therefore, we employed ElliPro (http://tools.iedb.org/ellipro/) for discontinuous B cell epitope predictions.
Molecular docking of the vaccine construct with TLR-4 and several HLA alleles
Molecular docking is an important tool for studying interactions amongst biological molecules. We employed molecular docking tools to find out the effect of vaccine construct with TLR-4 and HLA alleles. Since the majority of adjuvants originate from microbial components known as PAMPs [pathogen-associated molecular patterns], the immune system responds to these PAMPs by using Toll-like receptors [TLRs]96.
For docking assessment, the 3D structures of different MHC molecules and human TLR-4 [PDB ID: 4G8A] were retrieved from RCSB PDB. It is observed that specific varieties of Human Leukocyte Antigen [HLA] alleles are predominant in the South and Central American region. Therefore, we focused on these specific classes of HLA in interaction studies. Molecular interactions between various HLA molecules namely HLA-A, HLA-B7 [3VCL], HLA-DRB1*01:01 [2fse], HLA-DRB1*03:01 [1a6a], HLA-DRB5*01:01 [1h15] and the designed vaccine construct was performed. Various online tools for protein–protein docking were employed to calculate the binding affinity of designing a vaccine construct with different HLA alleles and TLR-4 immune receptors. The tools include ClusPro 2.097, HDOCK98, and PatchDock99. PatchDock generated numerous possible solutions that were further subjected to the refinement of the complexes using FireDock100.
Codon optimization of the chimeric protein
Java Codon Adaptation Tool or JCat server101 [http://www.jcat.de] was employed for codon optimization of the predicted vaccine construct. It involves the reverse transcription of the chimeric protein sequence to the nearest obtainable DNA sequence, which should contain specific genes responsible for encoding the target vaccine construct. This reverse-transcribed DNA sequence [RT-DNA] obtained is incorporated into the multiple cloning site of the pET-28a [+] vector using the SnapGene tool102 following our previous strategy. This was done to adapt the DNA sequence in the model organism [E. coli strain K12] so that this RT-DNA undergoes cellular adaptations within the model organism and the codons of RT-DNA are utilized by the model organism to produce the desired vaccine construct. This is a crucial step in vaccine construction, due to the effect of the degeneracy of codons, which can vary from one organism to another, including the cellular mechanisms that exist. To circumvent issues of glycosylation in the bacterial system, we also performed codon optimization using the yeast model (Supplementary File Y_F in Supplementary File 5).
Characterization of the immune profile of the vaccine construct
The simulation of the actual response of an immune system to our final vaccine construct was obtained using the C-ImmSim immune simulator [http://150.146.2.1/C-IMMSIM/index.php]. The tool was run with default parameters with three-time steps [1, 42, and 84] and without Lipopolysaccharide [LPS]. It works on Position-Specific Scoring Framework [PSSM] to simulate and predict immune interactions along with immunogenic epitopes.
Evaluation of genetic diversity
In order to develop a broad-spectrum T. cruzi vaccine, the prioritized proteins were scrutinized for their genetic diversity among fully annotated proteomes of 13 T. cruzi strains and different species (Supplementary Table 10). Protein sequences from these strains which are positive for that particular protein, were downloaded from NCBI RefSeq103 and aligned to predict conserved regions using CLC Main Workbench 21.0.2 (QIAGEN). Evolutionary distances (p-distances) among variant sites were also calculated for prioritized proteins using Mega 6.0104. The predicted epitopes were also checked for their sequence divergence among different strains and species of Trypanosoma. Each predicted epitope was further checked for antigenicity using VaxiJen (threshold value = 0.4)54. In addition, we also mapped epitopes to genomic sequences. For this purpose, we first reverse translated the epitope sequences and thereafter used pairwise alignment tools for mapping. We also checked the conservancy of epitopes through IEDB conservancy analysis tool105.
Results
Defining a potential vaccine candidate (PVC)
A Potential Vaccine Candidate (PVC) could be defined as the protein or corresponding DNA/RNA sequence that possesses properties of an “ideal vaccine” such as non-homology with the host (i.e., human) proteins to avoid the generation of a potential autoimmune response106, the lack of transmembrane regions to facilitate expression, antigenicity, adhesion-like properties, immunogenicity, a molecular weight of < 110 kDa, non-homology with the gut flora proteome, surface-exposure/secretion, and the presence of anchoring and/or secretion signals. Based on sequence similarity, proteins relevant to microbial pathogenesis would also be highly ranked. For our model, we label these desirable properties Pi [i = 1, 2, 3….n] (Supplementary Table 9).
Selection, ranking, and filtering of PVCs
To understand the distribution of properties in the T. cruzi CL-Brenner (TC-CLB) proteome, we used python-based scripts to characterize the whole proteome using various bioinformatics tools. During the analysis, we found that 91.46% of all proteins [i.e., 19,602] have a molecular weight < 110 kDa, 13.20% of proteins are secretory and 7.12% are extracellular. Also, 84.80% of the proteome is dissimilar to human proteins. Likewise, we observed similar trends in proteomes of four related species and thirteen different strains of Trypanosoma (Table 2). In addition, we computed distributions of properties in other pathogens for comparative purposes (Supplementary Tables 11a–11c).
Identification of subcellular location of the proteins
Using the PSORTb tool (Strategy 1A), we screened 19,602 proteins of the reference proteome of TC-CLB [Accession ID: NZ_AAHK00000000] and found that 1846 proteins were predicted to be localized in the periplasm, extracellular matrix, and outer membrane of the cell. Next, we used the PSORTb score (threshold set to 9.5) as an additional filter to shortlist 653 proteins. Alternatively, WoLF-PSORT (Strategy 1B) predicted 7274 proteins, localized in the plasma membrane and extracellular matrix. Despite using two different approaches (1A and 1B), we observed that most of the proteins (i.e., mucin TcMUCII, Mucin Associated Surface Protein (MASP), trans-sialidase, hypothetical protein, dispersed gene family (DGF-1) and subtilisin-like peptidase) were present in the top-ranking filtered list of both the approaches.
Identification of TC-CLB proteins that are non-homologous to human proteins
To prevent undesired cross-reactivity of vaccines with the human host, the proposed vaccine candidate must be different from human proteins. Therefore, we used BLASTp to identify the 572 such non-homologous proteins out of the 653 proteins identified through PSORTb.
Instability analysis
Protein stability is of crucial importance for the efficient presentation of antigenic peptides on MHC, which plays a decisive role in triggering strong immune reactions. Using ProtParam, the protein instability index was determined and proteins having an Instability Index (II) less than 40 were selected. This led to shortlisting of 138 proteins (out of 572) that were predicted to be stable.
Non-allergenicity analysis
To find out non-allergenic proteins in our list, we performed a BLASTp search against the Allergen Online database and found 137 proteins to be non-allergenic.
Evaluation of antigenicity
To determine the antigenicity of the shortlisted proteins for vaccine construction, VaxiJen 2.0 was employed. Proteins having antigenicity greater than 0.5 were selected for subsequent analysis. We identified 122 antigenic proteins out of 137 proteins using this tool.
Adhesion prediction
Next, we performed adhesion prediction using FungalRV with a threshold value of greater or equal to − 1.2. Several studies have shown that adhesins are vital in initiating pathogen-based infections107. Therefore, it seemed practical to target these proteins for vaccine development. A total of 100 proteins (out of 122) were predicted to possess desired properties similar to adhesin proteins. We used these top 100 proteins for subsequent analysis as a filtered list. It was also found that several hits belonging to the same gene/protein family such as trans-sialidases, and mucin-associated surface protein were present in the top 100 list. In VAX-Elan, we have also included an option to filter (or include) multi-copy genes/proteins for subsequent analysis108.
Shortlisted potential vaccine candidates (PVCs)
The top 100 shortlisted proteins were analysed further to evaluate the presence of additional criteria (TM α-helices, signal peptides, essentiality, and virulence) to narrow down the best eight proteins as PVCs. These include Dispersed gene family [XP_813527.1], subtilisin-like serine peptidase [XP_809835.1], DNAJ Chaperone protein [XP_806816.1], Mucin-associated Surface Protein [MASP] [XP_809166.1], Mucin TcMUCII [XP_816522.1], Trans-sialidase [XP_818708.1], 90 kDa surface protein [XP_815016.1] and a hypothetical protein [XP_821916.1], each belonging to different protein families (Table 3). We also used alternative strategies (see “Methods”) which also reported these PVCs in their top-ranking lists. Next, we independently checked these proteins as PVCs from scientific literature using text mining and manual curation approaches (Supplementary Table 12).
