Abstract
We identified biologically relevant moderators of response to tumour necrosis factor (TNF)-α inhibitor, infliximab, among 60 individuals with bipolar depression. Data were derived from a 12-week, randomized, placebo-controlled clinical trial secondarily evaluating the efficacy of infliximab on a measure of anhedonia (ie, Snaith Hamilton Pleasure Scale). Three inflammatory biotypes were derived from peripheral cytokine measurements using an iterative, machine learning-based approach. Infliximab-randomized participants classified as biotype 3 exhibited lower baseline concentrations of pro- and anti-inflammatory cytokines and soluble TNF receptor-1 and reported greater anti-anhedonic improvements, relative to those classified as biotype 1 or 2. Pre-treatment biotypes also moderated changes in neuroinflammatory substrates relevant to infliximab’s hypothesized mechanism of action. Neuronal origin-enriched extracellular vesicle (NEV) protein concentrations were reduced to two factors using principal axis factoring: phosphorylated nuclear factorκB (p-NFκB), Fas-associated death domain (p-FADD), and IκB kinase (p-IKKα/β) and TNF receptor-1 (TNFR1) comprised factor “NEV1,” whereas phosphorylated insulin receptor substrate-1 (p-IRS1), p38 mitogen-activated protein kinase (p-p38), and c-Jun N-terminal kinase (p-JNK) constituted “NEV2.” Among infliximab-randomized subjects classified as biotype 3, NEV1 scores were decreased at weeks 2 and 6 and increased at week 12, relative to baseline, and NEV2 scores increased over time. Decreases in NEV1 scores and increases in NEV2 scores were associated with greater reductions in anhedonic symptoms in our classification and regression tree model (r2=0.22, RMSE=0.08). Our findings provide preliminary evidence supporting the hypothesis that the anti-anhedonic effects of infliximab require modulation of multiple TNF-α signalling pathways, including NF-κB, IRS1, and MAPK.
Introduction
Bipolar depression is a debilitating brain-based disorder; despite the availability of agents with demonstrated antidepressant efficacy in randomized clinical trials, health and functional outcomes remain woefully inadequate in real-world patient populations (1,2). A contributing factor to the tremendous human costs of bipolar depression is that the pathoetiology of bipolar disorder and the neurobiology of treatments remain poorly understood (3). Consequently, current treatment selection and sequencing strategies largely constitute a process of trial-and-error, wherein the risk of treatment resistance rises and the odds of full functional recovery falls with each unsuccessful treatment attempt (5–9). The foregoing set of observations instantiates the need for a disease model-informed framework for conceptualizing mechanistically relevant, and clinically useful, biomarkers for the treatment of bipolar depression.
The immune system is a pertinent therapeutic target for a subpopulation of individuals with depression (10–12). Infliximab is a tumour necrosis factor (TNF)-α inhibitor that has exhibited antidepressant and anti-anhedonic effects in preliminary studies among depressed individuals with aberrant immune-inflammatory activation (13–15). Cellular targets of TNF-α stimulation include, but are not limited to, pathways involving nuclear factor-κB (NF-κB), Fas-associated death domain (FADD), mitogen-activated protein kinase (MAPK), and insulin receptor substrate-1 (IRS1) (16–18). Taken together, immune and inflammatory processes are relevant to the neurobiology and treatment of depression; moreover, inflammatory biotypes can inform disease models and aid clinicians in selecting interventions for patients with bipolar depression (19–21).
Anhedonia is a transdiagnostic neuropsychiatric construct, defined as loss of interest and/or pleasure in nearly all day-to-day activities, and observed in bipolar disorders, major depressive disorders, Parkinson’s disease, diabetes mellitus, Alzheimer’s disease, and many other medical conditions affecting the brain (22,23). Herein, we aimed to develop an empirically driven model for understanding the mechanism of action of infliximab, an anti-inflammatory agent that has preliminarily demonstrated anti-anhedonic efficacy among individuals with bipolar I/II depression (14). The primary outcome of interest, in the present post hoc analysis, was improvement in hedonic capacity (i.e., reduction in anhedonic symptoms), which was operationalized using the Snaith Hamilton Pleasure Scale (SHAPS) total score (range 14–56), with higher scores indicative of greater hedonic capacity (24,25). Our secondary outcome of interest was reduction in overall depressive symptom severity, which was operationalized using the Montgomery-Asberg Depression Rating Scale (MADRS) total score (range 0–60), with higher scores indicative of greater depressive symptom severity.