Table 3.
Ranking of unique proteins with the highest antigenic score. Here the hypothetical protein has displayed similarity with regulator sigma E protease during the Blast search.
| Top proteins (unique) after filtration | VaxiJen score |
|---|---|
| XP_813527.1 (DGF-1) | 0.61 |
| XP_809835.1 (substilin-like serine peptidase) | 0.65 |
| XP_806816.1 (DNAJ Chaperone protein) | 0.51 |
| XP_809166.1 (MASP) | 1.41 |
| XP_816522.1 (Mucin TcMUCII) | 1.16 |
| XP_818708.1(Trans-sialidase) | 0.80 |
| XP_815016.1 (Surface Protein) | 0.85 |
| XP_821916.1 (hypothetical protein) | 0.76 |
Epitope predictions
Linear B-cell epitopes identification
We identified a total of 1173 linear B cell epitopes in 8 PVCs using different prediction servers (ABCPred, BCEPRED & Bepipred). The maximum number of epitopes [510 epitopes] were found in Dispersed Gene Family protein [XP_813527.1] whereas the minimum number of epitopes [34 epitopes] were identified for Hypothetical Protein [XP_821916.1]. We ranked the epitopes based upon antigenicity value generated by the VaxiJen 2.0 tool (threshold: 0.5; target organism used as ‘Parasite’). Further, we found that approximately 295 epitopes were predicted by multiple servers. In Table 4, we show the highest-ranked epitope found in each protein, shortlisted for further analysis.
Table 4.
Predicted linear B-cell epitopes in the selected proteins for designing vaccine constructs.
| S. no. | Protein ID | Top BCL epitopes | VaxiJen score |
|---|---|---|---|
| 1 | XP_813527.1 | GSCGCRC | 3.51 |
| 2 | XP_809835.1 | PLLLFVFF | 3.06 |
| 3 | XP_806816.1 | VHINLKQ | 1.49 |
| 4 | XP_809166.1 | TSPLFPLLLVVAC | 1.23 |
| 5 | XP_816522.1 | MTCRLLCALLVLALCCCPSVCVT | 0.77 |
| 6 | XP_818708.1 | SLWSVRL | 1.61 |
| 7 | XP_815016.1 | DVPPSSLP | 0.89 |
| 8 | XP_821916.1 | EKPQCLLLSSGILVDVLMR | 1.15 |
T-cell epitopes [CTL] prediction
First, we identified 16,385 CTL epitopes in the eight shortlisted proteins. Second, we found 221 epitopes (out of 16,385) that were predicted by four different prediction tools namely NETMHC, EpiJen, Propred1, and NetCTL. Third, we selected eight high-scoring epitopes for subsequent work (Table 5).
Table 5.
Predicted linear cytotoxic T-lymphocyte epitopes in the selected proteins for designing vaccine constructs.
| S. no. | Protein ID | Top CTL epitopes | VaxiJen score |
|---|---|---|---|
| 1 | XP_813527.1 (DGF-1) | DAALLGGDY | 2.09 |
| 2 | XP_809835.1 (subtilisin-like serine peptidase) | GVDFDSCFF | 1.84 |
| 3 | XP_806816.1 (DNAJ Chaperone protein) | KTGRNGDMY | 1.81 |
| 4 | XP_809166.1 (MASP) | STDDHATGS | 1.75 |
| 5 | XP_816522.1 (Mucin TcMUCII) | GTDGVTGTT | 1.48 |
| 6 | XP_818708.1 (Trans-sialidase) | SSDADPTVV | 1.03 |
| 7 | XP_815016.1 (Surface Protein) | LLVLAALTY | 0.94 |
| 8 | XP_821916.1 (hypothetical protein) | YTCGTSCAV | 0.75 |
Helper T lymphocytes [HTL] prediction
With the IEDB MHC-II prediction tool, HTL cell epitopes were predicted with the highest binding corresponding to the alleles from the human 7-allele reference set i.e., HLA-DRB alleles. Based on the percentile rank as well as IC50 value [< 50 nM], 41 epitopes were selected for further analysis. Out of those, a total of 8 HTL epitopes were chosen for the vaccine construct (Table 6).
Table 6.
Predicted helper T-lymphocyte epitopes in the selected proteins for designing vaccine constructs.
| S. no. | Protein ID | Top HTL epitopes | VaxiJen score |
|---|---|---|---|
| 1 | XP_813527.1 (DGF-1) | GSFVMDGTVALGGAG | 1.75 |
| 2 | XP_809835.1 (subtilisin-like serine peptidase) | KAPRGRIIRLQYLRF | 1.68 |
| 3 | XP_806816.1 (DNAJ Chaperone protein) | TGVSKNGRQLRVSGK | 1.79 |
| 4 | XP_809166.1 (MASP) | ASGVLGENGSHMPDG | 1.45 |
| 5 | XP_816522.1 (Mucin TcMUCII) | STSGSAEPTKKVQEQ | 1.23 |
| 6 | XP_818708.1 (Trans-sialidase) | MLVGKYSRNAAAGAR | 1.1 |
| 7 | XP_815016.1 (Surface Protein) | LKSWWQRNVETKAVT | 1.32 |
| 8 | XP_821916.1 (hypothetical protein) | SGILVDVLMRTSAHR | 1.01 |
The assemblage of multi-epitope subunit vaccine construct
The vaccine (V1) was constructed from high-scoring CTLs, B-cell epitopes, and HTL epitopes. To enhance its immunogenicity, a Beta-defensin adjuvant [Accession ID: AGV15514.1] was obtained from NCBI and incorporated into V1 (Fig. 6).
Figure 6.
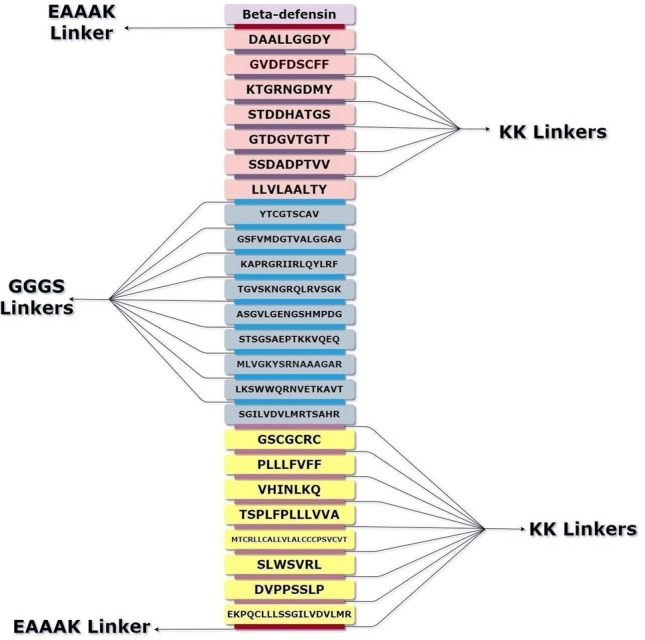
Multi-epitope vaccine constructs for Chagas disease. The vaccine construct consists of 24 epitope sequences, belonging to CTL, HTL and BCL epitopes of 8 T. cruzi proteins. Beta-defensin (Light purple) was used as the adjuvant and is linked to the top CTL epitope (light pink) using an EAAAK (maroon) linker. The other CTL epitopes were linked to each other using GGGS (violet) linkers. The last CTL epitope and the first HTL epitope (blue), as well as the other HTL epitopes were connected through an AAY (sky blue) linker. The last HTL epitope and the first BCL epitope (yellow) as well as the other BCL epitopes were connected through a KK (purple) linker. An EAAAK (maroon) linker was added at the end of the sequence for increasing stability (Draw.io—https://www.diagrams.net/-14.6.10).
Evaluation of antigenicity and allergenicity of the vaccine constructs
The predicted vaccine constructs were labelled as non-allergenic as predicted by AlgPred and AllerTop tools. The antigenicity value of the vaccine constructs was observed highest for V1 (1.06) as evaluated by Vaxijen 2.0 (Table 7).
Table 7.