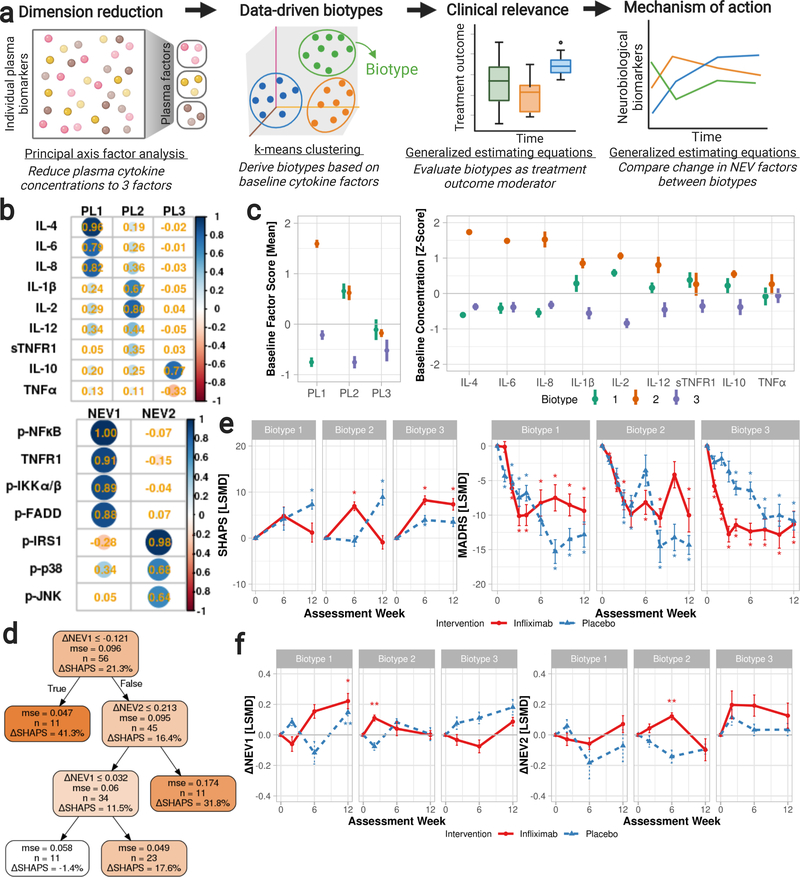
We used an iterative, machine learning-based approach to investigate peripheral markers of inflammatory activation relevant to infliximab’s hypothesized mechanism of action (Figure 1a). Plasma cytokine and neuronal origin-enriched extracellular vesicle (NEV) protein concentrations, assessed at weeks 0, 2, 6, and 12, were evaluated to identify mechanistically relevant inflammatory biotypes. Principal axis factor analyses of plasma cytokine concentrations reduced the number of cytokine measurements to three factors. Next, k-means clustering of cytokine factor scores stratified subjects by baseline cytokine concentrations and yielded three biotypes. Generalized estimating equations were used to evaluate to what extent these three biotypes moderate infliximab’s effects on neurobiological substrates (i.e., by comparing change in NEV concentrations across time, treatment groups, and biotypes). Similarly, we evaluated the biotypes’ clinical utility by evaluating their role in moderating change in SHAPS and MADRS total scores using generalized estimating equations. Finally, classification and regression trees assessed the predictive utility of the three data-driven biotypes.
Figure 1.
We used an iterative, machine learning-based approach to investigate peripheral markers of inflammatory activation relevant to infliximab’s hypothesized mechanism of action. a. Plasma cytokine and neuronal origin-enriched extracellular vesicle (NEV) protein concentrations, assessed at weeks 0, 2, 6, and 12, were evaluated to identify mechanistically relevant inflammatory biotypes: principal axis factor analyses of plasma cytokine concentrations reduced the number of cytokine measurements to three factors; next, k-means clustering of cytokine factor scores stratified subjects by baseline cytokine concentrations and yielded three biotypes. Generalized estimating equations were used to evaluate to what extent these three biotypes moderate infliximab’s effects on neurobiological substrates (i.e., by comparing change in NEV concentrations across time, treatment groups, and biotypes). Similarly, we evaluated the biotypes’ clinical utility by evaluating their role in moderating change in SHAPS and MADRS total scores using generalized estimating equations. Finally, classification and regression trees assessed the predictive utility of the three data-driven biotypes. Plasma NEV biomarker concentrations were reduced to two factors, given their interrelatedness, for improved interpretability.
Figure 1b. Principal axis factor analyses with varimax rotation were performed for peripheral cytokine measurements. Rotated factor loadings are presented as standardized z-scores. Fit indices: (left) KMO = 0.76, RMSEA = 0.12 (95%CI: 0.074, 0.16), SRMR = 0.03, TLI = 0.88; (right) KMO = 0.70, RMSEA = 0.29 (95%CI: 0.237, 0.346), SRMR = 0.04, TLI = 0.72.
Abbreviations: CI: confidence interval; IL: interleukin; KMO: Kaiser-Meyer-Olkin; NEV: neuronal origin-enriched extracellular vesicle factor; p-: phosphorylated; p-FADD:pS194-Fas-associated protein with death domain; p-IKKα/β: pS177/181-IκB kinase; p-IRS1: pS312-insulin receptor substrate-1; p-JNK: pT183/Y185-c-Jun N-terminal kinase; p-NFκB: pS536-nuclear factorκ-light-chain-enhancer of activated B cells; PL: plasma cytokine factor; p-p38: pT180/Y182-p38 mitogen-activated protein kinase;RMSEA: Root Mean Square Error of Approximation; SRMR: Standardized Root Mean Square Residual; sTNFR: soluble TNFR; TLI: Tucker-Lewis Index; TNF: tumour necrosis factor; TNFR: TNF receptor.
Figure 1c. Mean (left) standardized cytokine factor scores and (right) z-scores of individual plasma biomarker measurements are presented by cluster biotypes at baseline. Subjects with bipolar disorder, meeting criteria for a current major depressive episode at the time of study enrollment, were stratified by baseline cytokine factor scores into three biotypes through k-means clustering (2000-bootstrapped Jaccard coefficients = 0.79, 0.92, 0.77). Abbreviations: IL: interleukin; PL: plasma factor; sTNFR: soluble TNF receptor; TNF: tumour necrosis factor.
Figure 1d. Classification and regression trees were used to predict change in anhedonic symptom severity from baseline-to-endpoint as a proportion of baseline severity: (SHAPSWeek 6 or 12 −SHAPSWeek 0)/SHAPSWeek 0. Fit indices: r2 = 0.22, RMSE = 0.08. Abbreviations: mse: mean squared error; NEV: exosome factor 1 (p-NFκB, p-FADD, p-IKKα/β, TNFR1), 2 (p-IRS1, p-JNK, p-p38); RMSE: root-mean-square error; SHAPS: Snaith-Hamilton Pleasure Scale.
Figure 1e. Differences in mean change in (a) SHAPS and (b) MADRS total scores between infliximab- and placebo-randomized subjects with bipolar I/II depression, stratified by baseline cytokine biotypes. Increases in SHAPS and decreases in MADRS total scores indicate improvements in hedonic capacity and reductions in overall depressive symptom severity, respectively. Abbreviations: LSMD: least squares mean difference; MADRS: Montgomery-Asberg Depression Rating Scale (range 0 to 60); SHAPS: Snaith-Hamilton Pleasure Scale (14 to 56); *: Significant (p<0.05) baseline-to-endpoint change within the treatment-biotype group.