The top three vaccine constructs V1, V2, and V3 made using Beta-defensin, L7/L12 Ribosomal protein, and Gaba protein adjuvants along with the top BCL, HTL, and CTL epitope sequences.
| Vaccine | Sequence | Antigenic propensity |
|---|---|---|
| V1 | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKDAALLGGDYGGGSGVDFDSCFFGGGSKTGRNGDMYGGGSSTDDHATGSGGGSGTDGVTGTTGGGSSSDADPTVVGGGSLLVLAALTYGGGSYTCGTSCAVAAYGSFVMDGTVALGGAGAAYKAPRGRIIRLQYLRFAAYTGVSKNGRQLRVSGKAAYASGVLGENGSHMPDGAAYSTSGSAEPTKKVQEQAAYMLVGKYSRNAAAGARAAYLKSWWQRNVETKAVTAAYSGILVDVLMRTSAHRKKGSCGCRCKKPLLLFVFFKKVHINLKQKKTSPLFPLLLVVAKKMTCRLLCALLVLALCCCPSVCVTKKSLWSVRLKKDVPPSSLPKK EKPQCLLLSSGILVDVLMREAAAK | 1.06 |
| V2 | MSDINKLAETLVNLKIVEVNDLAKILKEKYGLDPSANLAIPSLPKAEILDKSKEKTSFDLILKGAGSAKLTVVKRIKDLIGLGLKESKDLVDNVPKHLKKGLSKEEAESLKKQLEEVGAEVELKEAAAKDAALLGGDYGGGSGVDFDSCFFGGGSKTGRNGDMYGGGSSTDDHATGSGGGSGTDGVTGTTGGGSSSDADPTVVGGGSLLVLAALTYGGGSYTCGTSCAVAAYGSFVMDGTVALGGAGAAYKAPRGRIIRLQYLRFAAYTGVSKNGRQLRVSGKAAYASGVLGENGSHMPDGAAYSTSGSAEPTKKVQEQAAYMLVGKYSRNAAAGARAAYLKSWWQRNVETKAVTAAYSGILVDVLMRTSAHRKKGSCGCRCKKPLLLFVFFKKVHINLKQKKTSPLFPLLLVVAKKMTCRLLCALLVLALCCCPSVCVTKKSLWSVRLKKDVPPSSLPKKEKPQCLLLSSGILVDVLMREAAAK | 0.83 |
| V3 | MAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKDAALLGGDYGGGSGVDFDSCFFGGGSKTGRNGDMYGGGSSTDDHATGSGGGSGTDGVTGTTGGGSSSDADPTVVGGGSLLVLAALTYGGGSYTCGTSCAVAAYGSFVMDGTVALGGAGAAYKAPRGRIIRLQYLRFAAYTGVSKNGRQLRVSGKAAYASGVLGENGSHMPDGAAYSTSGSAEPTKKVQEQAAYMLVGKYSRNAAAGARAAYLKSWWQRNVETKAVTAAYSGILVDVLMRTSAHRKKGSCGCRCKKPLLLFVFFKKVHINLKQKKTSPLFPLLLVVAKKMTCRLLCALLVLALCCCPSVCVTKKSLWSVRLKKDVPPSSLPKKEKPQCLLLSSGILVDVLMREAAAK | 0.99 |
Analysis of solubility and physicochemical properties
Using ProtParam, the theoretical molecular weight of the vaccine construct V1 was found to be 42.3 kDa constructed with Beta-defensin as an adjuvant (406 amino acids) whereas the theoretical isoelectric point [pI] of the protein was found to be 9.70 which suggest that the vaccine construct is highly charged. The instability index [II] was estimated to be 30.95, indicating that the vaccine construct is stable (II < 40 indicates stability). V1 was predicted to be thermostable (Aliphatic index—78.37). V1 was also found to be hydrophilic (the predicted hydropathicity or GRAVY came out to be − 0.062). The presence of negative value scores suggests hydrophilic epitopes that are likely to be present in the outer surface and have more chance to elicit the high immunogenicity in the host cell. Furthermore, the solubility value of the vaccine construct is 0.651 as predicted by the Protein-Sol tool which has a threshold value of 0.45 indicating that the vaccine construct has a higher solubility than the average soluble E. coli protein from the experimental dataset utilized by this tool (Table 8). The half-life was estimated to be 30 h in mammalian reticulocytes in vitro, and > 20 h in yeast in vivo, and > 10 h in E. coli in vivo.
Table 8.
Comparison of physicochemical and solubility properties of different vaccine constructs.
| Physicochemical properties | Vaccine 1 | Vaccine 2 | Vaccine 3 |
|---|---|---|---|
| Antigenic propensity | 1.062 | 0.83 | 0.992 |
| Solubility | 0.651 | 0.601 | 0.472 |
| Molecular weight | 42.3 kDa | 50.70 kDa | 54.7 kDa |
| Allergenicity | Non-allergenic | Non-allergenic | Non allergenic |
| Hydropathicity | − 0.062 | − 0.056 | − 0.14 |
| Amino acids | 406 | 485 | 520 |
| Theoretical isoelectric point (pI) | 9.7 | 9.45 | 9.05 |
| Aliphatic index | 78.37 | 89.94 | 83.23 |
| Instability index (II) | 30.95 | 27.71 | 32.94 |
Secondary structure analysis
Using the CFSSP tool and PSIPRED, we found that V1 consists of 55.2% helix, 14.0% turns and 40.9% of sheets (Fig. 7a–d). The presence of random coils in the vaccine construct suggests the existence of natively unfolded protein regions that can be identified by antibodies that are produced in response to infection109.
Figure 7.
Secondary structure prediction of the final vaccine sequence using (a) CFSSP, (b) and (c) PSIPRED. (d) Graph of normalized B-factor predicted by I-TASSER.
Tertiary structure assessment of the vaccine construct
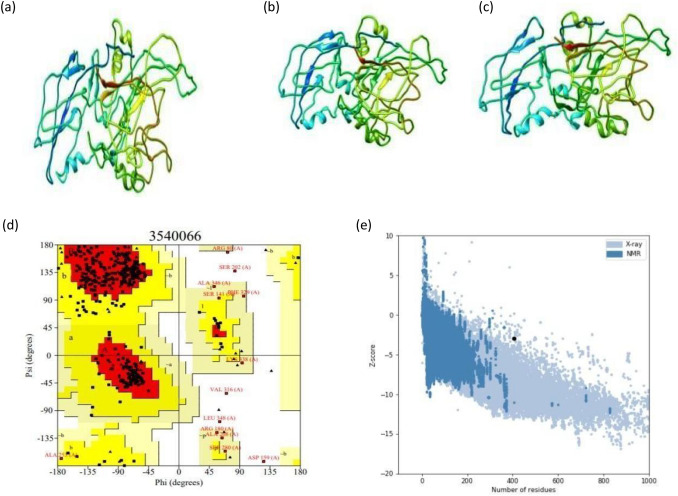
The tertiary structure models of the chimeric construct were predicted by the I-TASSER server by employing several threading templates [1kj6, 5nf2A, 1kj6A, 5ke1, 4om9A, 5ke1A, 4kh3A]. Out of 5 predicted results, model 1 was found to be the best one based upon the scores. In this study, the highest C-score model, derived from the homology modelling was selected for subsequent refinement protocol (Fig. 8a). The TM-score is defined to assess the topological similarity of the two protein structures. The TM-Score for our vaccine construct was found to be 0.56 ± 0.15 and the RMSD value was 9.6 ± 4.6 Å. It has been reported that a model with a TM score greater than 0.5, shows accurate topology, whereas a model with a TM score less than 0.17 indicates nonspecific similarity.
Figure 8.
Modeling, refinement and validation of tertiary structures. (a) Multi-epitope vaccine chimeric protein 3D model generated using homology modelling (Chimera 1.15—https://www.cgl.ucsf.edu/chimera/download.html). (b) Refined model using 3Drefine (Chimera 1.15—https://www.cgl.ucsf.edu/chimera/download.html). (c) GalaxyRefine generated refined 3D structure (Chimera 1.15—https://www.cgl.ucsf.edu/chimera/download.html). (d) Ramachandran plot of vaccine construct V1. (e) Prosa-Web giving a Z- score of -2.9.
Refinement of the tertiary structure
The putative chimeric vaccine model was refined by the 3Drefine server (http://sysbio.rnet.missouri.edu/3Drefine/) and subsequently by GalaxyRefine (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). The 3D-refine server-generated five models, out of which top-ranking model having favourable parameters such as lowest 3Drefine score (29,567.2), GDT-HA (0.96), RMSD (0.37 Å), lowest RWPlus score (− 63,611.70), and MolProbity (3.55). We also shortlisted Model 1 (from GalaxyRefine server) using a clash score (20.8), a score of poor rotamers (0.3), and the Ramachandran plot with a statistical score (89.4%) for downstream validation studies (Fig. 8b,c).
Validation of model stability
Ramachandran plot analysis of the protein model by ProCHECK-web predicted that 82.4% of amino acids were present in favoured regions. Moreover, 13.7% of the residues were present in the allowed regions, and only 1.5% of proteins were present in the disallowed or outlier boundary (Fig. 8d) indicating the quality of the model. The ProSA-web server authenticated the overall quality and errors that may potentially arise in the refined model. The refined model (obtained in this study) was considered to be appropriate with a Z-score of − 2.9 (Fig. 8e).
Prediction of discontinuous B-cell epitopes
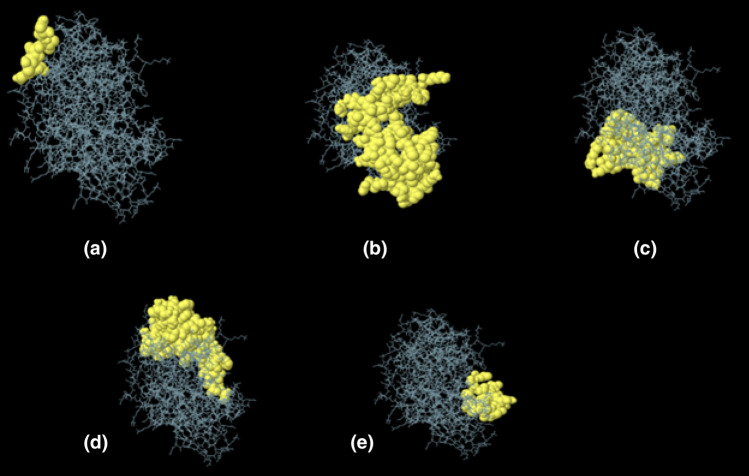
Ellipro estimated the five discontinuous B-cell epitopes and revealed the presence of 221 total residues among them (with score variation from 0.61 to 0.75) (Table 9, Fig. 9).
Table 9.
Discontinuous B-cell epitopes predicted by the ElliPro. Two hundred and twenty-one residues were found to be located in five discontinuous B-cell epitopes of the refined vaccine model.
| S. no. | Residues | Number of residues | Score |
|---|---|---|---|
| 1 | A: N269, A: V270, A: E271, A: T272, A: K273 | 5 | 0.75 |
| 2 | A:R14, A:G15, A:G16, A:R17, A:V20, A:S22, A:C23, A:L24, A:P25, A:K26, A:E27, A:E28, A:Q29, A:I30, A:G31, A:K32, A:C33, A:S34, A:T35, A:R36, A:G37, A:R38, A:K39, A:C40, A:C41, A:R42, A:R43, A:K45, A:E46, A:A47, A:A48, A:A49, A:K50, A:D51, A:A52, A:A53, A:L54, A:L55, A:G56, A:G57, A:D58, A:Y59, A:G60, A:G61, A:G62, A:S63, A:G64, A:V65, A:D66, A:F67, A:D68, A:S69, A:N81, A:G82, A:D83, A:M84, A:G86, A:G87, A:G88, A:S89, A:D92, A:D93, A:L135, A:G138, A:G139, A:G140, A:S141, A:Y142, A:T143, A:C144, A:G145, A:T146, A:S147, A:C148, A:A149, A:A151, A:A152, A:Y153 | 78 | 0.70 |
| 3 | A:S226, A:T227, A:S228, A:G229, A:S230, A:A231, A:E232, A:P233, A:T234, A:K235, A:K236, A:V237, A:E239, A:Q240, A:R302, A:C354, A:P356, A:C359, A:V360, A:T361, A:K362, A:K363, A:S364, A:L365, A:W366, A:S367, A:V368, A:R369, A:L370, A:K371, A:D373, A:V374, A:P375, A:P376, A:S377, A:S378, A:L379, A:P380, A:K381, A:E383, A:K384, A:P385, A:Q386, A:C387 | 44 | 0.65 |
| 4 | A:I3, A:N4, A:T5, A:L6, A:Q7, A:K8, A:Y10, A:G166, A:A167, A:G168, A:A169, A:A170, A:Y171, A:K172, A:A173, A:P174, A:R175, A:G176, A:R177, A:I178, A:I179, A:R180, A:L181, A:Q182, A:Y183, A:L184, A:R185, A:F186, A:A187, A:A188, A:Y189, A:T190, A:V192, A:S193, A:K194, A:A260, A:Y261, A:L262, A:K263, A:S264, A:W265, A:W266, A:Q267, A:R268, A:R294, A:V311, A:F312, A:F313, A:K314, A:K315, A:V316, A:H317, A:I318, A:N319, A:L320, A:K321, A:Q322, A:K324, A:T325, A:S326, A:P327, A:L328, A:F329, A:P330, A:L347, A:L348, A:L389, A:L390, A:S391, A:S392, A:G393, A:I394, A:D397, A:V398 | 74 | 0.65 |
| 5 | A:V211, A:L212, A:G213, A:E214, A:N215, A:G216, A:S217, A:P220, A:R289, A:T290, A:S291, A:A292, A:H293, A:K295, A:K296, A:G297, A:S298, A:C299, A:L331, A:L332 | 20 | 0.61 |
Figure 9.
Discontinuous B-cell epitopes predicted by ElliPro. (A–E): 3D representation of conformational or discontinuous epitopes of the most antigenic chimeric protein from T. cruzi CL Brenner. Epitopes are shown as yellow surfaces, and the bulk of the protein is represented in grey sticks (JSmol 13.3.9—https://sourceforge.net/projects/jsmol/).
Molecular docking of the chimeric protein with TLR-4
The CastP110 server was employed for determining protein binding and hydrophobic contact sites on the protein surface. One of the potential binding pockets (A) was identified for the interaction with a TLR-4 receptor. It was found that the molecular surface area of the pocket ‘A’ was 6008.1 Å2 with a molecular surface volume of 39,003.9 Å3, the mouth molecular area was about 1088.07 Å2, and the molecular surface sum was calculated to be 1888.9 Å. CPORT predicted G1, A19, L21, C33 as active amino acid residues in the adjuvant sequences; A52, L54, L55, G56, G57, D58, T59, G60, D68, S69, C70, F72, M84, G87, T137, G138, G140, S141, Y142, T143, C144, G145, T146, C148, P174, G176, I178, I179, R180, L181, Y183, L184, R185, F186, A187, Y189, N215, A255, G300, C301, P306, L307, L308, L309, F310, V311, F312, F313, K314, K315, V316, H317, I318, N319, L320, K321, S326, L328, F329, P330, L333, C345, L348, V349, L350, A351, L352, C353, C354, C355, P356, S357, D373, L388, L389, L390, S391, S392, G393, I394, L395, V396, V398, L399 from the chimeric protein joined with linker sequences111.
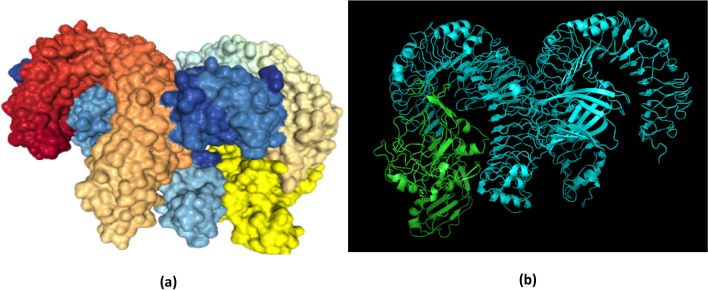
For the highest-ranking docked complex, the ClusPro tool revealed the lowest total intermolecular energy (− 973.2 kcal/mol), indicating a good interaction between V1 and TLR-4. The HDOCK server predicted the binding energy for the protein–protein complex as − 314.02 kcal/mol (Fig. 10). The refinement of PatchDock docking results, as obtained by the Firedock result also showed the lowest global energy values (Table 10).
Figure 10.
Molecular docking of subunit vaccine with the immune receptor—TLR4. (a) Docked image of the chimeric protein generated by HDOCK server having a binding energy score of -314.02. The rainbow-colored complex represents the TLR4 receptor molecule, while the golden-yellow colour denotes vaccine construct V1. (b) ClusPro generated model 5, which represents the protein–ligand complex (cyan–green). The lowest binding energy of -973.2 kcal was achieved for this model (Chimera 1.15—https://www.cgl.ucsf.edu/chimera/download.html).
Table 10.
Molecular docking results using the PatchDock server.
| Vaccine construct | PDB ID of the HLA alleles | Solution no. | Global energy | Hydrogen bond energy | ACE | Score | Area |
|---|---|---|---|---|---|---|---|
| VACCINE 1 | 3vcl1 | 28 | 8.99 | 0.0 | − 136.02 | 14,722 | 2001.1 |
| 1a6a | 82 | 8.82 | − 1.63 | − 284.02 | 14,114 | 2888.6 | |
| 1h15 | 48 | 4.17 | − 2.06 | − 264.30 | 15,388 | 3810.5 | |
| 2fse | 9 | − 29.11 | − 2.32 | − 192.83 | 16,506 | 2909.7 | |
| TLR4 | 83 | − 19.85 | − 3.35 | − 152.69 | 15,728 | 2531.6 |
The model was refined further using Firedock server.