Figure 1f. Mean difference in change in NEV factor scores between infliximab- and placebo-randomized patients with bipolar I/II depression, stratified by baseline biotype. Abbreviations: NEV: exosome factor 1 (p-NFκB, p-FADD, p-IKKα/β, TNFR1), 2 (p-IRS1, p-JNK, p-p38); LSMD: least squares mean difference; *: Significant (p < 0.05) baseline-to-endpoint change within the treatment-biotype group.
Materials and methods
Study Design and Clinical Trial Data
Data were derived from a 12-week, randomized, double-blind, placebo-controlled clinical trial evaluating the antidepressant efficacy of infliximab in 60 adults (ages 18–65) with bipolar I/II disorder (13). We have previously reported that infliximab-randomized participants exhibited significant reductions in anhedonic symptoms at week 6; however, the beneficial effects on anhedonia were not sustained at week 12 in the overall study sample (14). Complete inclusion and exclusion criteria have been published (13). Baseline participant characteristics are summarized in Table S1.
Sixty subjects were enrolled between October 1, 2015 and April 30, 2018 at the Mood Disorders Psychopharmacology Unit (MDPU), University Health Network (Toronto, Ontario, Canada) and the Department of Psychiatry and Behavioral Sciences, School of Medicine, Stanford University (Palo Alto, California, USA). Participants met diagnostic criteria for a current major depressive episode according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5). The study population was enriched for a pre-treatment systemic pro-inflammatory phenotype, as determined by meeting at least one of the following metabolic/inflammatory criteria: obesity and dyslipidemia, obesity and hypertension, daily cigarette smoking, diabetes mellitus, migraine, inflammatory bowel disease, and/or C-reactive protein level of ≥5 mg/L (26).
Participants were randomized to intravenously receive 5 mg/kg infliximab (n=29) or saline placebo (n=31) at weeks 0, 2, and 6. Subjects were assessed weekly for the first 4 weeks (i.e., at weeks 1, 2, 3, 4) and bi-weekly thereafter (i.e., at weeks 6, 8, 10, 12). Saline was matched to infliximab in colour and consistency. All study participants, clinicians, investigators, infusion nurses, and outcome assessors were masked to treatment allocation. The institutional ethics boards at the University Health Network and Stanford University approved the study; all participants provided written, informed consent.
Dataset Description
We primarily evaluated predictors and moderators of change in the Snaith Hamilton Pleasure Scale (SHAPS) total score from baseline to weeks 6 and 12. Our primary analysis included data from participants that received at least one infusion and completed at least one post-baseline SHAPS assessment. Data from 25 infliximab- and 27 placebo-randomized subjects were included. Week 6 SHAPS data were available only in subjects enrolled in the study after a protocol amendment in December 2016. Eleven infliximab- and 11 placebo-randomized subjects who completed the 12-week study were missing SHAPS data at week 6. Week 6 SHAPS data were available for 4 infliximab- and 1 placebo-randomized subjects without week 12 data.
We secondarily evaluated the change in Montgomery-Asberg Depression Scale (MADRS) total score from baseline to endpoint. At least one post-baseline MADRS assessment data was available for 29 infliximab- and 30 placebo-randomized participants. Week 1, 2, 3, 4, 5, 6, 8, 10, and 12 data were available for 29, 28, 27, 28, 26, 23, 21, and 22 infliximab-randomized subjects and 30, 28, 30, 29, 26, 27, 24, 26, and 26 placebo-randomized subjects, respectively.
Plasma Sample Collection and Analyses
Blood samples were collected after 12-hour fasts at baseline and weeks 2, 6, and 12 in ethylenediaminetetraacetic acid (EDTA)-coated tubes, centrifuged at 1000 g for 15 minutes at 4 degrees Celsius, and stored at −80 degrees Celsius until analysis. All personnel involved in sample analyses were masked to treatment allocation.
Cytokine Analysis
Plasma concentrations of TNF-α, sTNFR1, interleukin (IL)-1β, IL-2, IL-4, IL-6, IL-8, IL-10, and IL-12 were quantified by personnel at Eve Technology (Calgary, AB, Canada) using BioPlex-200 (Luminex Corporation, Austin, TX, USA) and Millipore MILLIPLEX panels Human Cytokine Array Proinflammatory Focused 13-plex and Human Soluble Cytokine Receptor Array 14-plex (MilliporeSigma Corporation, Billerica, MA, USA). Baseline and endpoint samples were analysed in a single replication on the same day; analyte concentrations were calculated using a standard curve as per the manufacturer’s instructions.
Minimum detectable concentrations (pg/mL) were: TNF-α (0.1), sTNFR1 (12), IL-1β (0.4), IL-2 (0.3), IL-4 (0.6), IL-6 (0.3), IL-8 (0.2), IL-10 (0.3), and IL-12 (0.4). The mean intra-assay coefficients of variation (CVs) were <10% for sTNFR1 and <5% for TNF-α, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, and IL-12; the mean inter-assay coefficients of variation were <15% for TNF-α, sTNFR1, IL-1β, IL-2, IL-4, and IL-8 and <20% for IL-6, IL-10, and IL-12.
Neuronal Origin-enriched Extracellular Vesicle (NEV) Analysis
Investigators at the Laboratory of Clinical Investigation, Intramural Research Program, National Institute on Aging, National Institutes of Health (Baltimore, MD, USA) quantified plasma concentrations of NEVs in accordance with published methods. A detailed description of the procedures and evidence supporting its use are available elsewhere (27–31).
We quantified NEV protein concentrations of pS312-insulin receptor substrate-1 (p-IRS1), pT183/Y185-c-Jun N-terminal kinase (p-JNK), and pT180/Y182-p38 mitogen-activated protein kinase (p-p38) using the MESO SCALE DISCOVERY (MSD) Phospho-IRS1 and Mitogen-Activated Protein Kinase (MAPK) Phosphoprotein Assay Whole Cell Lysate Kits (catalogue IDs: K150HLD, K15101D). We quantified phosphorylated nuclear factor κ-light-chain-enhancer of activated B cells (p-NFκB; Ser536), Fas-associated protein with death domain (p-FADD; Ser194), and IκB kinase (IKKα/β; Ser177/Ser181), as well as total protein levels of TNFR1 using the MILLIPLEX MAP 6-Plex NF-κB Magnetic Bead Signaling kit (48–630MAG; MilliporeSigma Corporation, Billerica, MA, USA). We quantified Alix concentrations using the Human Programmed cell death 6-interacting protein (PDCD6IP) ELISA kit (CSB-EL017673HU; Cusabio Biotech Co., LTD, Houston, TX, USA).