Codon optimization of the chimeric protein
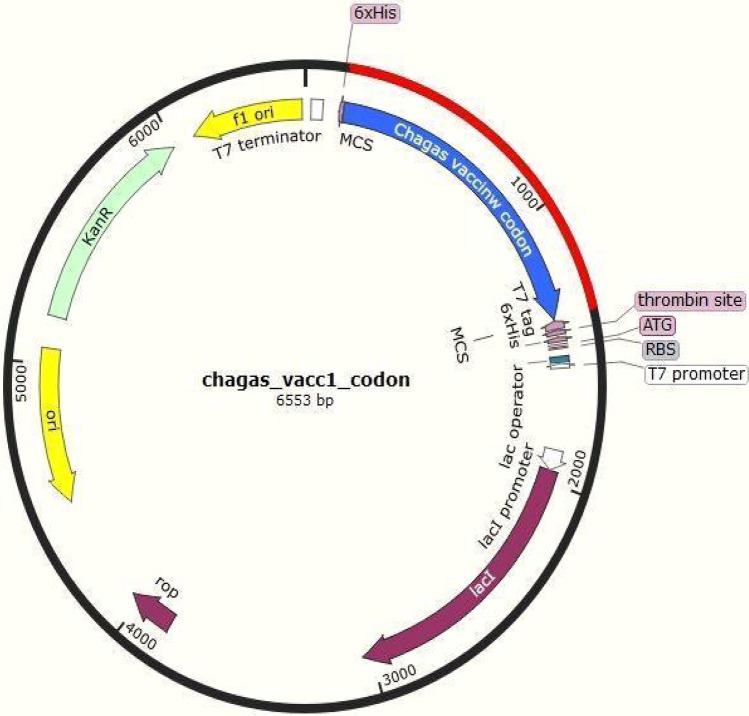
JCAT results revealed that the optimized codon sequence has a length of 1308 nucleotides and its CAI (Codon Adaptation Index) was predicted to be 0.98, with an average of 51.88% GC for the adapted sequence. These values indicate stable expression of the designed vaccine construct in the selected microbial host. For optimal gene expression, SnapGene software was employed, the designed chimeric protein sequence was integrated into the E. coli pET-28a [+] vector by incorporating restriction sites which were followed by cloning into the vector using published methods (Figs. 11 and 12).
Figure 11.
Codon optimization of the vaccine construct V1. Here, CAI of the optimized codon and average GC content were 0.98 and 51.8% respectively.
Figure 12.
In silico cloning of optimized codons encoding vaccine protein into pET28a (+) vector to ensure expression in microbial systems. The DNA sequence was inserted into the multiple cloning-site of the cloning vector. Here, the red portion denotes the gene sequence of our designed vaccine construct while the black portion denotes the backbone of the vector. All colored arrows denote the location and direction of the expression of gene. The blue portion shows vaccine codon sequence while green denotes kanamycin resistance gene, violet represents vector genes and yellow denotes origin of replication (SnapGene 5.2.5.1—https://www.snapgene.com/).
Characterization of the immune profile of the vaccine construct
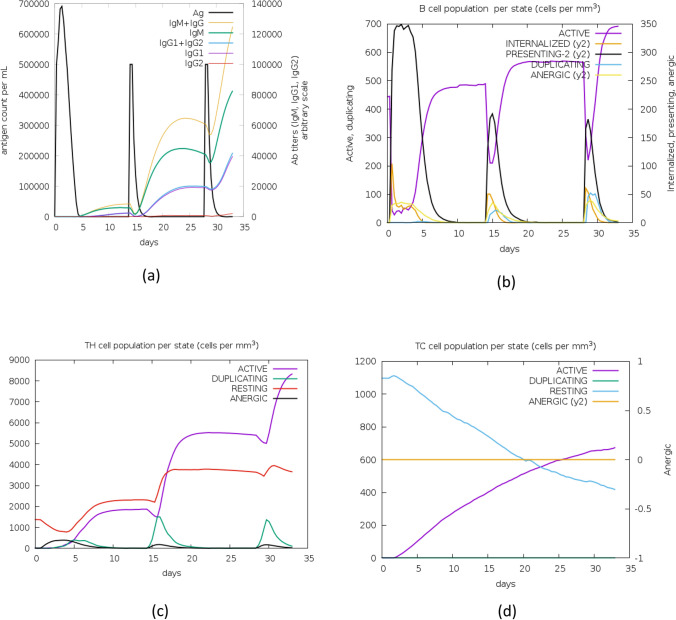
With C-ImmSim, the immune response of the final vaccine construct was analysed. Results of the simulated immune responses indicated an increased surge in the induction of secondary and tertiary immune responses. At the first dose, a high surge of IgM and IgG1 antibodies was predicted. However, these titters increased exponentially with the second and third dose. Furthermore, an increase in active B-cell, CTL, and HTL cell populations was predicted for all doses (Fig. 13).
Figure 13.
Molecular simulations of the chimeric protein. (a) Successive antigen injections leading to immunoglobulins production (Colored peaks indicate black vertical lines, along with other sub-classes of immune cells). (b) Post-insertion development in B-cell population per state as per observation after inserting three injections. (c) Development of T-helper cells (d) T-cytotoxic cell population per state after injections. The resting phase denotes those cells that are not presented with the antigens and energy state denotes cells that are tolerant to antigens due to repeated exposure, thereby indicating a lack of immune responsiveness.
Evaluation of genetic diversity
Protein sequences of the prioritized proteins were extracted from 13 T. cruzi annotated proteomes which were aligned to predict conserved regions (Supplementary Table 13). Five proteins namely DNAJ chaperon protein, subtilisin-like serine peptidase, DGF-1, MASP, and trans-sialidase displayed strong homology (above 80%) across 13 different strains of T. cruzi.
In the context of the DNAJ protein, the estimated evolutionary distance (p-distance) was found to be 0.005 (across 13 strains) and 0.746 (across species). Whereas for TS, p-distance was found to be 0.234 (across strains) and 0.795 (across species). Next, we extracted all the copies of TS from TC-CLB proteome and computed evolutionary divergence (0.616) as well (Supplementary Tables 14a–14c).
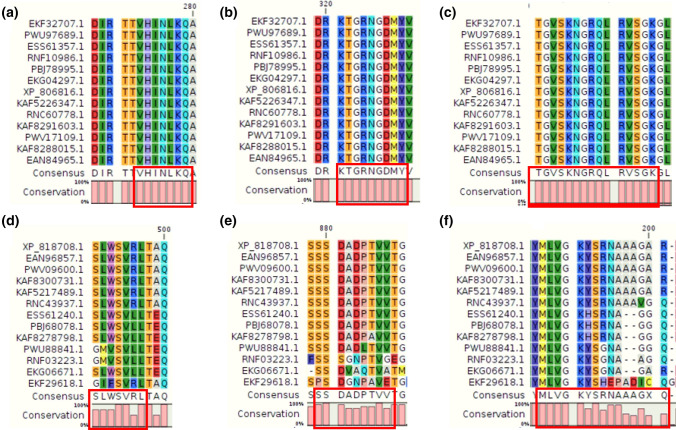
Estimates of evolutionary divergence between sequences and the number of amino acid differences per site among sequences are shown along with the standard error in Supplementary Tables 14a–14c. We found that most of the epitopes (belonging to the top eight proteins) were mapped/aligned to the conserved regions. For example, a 15-mer HTL epitope, "TGVSKNGRQLRVSGK" (from DNAJ protein), was found to be completely conserved (100%) across 13 different strains and four species (see Fig. 14 and Supplementary Table 15). Next, a predicted CTL epitope (‘SSDADPTVV’) from trans-sialidase protein sequences was also found to be conserved (Fig. 14). Likewise, we performed epitope conservancy analysis using the IEDB tool and observed that all the predicted epitopes were conserved across different strains of T. cruzi (Supplementary Table 16, Supplementary File 4). In addition, we also mapped epitopes (after reverse translation) on genomic sequences of Trypanosoma strains and species to check the conservation at the genomic level (See “Supplementary Website”). Further, we extracted 5750 copies of TS from different proteomes of T cruzi. Thereafter, we searched for the presence of epitopes in variants of TS using the Smith Water-Mann algorithm as well as using the IEDB conservancy tool. We found that the epitopes were present in the proteins with varying levels of conservation (See Supplementary Files Y_G and H in Supplementary File 5).