The samples were precipitated with Exoquick (System Biosciences, Inc., Mountainview, CA, USA) to enrich for extracellular vesicle concentrations and mouse anti-human CD171 (L1CAM) biotinylated antibody (clone 5G3; Thermo Scientific, Inc., Waltham, MA) to isolate neuronal surface antigen L1CAM. Extracellular vesicles were lysed with Mammalian Protein Extraction Reagent (M-PER; Thermo Scientific, Inc.). Plates with the MSD phosphoassays were read using the MESO QuickPlex SQ120 imager (Meso Scale Discovery, Rockville, MD, USA); plates with the MILLIPLEX panel were read using the Luminex 200 System (Luminex Corporation, Austin, TX, USA). We analyzed electrochemiluminescence signals for the MSD phosphoassays and fluorescence signals for the MILLIPLEX panel as a standard curve could not be constructed. Alix plates were read using the Synergy H1 microplate reader, which was set to 450 nm as per the manufacturer’s instructions, and Gen5 Microplate Data Collection Software (BioTek Instruments, Winooski, VT, USA).
All assays were conducted in duplicate. The mean intra-assay and inter-assay CVs were <15% and <20%, respectively, for all NEV analytes. The limits of detection (LODs) were, in ng/mL: 84.08 (p-IRS1), 119.42 (p-JNK), 158.86 (p-p38), 48.16 (p-NFκB), 52.68 (p-FADD), 46.42 (p-IKKα/β), 34.43 (TNFR1), and 168.71 (Alix). We excluded from our analyses samples that had a mean CV ≥15% (42 p-IRS1, 11 p-JNK, 9 p-p38, 35 p-NFκB, 32 p-FADD, 32 p-IKKα/β, 34 TNFR1, 5 Alix) or were below the LOD (3 p-IRS1, 0 p-JNK, 0 p-p38, 1 p-NFκB, 10 p-FADD, 0 p-IKKα/β, 1 TNFR1, 0 Alix).
Dimension Reduction
Principal axis factor analyses of ln-transformed plasma cytokine and NEV concentrations were conducted, pooling data from weeks 0, 2, 6, and 12 to maximize sample size (cytokines, n = 216: nWeek 0 = 58, n2 = 58, n6 = 52, n12 = 48; NEVs, n = 164: nWeek 0 = 44, n2 = 41, n6 = 42, n12 = 37). Peripheral cytokine and NEV measurements were analyzed separately to avoid stratification due to differences in methodologies and to include cases missing NEV data in cytokine analyses. There was adequate sampling for the analysis, as determined using the Kaiser-Meyer-Olkin method (KMO = 0.76 and 0.70 for the cytokine and NEV factor analyses, respectively). The rotated factor scores of cytokine biomarkers were then analyzed using k-means clustering. The resultant clusters were used to stratify subjects by baseline cytokine measurements.
Model Development
We evaluated baseline cytokine biotype, intervention assignment, and week as predictors of pro-hedonic efficacy using classification and regression trees (CART). Classification and regression trees are a versatile, nonparametric type of decision tree (supervised) learning algorithm capable of predicting and explaining non-linear relationships and high-order interactions (32). Change in hedonic capacity was operationalized as change in SHAPS total score from baseline as a proportion of baseline SHAPS total score: (SHAPSWeek 6 or 12 − SHAPSWeek 0)/SHAPSWeek 0.
Data were pooled across visits to maximize sample size (infliximab: nWeek 6 = 13, n12 = 21; placebo: nWeek 6 = 16, n12 = 25). A randomized search was used to optimize hyperparameters (i.e., splitting criterion [search range: random, best]; maximum tree depth [2:6, None], number of leaf nodes [10:24, None], number of features to consider for each split [auto, None, log2]; minimum numbers of observations per leaf [1:19], required to split an internal node [2:20]) (33). We used cross-validation on the entire dataset and, to increase explanatory power, we did not reserve a separate test set, as our overarching objective was to characterize biomarkers with clinical relevance (i.e., pro-hedonic intervention efficacy), rather than to maximize predictive capability at the expense of elucidating mechanisms or moderators of treatment response (34).
Generalized estimating equations were used to evaluate the effect of baseline cytokine cluster biotypes on change in treatment outcome measures. We used negative binomial and gamma distributions, as appropriate. We included ln-transformed Alix concentration as a moderator in generalized estimating equations where an NEV concentration was the outcome of interest.
Statistical Analysis
Factor and cluster analyses were conducted on R version 3.6.2 (35). Factor analyses with the principal axis method, varimax rotation, and 100 iterations were performed using the fa function from the package psych and varimax from stats. Scree plots identified 3 and 2 as the optimal numbers of factors for plasma cytokine and NEV factor analyses, respectively. There was adequate sampling for the analyses (Kaiser-Meyer-Olkin [KMO] = 0.76 for cytokines, 0.70 for NEVs). Factor scores were predicted using the regression method. K-means clustering analyses were conducted with 1000 iterations, 2000 bootstraps, and an α value of 0.05 using clusterboot and kmeansCBI from fpc (36,37). The Elbow method determined 3 as the optimal number of clusters. The Jaccard coefficients were examined to evaluate clusterwise stability (38).
Classification and regression trees were developed on Python 3.6.8 using the module sklearn (39). We ran DecisionTreeRegressor with mean squared error splitting criteria, a maximum tree depth of 3, a minimum of 11 samples at each node, and a minimum of 18 samples required to split an internal node. Hyperparameters were determined by results from a randomized search with 10-fold cross-validation, 600 iterations, and r2 scoring criteria using RandomizedSearchCV. Generalized estimating equations were conducted on SPSS Statistics 26 for Windows. Missing data were omitted (i.e., pairwise deletion).