Figure 14.
Aligned regions showing conserved epitopes among various strains of T. cruzi. Individual targeted proteins (DNAJ chaperon and trans-sialidase proteins) among the 12 strains are aligned using CLC Main workbench and the regions with conserved epitopes (sequences) have been shown in the red boxes. (a) DNAJ BCL epitope; (b) DNAJ CTL epitope; (c) DNAJ HTL epitope; (d) trans-sialidase BCL epitope; (e) trans-sialidase CTL epitope; and (f) trans-sialidase HTL epitope.
Discussion
The study reported here comprises a comprehensive approach to utilize informatics and computer algorithms towards the prediction of vaccine targets in pathogens. Our work combines immuno-informatics approaches and reverse vaccinology methods to design an in-silico multi-epitope subunit vaccine that can offer protection against CD. The datasets and frameworks are also used to develop a new machine learning and deep learning system for the prediction of vaccine candidates in general. We have created a resource base for the scientific community working in the area of CD vaccine design [https://tinyurl.com/CDWork800]. We used several strategies to shortlist potential vaccine candidates. The goal was to obtain non-allergenic, antigenic, non-toxic, conserved B-cell, CD8+ and CD4+ epitopes that were assembled into three separate vaccine constructs, V1, V2, and V3. Our major findings include several unique vaccine antigens that are antigenic, immunogenic, and safe (showing no homology with human proteins and the proteome of the gut flora). Further, the designed vaccine constructs are also found to be, theoretically, soluble, thermostable, amenable for expression in model systems, and likely to interact with other proteins. Structurally, the designed constructs show a likelihood of favourable interactions with the TLR-4 on professional antigen-presenting cells. Our vaccine construct consists of epitopes derived from multiple protein molecules (PVCs) which have exhibited the potential to be PVCs in various independent experimental studies. The designed vaccine construct is likely to offer cross-protection since the selected proteins and predicted epitopes used in generating the cocktail vaccine exhibited considerable conservation across the related Trypanosoma species/strains.
In the past decade, different research groups have used several strategies ranging from stages of pathogenesis112; immunogenic assays109, subtractive proteomics9, and as well as properties/filters (Supplementary Table 9) to determine candidates for their respective pathogens. Different authors have used different orders of these properties [P1, P2…. Pn] as a combined filter to reach the final list of PVCs. Our study explains the impact of the order of applications of these properties on the outcome. Since no proteome-wide studies have been conducted to find the distribution of properties, we decided to apply multiple strategies to rank or filter TC-CLB proteins by randomizing the order, changing the number of filters, etc. For instance, in one of the strategies, we randomized the order of applications for properties [Pn] on TC-CLB. In another strategy, we removed the P1 [extracellular/secretory] filter which allowed an additional set of proteins [i.e., intracellular] to appear as PVCs. The objective was to screen proteomes diversely to select all best-ranking protein molecules (i.e., PVCs) with desired properties. One of the unique highlights is that we have examined the distribution of different properties across the pathogens’ proteome as well as on positive and control datasets. Further, we also applied Vax-ELAN on recently sequenced Y strain. We observed that top ranking candidates (in both CLB and Y strains) includes TS, Mucin, and Mucin associated surface proteins.
Researchers have initiated several efforts to develop vaccines against CD but issues related to a variety of T. cruzi strains, the genetic variability of the host, complex genomic structure24, significant phenotypic variation, and variable behaviour of pathogen (in vitro and in vivo) in context of pathophysiology, virulence, tropism, and immunological responses, have created several obstacles113. Further, T. cruzi is known to be a complex organism with multiple developmental forms with transient expression of different antigens. The problem is compounded by a wide variety of strains, antigenic shifts during different life stages, making proper immunization against the parasite an improbable task. The ability of T. cruzi to modulate and evade host immune responses and influence host-parasite interactions allows the parasite to survive through novel mechanisms114.
Several vaccine candidates have been reported for CD vaccine development programs across the world. These include Tc24 [and its modified Tc24-C4 derivative], TSA-1, ASP-2, TS, TSSA CD8 epitope, Tc52, TcG1, TcG2, TcG4, TcVac2, TcVac4, and MASP25. It is interesting to note that several of these candidates appeared in the final protein list used for our final vaccine construct. In one of the research studies, Michel-Todo et al. extracted T. cruzi epitopes from several antigens using publicly available databases115. They prioritized a set of epitopes based on sequence conservation criteria, projected population coverage of Latin America population, and biological features using in-silico methods and selected CD8+ T cell, CD4+ T, and B-cell epitopes with < 70% identical to human or human microbiome protein sequences. As a benchmark, we also compared epitopes115 with epitopes identified in our study using the VaxiJen tool (Supplementary File-F).
The in-silico approach to design a multi-epitope vaccine construct for Chagas disease presents challenges as a protein-based vaccine given the complexities of producing such candidates as experimental soluble proteins109 suitable for scale-up production and purification. However, we have recently embarked upon an mRNA vaccination approach for Chagas disease that might obviate the need for expression and purification steps116. We are now working to incorporate the findings here into our mRNA vaccination program.
Conclusion
Therapeutic interventions for the prevention and elimination of Chagas disease require novel treatment and immunization methods that can protect people at risk and infected populations while providing them with a good quality of life. This study is aimed at developing putative multi-epitope vaccines against CD, a protozoan infection caused by T. cruzi. The disease is endemic in Latin America and has impacted other parts of the world. In this study, computational approaches and a reverse vaccinology pipeline were used to screen the complete genomic and proteomic sequences for predicting potential vaccine candidates and designing in-silico chimeric vaccine constructs against the T. cruzi CL Brenner. Multiple antigenic B-cell, CD8+, and CD4+ epitopes were assembled into three non-allergenic, antigenic, and non-toxic constructs that can act as a prophylactic potential multi-epitope vaccine construct. Appropriate linkers and adjuvant sequences were also used to enhance the stability, effectiveness, as well as immune response of the engineered vaccine constructs. The designed vaccine construct has suitable structural, physicochemical, and immunological properties which can strongly stimulate both humoral and cellular immune responses in humans. However, experimental validation for efficacy and safety is needed along with pre-clinical studies before human immunization. Planning for such studies in appropriate mouse models of T. cruzi and CCC is in progress.
Supplementary Information
Acknowledgements
This work was supported by the Robert J. Kleberg Jr. and Helen C. Kleberg Foundation. We are also thankful to Amity University for the support provided during the conduct of this study.
Author contributions
K.R., P.H., U.S., and M.E.B. designed research; K.R., B.A.A., R.S., S.Si., T.S., A.M., and K.M. performed research; K.R., R.S., D.S., and K.M. contributed new reagents/analytic tools; K.R., B.A.A., D.S., and S.So. analysed data; K.R., B.A.A., D.S., P.H., M.E.B. and U.S. wrote the paper; and K.R., A.K.D., T.S., P.G., A.C., S.G., P.P., P.K., PR.S., S.K.N., and P.S., compiled information from many different sources.
Funding
This work was supported by a Grant from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, USA, Texas Children’s Hospital and Baylor College of Medicine, Houston USA. We also acknowledge the grant provided by SERB, Department of Science and Technology, Government of India (File Number: CVD/2020/000842).
Data availability
All raw data were obtained from open sources and have been cited and deposited in Datasets S1 and also available on our website. Supplementary Data: https://tinyurl.com/2b2s927h. Software Pipeline: Vax-ELAN: https://vac.kamalrawal.in/vaxelan/, Vax-ELAN Version 2: https://vac.kamalrawal.in/vaxelan/v2, Vaxi-DL: https://vac.kamalrawal.in/vaxidl/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96863-x.
References
- 1.Bibi S, et al. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci. Rep. 2021;11:1249. doi: 10.1038/s41598-020-80899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashfaq UA, et al. Rational design of multi epitope-based subunit vaccine by exploring MERS-COV proteome: Reverse vaccinology and molecular docking approach. PLoS ONE. 2021;16:e0245072. doi: 10.1371/journal.pone.0245072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raeven RHM, van Riet E, Meiring HD, Metz B, Kersten GFA. Systems vaccinology and big data in the vaccine development chain. Immunology. 2019;156:33–46. doi: 10.1111/imm.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong E, Wong MU, Huffman A, He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. bioRxiv. 2020 doi: 10.1101/2020.03.20.000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monterrubio-López GP, Ribas-Aparicio RM. Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. Biomed Res. Int. 2015;2015:1–16. doi: 10.1155/2015/483150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science (80-). 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez Alberti A, et al. Engineered trivalent immunogen adjuvanted with a sting agonist confers protection against Trypanosoma cruzi infection. NPJ Vaccines. 2017;2:1–12. doi: 10.1038/s41541-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, et al. Reverse vaccinology approach for the identifications of potential vaccine candidates against Salmonella. Int. J. Med. Microbiol. 2021 doi: 10.1016/j.ijmm.2021.151508. [DOI] [PubMed] [Google Scholar]
- 9.Solanki V, Tiwari M, Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci. Rep. 2019;9:1–19. doi: 10.1038/s41598-019-41496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajialibeigi A, Amani J, Gargari SLM. Identification and evaluation of novel vaccine candidates against Shigella flexneri through reverse vaccinology approach. Appl. Microbiol. Biotechnol. 2021;105:1159–1173. doi: 10.1007/s00253-020-11054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bencurova E, Gupta SK, Oskoueian E, Bhide M, Dandekar T. Omics and bioinformatics applied to vaccine development against: Borrelia. Mol. Omi. 2018;14:330–340. doi: 10.1039/C8MO00130H. [DOI] [PubMed] [Google Scholar]
- 12.Goodswen SJ, Kennedy PJ, Ellis JT. A novel strategy for classifying the output from an in silico vaccine discovery pipeline for eukaryotic pathogens using machine learning algorithms. BMC Bioinform. 2013;14:315. doi: 10.1186/1471-2105-14-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhal AK, Pani A, Mahapatra RK, Yun SIL. An immunoinformatics approach for design and validation of multi-subunit vaccine against Cryptosporidium parvum. Immunobiology. 2019;224:747–757. doi: 10.1016/j.imbio.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Dhanda SK, et al. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalsass M, Brozzi A, Medini D, Rappuoli R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019;10:113. doi: 10.3389/fimmu.2019.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchhoff LV. Chagas disease: American Trypanosomiasis. Infect. Dis. Clin. N. Am. 1993;7:487–502. doi: 10.1016/S0891-5520(20)30539-0. [DOI] [PubMed] [Google Scholar]
- 18.Bivona AE, Alberti AS, Cerny N, Trinitario SN, Malchiodi EL. Chagas disease vaccine design: the search for an efficient Trypanosoma cruzi immune-mediated control. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2020;1866(5):165658. doi: 10.1016/j.bbadis.2019.165658. [DOI] [PubMed] [Google Scholar]
- 19.Cazorla SI, Frank FM, Malchiodi EL. Vaccination approaches against Trypanosoma cruzi infection. Expert Rev. Vaccines. 2009;8:921–935. doi: 10.1586/erv.09.45. [DOI] [PubMed] [Google Scholar]
- 20.Limon-Flores AY, et al. Effect of a combination DNA vaccine for the prevention and therapy of Trypanosoma cruzi infection in mice: Role of CD4+ and CD8+ T cells. Vaccine. 2010;28:7414–7419. doi: 10.1016/j.vaccine.2010.08.104. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez Alberti A, et al. Mucosal heterologous prime/boost vaccination induces polyfunctional systemic immunity, improving protection against Trypanosoma cruzi. Front. Immunol. 2020;11:128. doi: 10.3389/fimmu.2020.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonio Marin-Neto J, Rassi A, Avezum A, Mattos AC, Rassi A. The Benefit trial: Testing the hypothesis that trypanocidal therapy is beneficial for patients with chronic Chagas heart disease. Mem. Inst. Oswaldo Cruz. 2009;104:319–324. doi: 10.1590/S0074-02762009000900042. [DOI] [PubMed] [Google Scholar]
- 23.Marin-Neto JA, et al. Rationale and design of a randomized placebo-controlled trial assessing the effects of etiologic treatment in Chagas’ cardiomyopathy: The BENznidazole Evaluation For Interrupting Trypanosomiasis (BENEFIT) Am. Heart J. 2008;156:37–43. doi: 10.1016/j.ahj.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Arner E, et al. Database of Trypanosoma cruzi repeated genes: 20,000 additional gene variants. BMC Genom. 2007;8:391. doi: 10.1186/1471-2164-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaumier CM, et al. Status of vaccine research and development of vaccines for Chagas disease. Vaccine. 2016;34:2996–3000. doi: 10.1016/j.vaccine.2016.03.074. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelsson O, Nielsen H, Brunak S, Von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 27.Petersen TN, Brunak S, Von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 28.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook; 2005. pp. 571–607. [Google Scholar]
- 29.Nielsen M, Lundegaard C, Lund O, Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. FungalRV: Adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genom. 2011;12:192. doi: 10.1186/1471-2164-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: Application to the MHC class i system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann K, Stoffel W. TMbase: A database of membrane spanning protein segments. Biol. Chem. 1993;374:166. [Google Scholar]
- 33.Zhang R, Ou HY, Zhang CT. DEG: A database of essential genes. Nucleic Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(suppl_1):W325–W328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson WR. An introduction to sequence similarity (‘homology’) searching. Current Protocols in Bioinformatics, Chapter 3. 2013;42(1):1–3. doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins Struct. Funct. Genet. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 37.Armenteros, J. J. A., Salvatore, M., Emanuelsson, O., Winther, O., Von Heijne, G., Elofsson, A., & Nielsen, H. Detecting Novel Sequence Signals in Targeting Peptides Using Deep Learning. Life science alliance 2(5), e201900429 (2019). [DOI] [PMC free article] [PubMed]
- 38.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikeda M, Arai M, Okuno T, Shimizu T. TMPDB: A database of experimentally-characterized transmembrane topoligies. Nucleic Acids Res. 2003;31:406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calis JJA, et al. Properties of MHC Class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9:e1003266. doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 2007;35(suppl_1):W391–W394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naz K, Naz A, Ashraf ST, Rizwan M, Ahmad J, Baumbach J, Ali A. PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinform. 2019;20(1):1–10. doi: 10.1186/s12859-019-2713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solanki V, Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8(1):1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebenberg J, Pretorius A, Faber FE, Collins NE, Allsopp BA, Van Kleef M. Identification of Ehrlichia ruminantium proteins that activate cellular immune responses using a reverse vaccinology strategy. Vet. Immunol. Immunopathol. 2012;145(1–2):340–349. doi: 10.1016/j.vetimm.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Goodswen SJ, Kennedy PJ, Ellis JT. Vacceed: a high-throughput in silico vaccine candidate discovery pipeline for eukaryotic pathogens based on reverse vaccinology. Bioinformatics. 2014;30(16):2381–2383. doi: 10.1093/bioinformatics/btu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder J, Aebischer T. Vaccines for leishmaniasis: From proteome to vaccine candidates. Hum. Vaccin. 2011;7(sup1):10–15. doi: 10.4161/hv.7.0.14556. [DOI] [PubMed] [Google Scholar]
- 49.Dhanda SK, Usmani SS, Agrawal P, Nagpal G, Gautam A, Raghava GP. Novel in silico tools for designing peptide-based subunit vaccines and immunotherapeutics. Brief. Bioinform. 2017;18(3):467–478. doi: 10.1093/bib/bbw025. [DOI] [PubMed] [Google Scholar]
- 50.Muruato LA, Tapia D, Hatcher CL, Kalita M, Brett PJ, Gregory AE, Samuel JE, Titball RW, Torres AG. Use of Reverse Vaccinology in the Design and Construction of Nanoglycoconjugate Vaccines against Burkholderia pseudomallei. Clin. Vaccine Immunol. 2017;24(11):e00206-17. doi: 10.1128/CVI.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe, Y., Zenke, K., Itoh, N. & Yoshinaga, T. Functional analysis of the proteases overexpressed during the invasive and parasitic stages of Cryptocaryon irritans and their potential as vaccine antigens. Aquaculture. 540, 736657 (2021).