Results
Extraction of Component Factor Scores
Plasma cytokine concentrations were reduced to three factors henceforth named, “PL1,” “PL2,” and “PL3,” which were, respectively, characterized by (PL1:) high levels of interleukin (IL)-4, IL-6, and IL-8; (PL2:) elevations in IL-1β, IL-2, IL-12, and soluble TNF receptor-1 (sTNFR1); and (PL3:) high IL-10 with low TNF-α. Plasma NEVs produced an additional two factors. Phosphorylated nuclear factor-κB (p-NFκB), phosphorylated Fas-associated death domain (p-FADD), phosphorylated IκB kinase (p-IKKα/β), and TNF receptor1 (TNFR1) comprised factor “NEV1,” whereas phosphorylated insulin receptor substrate-1 (p-IRS1), p38 mitogen-activated protein kinase (p-p38), and c-Jun N-terminal kinase (p-JNK) constituted “NEV2.” Factor loadings and fit indices are depicted in Figure 1b. Factors PL1, PL2, and PL3 respectively explained 28.2%, 19.1%, and 8.0% of the variance in plasma cytokine concentrations; factors NEV1 and NEV2 respectively accounted for 50.8% and 26.1% of the variance in NEV concentrations.
Plasma Cytokine Concentrations Define Three Biotypes
We grouped participants by plasma cytokine measurements using k-means clustering. Our analysis of cytokine factor scores identified three clusters defined by distinct patterns of plasma cytokine levels, comprising 38.4%, 22.7%, and 38.9% of the 216 samples. The 2000- bootstrapped Jaccard coefficients were >0.75, indicating the clusters were valid and stable (Jaccard = 0.79, 0.92, 0.77). The mean PL1, PL2, and PL3 scores were, by cluster: −0.77, 0.54, 0.23 (cluster 1); 1.55, 0.56, −0.01 (cluster 2); and −0.14, −0.87, −0.22 (cluster 3).
Of the 56 subjects with at least one post-baseline assessment of SHAPS: 16 subjects (nInfliximab= 8, nPlacebo= 8) were classified as cluster 1 at baseline–hereafter referred to as biotype 1–with lowerPL1 and higher PL2 and PL3 scores, indicative of lower IL-4, IL-6, IL-8, and TNF-αand higher IL-1β, IL-2, IL-12, sTNFR1, and IL-10 levels prior to the first infusion. Eleven subjects (nInfliximab= 4,nPlacebo= 7) exhibited elevations in all cytokines at baseline and were classified as biotype 2 (Figure 1c). Twenty-five subjects (nInfliximab= 13,nPlacebo= 12) were classified as biotype 3,with lower PL1, PL2, and PL3 scores.
Baseline Biotypes Predict Treatment Outcomes
Baseline cytokine biotype, intervention allocation, week, as well as baseline and change in NEV1 and NEV2 factor scores, predicted baseline-to-endpoint reduction in anhedonic symptoms (r2 = 0.22, RMSE = 0.08). Decreases in NEV1 factor scores and increases in NEV2 factor scores were associated with greater reductions in anhedonic symptoms (Figure 1d).
We used generalized estimating equations to further characterize changes in hedonic capacity over time across intervention and baseline biotype groups. Baseline biotype significantly moderated change in SHAPS total score (Table 1). Biotype 3 subjects exhibited greater improvements in hedonic capacity with infliximab (vs placebo) at weeks 6 and 12. Biotype 2 subjects also derived greater pro-hedonic benefits with infliximab relative to placebo at week 6; however, the effects were not sustained 6 weeks past the last infusion (Figure 1e). In contrast, baseline biotype did not significantly moderate changes in MADRS total score (Table 1), although a similar stratification of antidepressant response was observed across baseline biotypes (Figure 1e).
Table 1.
Model effects of baseline cytokine biotypes on measures of anhedonia (SHAPS) and overall depressive symptom severity (MADRS) in patients with bipolar I/II disorder who were experiencing a current major depressive episode at the time of study enrollment. A generalized estimating equation with a negative binomial distribution and autoregressive covariance structure was used.
| Model effects | SHAPS | MADRS | ||||
|---|---|---|---|---|---|---|
| df | χ 2 | p | df | χ 2 | p | |
|
| ||||||
| Time | 2 | 19.89 | <0.001 | 3 | 73.66 | <0.001 |
| Treatment | 1 | 0.005 | 0.946 | 1 | 0.29 | 0.593 |
| Biotype | 2 | 0.003 | 0.999 | 2 | 1.55 | 0.460 |
| Time × Treatment | 2 | 8.26 | 0.016 | 3 | 7.22 | 0.065 |
| Time × Biotype | 4 | 2.31 | 0.679 | 6 | 1.36 | 0.968 |
| Treatment × Biotype | 2 | 0.06 | 0.972 | 2 | 0.87 | 0.649 |
| Time × Treatment × Biotype | 4 | 10.40 | 0.034 | 6 | 5.13 | 0.528 |
Abbreviations: df: degree of freedom; MADRS: Montgomery-Asberg Depression Rating Scale (0 to 60); SHAPS: Snaith-Hamilton Pleasure Scale (14 to 56).
Baseline Biotypes Moderate Treatment Effects on Plasma Cytokines and Neuronal Origin-enriched Extracellular Vesicles
Given that the baseline biotypes were associated with disparate treatment trajectories, we hypothesized that the baseline biotypes would also moderate divergent changes in peripheral inflammatory biomarkers in response to treatment with infliximab or placebo. Furthermore, we hypothesized that individuals randomized to infliximab who were classified as biotype 3 would exhibit cellular and molecular changes in the immune-inflammatory system targeted by infliximab that would be absent or differentially modulated in individuals classified as biotype 1 or biotype 2. To investigate to what extent baseline biotypes moderate the effects of treatment on the immune-inflammatory system, we assessed and compared changes in cytokine factor scores and individual NEV markers between intervention groups and baseline biotypes using generalized estimating equations.
Overall, individuals classified as biotype 3 exhibited significant reductions in PL1 (i.e., IL-4, IL-6, IL-8) and increases in PL2 (i.e., IL-1β, IL-2, IL-12, sTNFR1) factor scores with infliximab, whereas those classified as biotype 1 or biotype 3 exhibited different patterns of change in PL1 and PL2 factor scores (Figure S1). Participants randomized to infliximab exhibited significant increases in PL3 (i.e., increased IL-10, reduced TNF-α) factor scores across all biotypes when compared to participants receiving placebo. The results of the generalized estimating equations are summarized in Table 2 and detailed in the Supplementary Results.
Table 2.
Model effects of baseline cytokine biotypes on plasma cytokine and NEV factor scores. A generalized estimating equation with a gamma distribution and exchangeable covariance structure was used.
| Model effects | df | PL1 | PL2 | PL3 | NEV1 | NEV2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | p | χ2 | p | χ2 | p | χ2 | p | χ2 | p | ||
|
| |||||||||||
| Time | 3 | 6.86 | 0.08 | 1.35 | 0.72 | 39.77 | <0.01 | 16.85 | <0.01 | 3.80 | 0.28 |
| Treatment | 1 | 0.03 | 0.86 | 0.56 | 0.45 | 24.32 | <0.01 | 1.36 | 0.24 | 0.02 | 0.88 |
| Biotype | 2 | 437.12 | <0.01 | 48.88 | <0.01 | 0.44 | 0.80 | 9.62 | <0.01 | 3.50 | 0.17 |
| Time × Treatment | 3 | 4.88 | 0.18 | 1.20 | 0.75 | 14.81 | <0.01 | 0.85 | 0.84 | 4.26 | 0.24 |
| Time × Biotype | 6 | 22.33 | <0.01 | 12.22 | 0.06 | 13.79 | 0.03 | 13.48 | 0.04 | 5.68 | 0.46 |
| Treatment × Biotype | 2 | 3.02 | 0.22 | 2.23 | 0.33 | 0.40 | 0.82 | 1.32 | 0.52 | 0.16 | 0.92 |
| Time × Treatment × Biotype | 6 | 17.02 | <0.01 | 3.73 | 0.71 | 10.74 | 0.10 | 18.21 | <0.01 | 6.57 | 0.36 |
Abbreviations: df: degree of freedom; NEV: neuronal origin-enriched extracellular vesicle factor; SHAPS: Snaith-Hamilton Pleasure Scale; PL: plasma cytokine factor; ΔSHAPS: (SHAPSWeek 6 or 12 − SHAPSWeek 0)/SHAPSWeek 0
The baseline biotypes moderated changes in NEV markers of inflammation (Figure 1f), notably NEV1 factor scores (i.e., p-NFκB, p-FADD, p-IKKα/β, TNFR1). Among infliximab-randomized subjects classified as biotype 3, NEV1 scores decreased after the first and second infusions (i.e., from baseline to weeks 2 and 6) and increased after the third infusion (i.e., from baseline to week 12). In contrast, among infliximab-randomized subjects classified as biotype 1, NEV1 scores were decreased at week 2 and increased at weeks 6 and 12, relative to baseline levels. Similarly, among subjects classified as biotype 2, NEV1 scores increased after one infusion of infliximab and gradually returned to baseline levels thereafter. Baseline NEV1 factor scores were higher among those classified as biotype 2, relative to those classified as biotype 1 or biotype 3; the foregoing differences between biotypes were not significant within the infliximab group (χ2 = 2.64, p = 0.27) but were significant within the placebo group (χ2 = 9.75, p < 0.01).
Within biotype 3, NEV2 scores increased over time with infliximab, denoting increases in the NEV markers p-IRS1, p-JNK, and p-p38, relative to baseline concentrations. Among infliximab-randomized subjects classified as biotype 1, NEV2 scores decreased at weeks 2 and 6 and increased at week 12, relative to baseline levels. Among those classified as biotype 2, NEV2 scores increased at weeks 2 and 6 and decreased at week 12 with infliximab. Baseline NEV2 scores were numerically higher among baseline NEV2 scores were numerically higher among participants classified as biotype 2 when compared to participants classified as biotype1 or biotype 3; however, the differences were not significant in either treatment group (infliximab: χ2= 1.35, p= 0.51; placebo: χ2= 2.30, p= 0.32).
Discussion
We used an iterative approach to identify and characterize biologically relevant predictors of treatment response among adult patients with bipolar depression. Pre-treatment biotypes, derived from peripheral cytokine measurements, were capable of predicting anti-anhedonic efficacy with the anti-inflammatory biologic drug, infliximab. In our sample, which was enriched for elevated baseline inflammatory activity, participants classified as biotype 3–who exhibited lower plasma concentrations of pro- and anti-inflammatory cytokines and sTNFR1 prior to randomization relative to those classified as biotype 1 or biotype 2–were more likely to report reductions in anhedonic severity with infliximab than with placebo. Participants classified as biotype 1 or biotype 2, who exhibited higher PL2 factor scores relative to those classified as biotype 3, exhibited a less favourable response trajectory with infliximab when compared to those classified as biotype 3.
The pre-treatment inflammatory biotypes, informed solely by peripheral cytokine measurements, significantly moderated the observed changes in markers of neuroinflammation with infliximab (vs. placebo). Differences across biotypes in NEV1 scores may be explained by the aberrant increases in p-FADD and dysregulation of NF-κB substrates observed among infliximab-randomized participants classified as biotype 1 or 2 (Figure S2A–D). In contrast, p-FADD and p-IKKα/β decreased with infliximab treatment among those classified as biotype 3, particularly from weeks 2 to 6. Relative to baseline, NEV TNFR1 concentrations decreased with infliximab within biotype 3 and increased with infliximab within biotypes 1 and 2. The activation of TRADD by TNFR1 leads to the phosphorylation of FADD; increases in p-FADD are associated with NF-κB activation via the IKK complex (40).