- 52.Baseer S, Ahmad S, Ranaghan KE, Azam SS. Towards a peptide-based vaccine against Shigella sonnei: A subtractive reverse vaccinology based approach. Biologicals. 2017;50:87–99. doi: 10.1016/j.biologicals.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Hisham, Y. & Ashhab, Y. Identification of Cross-Protective Potential Antigens against Pathogenic Brucella spp. through Combining Pan-Genome Analysis with Reverse Vaccinology. J. Immunol. Res.2018, 1–15 (2018). [DOI] [PMC free article] [PubMed]
- 54.Naz A, et al. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect. Genet. Evol. 2015;32:280–291. doi: 10.1016/j.meegid.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 55.Pearce EJ, James SL, Hieny S, Lanar DE, Sher A. Induction of protective immunity against Schistosoma mansoni by vaccination with schistosome paramyosin (Sm97), a nonsurface parasite antigen. Proc. Natl. Acad. Sci. USA. 1988;85:5678–5682. doi: 10.1073/pnas.85.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biegel Carson SDB, Klebba PE, Newton SMC, Sparling PF. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 1999;181:2895–2901. doi: 10.1128/JB.181.9.2895-2901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nathaly Wieser S, Schnittger L, Florin-Christensen M, Delbecq S, Schetters T. Vaccination against babesiosis using recombinant GPI-anchored proteins. Int. J. Parasitol. 2019;49:175–181. doi: 10.1016/j.ijpara.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Cao J, Li JA, Li D, Tobin JF, Gimeno RE. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:19695–19700. doi: 10.1073/pnas.0609140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawat DS, et al. Identification, expression, modeled structure and serological characterization of Plasmodium vivax histone 2B. Gene. 2004;337:25–35. doi: 10.1016/j.gene.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 60.Favuzza P, Dreyer AM, Wittlin S, Matile H, Pluschke G. Cysteine-Rich Protective Antigen (CyRPA) as promising blood-stage candidate protein for inclusion in a malaria subunit vaccine. Malar. J. 2012;11:P30. doi: 10.1186/1475-2875-11-S1-P30. [DOI] [Google Scholar]
- 61.Gerbaba TK, Gedamu L. Cathepsin B gene disruption induced leishmania donovani proteome remodeling implies cathepsin B role in secretome regulation. PLoS ONE. 2013;8:79951. doi: 10.1371/journal.pone.0079951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goto Y, et al. Leishmania infantum sterol 24-c-methyltransferase formulated with MPL-SE induces cross-protection against L. major infection. Vaccine. 2009;27:2884–2890. doi: 10.1016/j.vaccine.2009.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daifalla NS, Bayih AG, Gedamu L. Immunogenicity of Leishmania donovani iron superoxide dismutase B1 and peroxidoxin 4 in BALB/c mice: The contribution of Toll-like receptor agonists as adjuvant. Exp. Parasitol. 2011;129:292–298. doi: 10.1016/j.exppara.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 64.Rahman MS, Rahman MK, Saha S, Kaykobad M, Rahman MS. Antigenic: An improved prediction model of protective antigens. Artif. Intell. Med. 2019;94:28–41. doi: 10.1016/j.artmed.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 65.Yang B, Sayers S, Xiang Z, He Y. Protegen: a Web-Based Protective Antigen Database and Analysis System. Nucleic Acids Res. 2011;39(suppl_1):W1073–W1078. doi: 10.1093/nar/gkq944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doytchinova IA, Flower DR. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kadam K, Karbhal R, Jayaraman VK, Sawant S, Kulkarni-Kale U. AllerBase: A comprehensive allergen knowledgebase. Database (Oxford). 2017;2017:1–12. doi: 10.1093/database/bax066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magnan CN, et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26:2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Groot AS, Moise L, McMurry W. Epitope-based immunome-derived vaccines: A strategy for improved design and safety. Clinical Applications of Immunomics. 2009;2:39–69. doi: 10.1007/978-0-387-79208-8_3. [DOI] [Google Scholar]
- 71.Hajissa K, Zakaria R, Suppian R, Mohamed Z. Epitope-based vaccine as a universal vaccination strategy against Toxoplasma gondii infection: A mini-review. J. Adv. Veterinary Animal Res. 2019;6:174–182. doi: 10.5455/javar.2019.f329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anthony DD, Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29:260–269. doi: 10.1016/S1046-2023(02)00348-1. [DOI] [PubMed] [Google Scholar]
- 73.Saha S, Raghava GPS. BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform.) 2004;3239:197–204. [Google Scholar]
- 74.Saha S, Raghava GPS. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Genet. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 75.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reche PA, Zhang H, Glutting JP, Reinherz EL. EPIMHC: A curated database of MHC-binding peptides for customized computational vaccinology. Bioinformatics. 2005;21:2140–2141. doi: 10.1093/bioinformatics/bti269. [DOI] [PubMed] [Google Scholar]
- 77.Toseland CP, et al. AntiJen: A quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data. Immunome Res. 2005;1:4. doi: 10.1186/1745-7580-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doytchinova IA, Guan P, Flower DR. EpiJen: A server for multistep T cell epitope prediction. BMC Bioinform. 2006;7:131. doi: 10.1186/1471-2105-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larsen MV, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh H, Raghava GPS. ProPred1: Prediction of promiscuous MHC class-I binding sites. Bioinformatics. 2003;19:1009–1014. doi: 10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- 81.Lundegaard C, Lamberth K, Harndahl M, Buus S, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36(suppl_2):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saha S, Raghava GPS. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(suppl_2):W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014;20(6):1–6. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 84.Gupta S, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8:e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein-Sol: A web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33:3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 87.Kumar TA. CFSSP: Chou and Fasman secondary structure prediction server. Wide Spectrum 1. 2013;1(9):15–19. [Google Scholar]
- 88.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhattacharya D, Nowotny J, Cao R, Cheng J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016;44:W406–W409. doi: 10.1093/nar/gkw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 92.Ferdous S, Kelm S, Baker TS, Shi J, Martin ACR. B-cell epitopes: Discontinuity and conformational analysis. Mol. Immunol. 2019;114:643–650. doi: 10.1016/j.molimm.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 93.Sweredoski MJ, Baldi P. PEPITO: Improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics. 2008;24:1459–1460. doi: 10.1093/bioinformatics/btn199. [DOI] [PubMed] [Google Scholar]
- 94.Ponomarenko J, et al. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rubinstein ND, Mayrose I, Martz E, Pupko T. Epitopia: A web-server for predicting B-cell epitopes. BMC Bioinform. 2009;10:287. doi: 10.1186/1471-2105-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mosaheb MM, Reiser ML, Wetzler LM. Toll-like receptor ligand-based vaccine adjuvants require intact MyD88 signaling in antigen-presenting cells for germinal center formation and antibody production. Front. Immunol. 2017;8:3. doi: 10.3389/fimmu.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kozakov D, et al. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(suppl_2):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Andrusier N, Nussinov R, Wolfson HJ. FireDock: Fast interaction refinement in molecular docking. Proteins Struct. Funct. Genet. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 101.Grote A, et al. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang Z, et al. A novel technique for constructing infectious cloning of type 3 porcine circovirus. Front. Microbiol. 2020;11:1067. doi: 10.3389/fmicb.2020.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bui H-H, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8(1):361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Urrutia-Baca VH, et al. Immunoinformatics approach to design a novel epitope-based oral vaccine against Helicobacter pylori. J. Comput. Biol. 2019;26:1177–1190. doi: 10.1089/cmb.2019.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wizemann TM, Adamou JE, Langermann S. Adhesins as targets for vaccine development. Emerg. Infect. Dis. 1999;5:395–403. doi: 10.3201/eid0503.990310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Centurion-Lara A, et al. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J. Exp. Med. 1999;189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shey RA, et al. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep. 2019;9:1–18. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Vries SJ, Bonvin AMJJ. Cport: A consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS ONE. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meza B, Ascencio F, Sierra-Beltrán AP, Torres J, Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: An in silico approach. Infect. Genet. Evol. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 113.Santi-Rocca J, et al. A multi-parametric analysis of Trypanosoma cruzi infection: Common pathophysiologic patterns beyond extreme heterogeneity of host responses. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-08086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nogueira RT, et al. Recombinant yellow fever viruses elicit CD8+ T cell responses and protective immunity against Trypanosoma cruzi. PLoS ONE. 2013;8:e59347. doi: 10.1371/journal.pone.0059347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Michel-Todó L, et al. In silico design of an epitope-based vaccine ensemble for chagas disease. Front. Immunol. 2019;10:2698. doi: 10.3389/fimmu.2019.02698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Versteeg L, Almutairi MM, Hotez PJ, Pollet J. Enlisting the mRNA Vaccine Platform to Combat Parasitic Infections. Vaccines. 2019;7(4):122. doi: 10.3390/vaccines7040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data were obtained from open sources and have been cited and deposited in Datasets S1 and also available on our website. Supplementary Data: https://tinyurl.com/2b2s927h. Software Pipeline: Vax-ELAN: https://vac.kamalrawal.in/vaxelan/, Vax-ELAN Version 2: https://vac.kamalrawal.in/vaxelan/v2, Vaxi-DL: https://vac.kamalrawal.in/vaxidl/.