Increase in p-NFκB in the context of moderately decreased p-FADD and p-IKKα/β, as was observed among participants classified as biotype 3 between weeks 2 and 6, suggests a shift from a pro-inflammatory state to an anti-inflammatory state may have occurred early with infliximab treatment (e.g., repression of pro-inflammatory cytokines, increased IL-10 expression) (41–45). The foregoing hypothesis is supported by the observed decrease in PL1 (i.e., IL-6, IL-8) and increase in PL3 (i.e., IL-10) scores among those classified as biotype 3. Similarly, genes relevant to TNF-related apoptosis (i.e., TNF superfamily member 12 [TNFSF12], TNF-like weak inducer of apoptosis [TWEAK]), NF-κB, and toll-like receptor (TLR) signalling have been reported to be differentially downregulated among individuals with major depressive disorder who exhibited improvements with infliximab, relative to those who did not respond favourably to infliximab (46).
Infliximab is a monoclonal antibody with high binding affinity for soluble, transmembrane, and receptor-bound TNF-α; moreover, infliximab is capable of lysing activated macrophages, which are the largest source of TNF-α (47–51). Phosphorylation of the p65 subunit of NF-κB and nuclear accumulation of IκBα have been associated with inflammatory resolution (52). Previous clinical trials investigating the efficacy of infliximab for the treatment of inflammatory bowel disease have reported that nuclear accumulation of mucosal p65 NF-κB, indicating immune reactivation, predicts non-response and relapse (53,54).
Differences in NEV2 scores between baseline biotypes are driven by disparate patterns of change in phosphorylated IRS1, p38, and JNK concentrations. We observed a gradual and sustained increase in NEV2 scores with infliximab among subjects classified as biotype 3, denoting significant increases in NEV concentrations of p-IRS1, p-p38, and p-JNK (Figure S2E–G). In contrast, changes in NEV2 scores with infliximab were not sustained among those classified as biotype 1 or biotype 2. Baseline NEV concentrations and factor scores are compared across biotypes in Figure S2H–I.
Tyrosine phosphorylation of IRS1 is required for the activation of insulin signalling cascades; insulin-evoked responses are attenuated by a negative feedback loop via serine phosphorylation of IRS1 (55–58). A previous clinical trial evaluating the effects of exenatide, a glucagon-like peptide 1 agonist, on NEV markers of insulin signalling also observed increased Ser312 and Ser616 phosphorylation of IRS1 and concomitant tyrosine phosphorylation of IRS1, as well as activation of downstream AKT and mTOR signaling (29). Similarly, altered glucose and lipid metabolism-related gene expression has been hypothesized to subserve the antidepressant effects of infliximab in patients with major depressive disorder (46). Mood disorders also increase cardiometabolic risk and patients often present with other medical comorbidities (e.g., atherosclerosis, obesity, diabetes mellitus) (58–60).
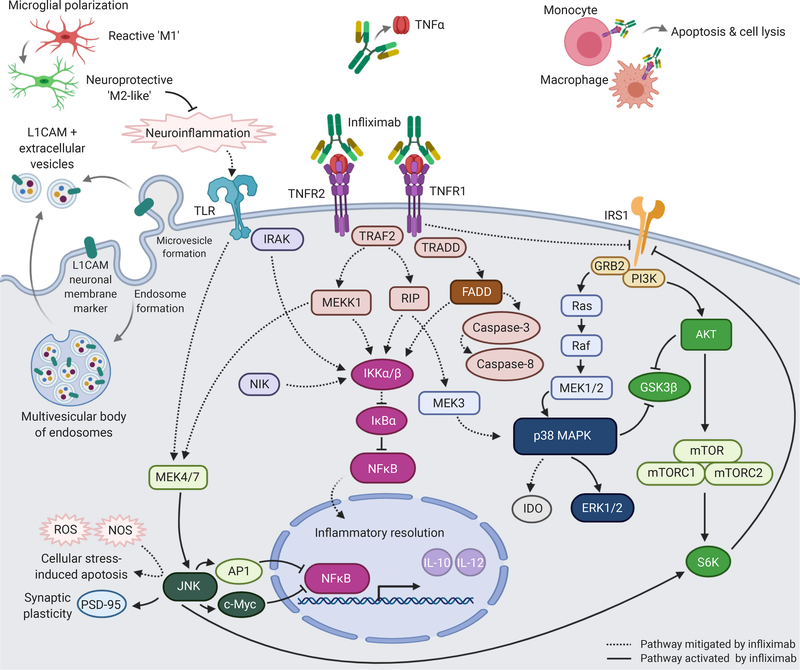
It may be hypothesized that the aberrant regulation and/or lack of sustained modulation of NF-κB and IRS1 pathways may at least partially explain the insufficient reductions in anhedonic symptoms observed in subjects classified as biotype 1 or biotype 2 (Figure 2). We observed a decrease in p-JNK, which regulates IRS1 via activation of p70S6 kinase and NF-κB via c-Myc, among those classified as biotype 2 (Figure S2F). The activation of p38 MAPK has also been reported to subserve infliximab’s beneficial anti-inflammatory effects in populations with inflammatory bowel disease (61–63).
Figure 2.
Hypothesized mechanism of action of infliximab in bipolar depression. Abbreviations: AP1: activator protein 1; AKT: protein kinase B; ERK1/2: extracellular signal-regulated kinases 1/2; FADD: Fas-associated protein with death domain; GRB2: growth factor receptor-bound protein 2; GSK3β: glycogen synthase kinase 3β; IκBα: inhibitor of NF-κB,α; IKKα/β: IκB kinase; IL: interleukin; IRAK: IL-1 receptor-associated kinase; IRS1: insulin receptor substrate-1; JNK: c-Jun N-terminal kinase; MAPK: mitogen activated protein kinase; MEK: MAPK/ERK kinase; MEKK1: MAPK/ERK kinase kinase 1; mTOR: mechanistic target of rapamycin; mTORC1: mTOR complex 1; mTORC2: mTOR complex 2; NFκB: nuclear factor κ-light-chain enhancer of activated B cells; NIK: NFκB inducing kinase; NOS: nitric oxide synthase; p38: p38 MAPK; PI3K: phosphoinositide 3-kinase; PSD-95: postsynaptic density protein 95; RIP: receptor-interacting protein; ROS: reactive oxygen species; S6K: p70S6 kinase; sTNFR: soluble TNFR; TLR: toll-like receptor; TNF: tumour necrosis factor; TNFR: TNF receptor; TRADD: TNFR-associated death domain; TRAF2: TNFR-associated factor 2. Original illustration created with BioRender.com by Yena Lee. The figure was exported under a paid subscription.
Taken together, our analysis integrating peripheral cytokine and neuronal origin-enriched biomarkers of inflammation provides preliminary evidence supporting the hypothesis that the anti-anhedonic effects of TNF-α antagonist infliximab may be subserved by modulation of NF-κB, IRS1, and MAPK signalling pathways. Our findings also support the clinical utility of inflammatory biomarkers for personalizing bipolar depression treatments and targeting anhedonia, for which specific treatments are unavailable. Moreover, the observation that baseline biotypes did not significantly predict changes in overall depressive symptom severity (i.e., MADRS total score) suggests that our results may be specific to the treatment of anhedonia, a discrete and transdiagnostic symptom dimension that correlates with, yet diverges from, overall depressive symptom severity. However, interpretations of our findings are limited by the secondary post hoc nature of the analysis and our relatively small sample size, which limit the generalizability of our results and warrant a prospectively designed replication study.
Supplementary Material
Acknowledgments
This study was funded by the Stanley Medical Research Institute (Grant 13T-012 to R.S.M. and T.S.). This research was supported in part by the Intramural Research Program of the National Institute on Aging, NIH (authors DK, FDP, SC, CNO).
Conflicts of interest
R.S.M. has received research grant support from the Stanley Medical Research Institute and the Canadian Institutes of Health Research/Global Alliance for Chronic Diseases/National Natural Science Foundation of China and speaker/consultation fees from Lundbeck, Janssen, Shire, Purdue, Pfizer, Otsuka, Allergan, Takeda, Neurocrine, Sunovion, and Minerva within the past 36 months. E.B. has been supported by Faculty of Health Sciences, Queen’s University and received honoraria as speaker/member of advisory board from Daiichi-Sankyo not related to the content of this study. J.D.R. has received research grant support from the University of Toronto, Canadian Cancer Society, Canadian Psychiatric Association, American Psychiatric Association, American Society of Psychopharmacology (New Investigator Award), University Health Network Centre for Mental Health, Joseph M. West Family Memorial Fund and Timeposters Fellowship and industry funding for speaker/consultation/research fees from Allergan, Lundbeck and COMPASS; and is the medical director of a private clinic providing off-label ketamine infusions for depression. B.I.G. receives grant or research support from the Brain and Behavior Research Foundation (NARSAD), Brain Canada, the Canadian Institutes of Health Research, the Heart and Stroke Foundation, National Institute of Mental Health, the Ontario Ministry of Research and Innovation, and the departments of psychiatry of Sunnybrook Health Sciences Centre and the University of Toronto. M.V. has received consultancy fees from Lundbeck, Sunovion and Janssen/Cilag within the last three years. All other authors declare no conflict of interest.
References
- 1.Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry [Internet]. 2006. November;163(11):1905–17. Available from: 10.1176/ajp.2006.163.11.1905 [DOI] [PubMed] [Google Scholar]
- 2.Fava M, Rush AJ, Wisniewski SR, Nierenberg AA, Alpert JE, McGrath PJ, et al. A comparison of mirtazapine and nortriptyline following two consecutive failed medication treatments for depressed outpatients: a STAR*D report. Am J Psychiatry [Internet]. 2006. July;163(7):1161–72. Available from: 10.1176/appi.ajp.163.7.1161 [DOI] [PubMed] [Google Scholar]
- 3.Insel TR, Cuthbert BN. Medicine. Brain disorders? Precisely. Science [Internet]. 2015. May 1;348(6234):499–500. Available from: 10.1126/science.aab2358 [DOI] [PubMed] [Google Scholar]
- 4.Trivedi MH, Morris DW, Wisniewski SR, Lesser I, Nierenberg AA, Daly E, et al. Increase in work productivity of depressed individuals with improvement in depressive symptom severity. Am J Psychiatry [Internet]. 2013. June;170(6):633–41. Available from: 10.1176/appi.ajp.2012.12020250 [DOI] [PubMed] [Google Scholar]
- 5.Webb CA, Trivedi MH, Cohen ZD, Dillon DG, Fournier JC, Goer F, et al. Personalized prediction of antidepressant v. placebo response: evidence from the EMBARC study. Psychol Med [Internet]. 2019. May;49(7):1118–27. Available from: 10.1017/S0033291718001708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Ragguett R-M, Mansur RB, Boutilier JJ, Rosenblat JD, Trevizol A, et al. Applications of machine learning algorithms to predict therapeutic outcomes in depression: A meta-analysis and systematic review. J Affect Disord [Internet]. 2018. December 1;241:519–32. Available from: 10.1016/j.jad.2018.08.073 [DOI] [PubMed] [Google Scholar]
- 7.Strawbridge R, Arnone D, Danese A, Papadopoulos A, Herane Vives A, Cleare AJ. Inflammation and clinical response to treatment in depression: A meta-analysis. Eur Neuropsychopharmacol [Internet]. 2015. October;25(10):1532–43. Available from: 10.1016/j.euroneuro.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 8.Benedetti F, Poletti S, Hoogenboezem TA, Locatelli C, de Wit H, Wijkhuijs AJM, et al. Higher Baseline Proinflammatory Cytokines Mark Poor Antidepressant Response in Bipolar Disorder. J Clin Psychiatry [Internet]. 2017;78(8):e986–93. Available from: 10.4088/JCP.16m11310 [DOI] [PubMed] [Google Scholar]
- 9.Arteaga-Henríquez G, Simon MS, Burger B, Weidinger E, Wijkhuijs A, Arolt V, et al. Low-Grade Inflammation as a Predictor of Antidepressant and Anti-Inflammatory Therapy Response in MDD Patients: A Systematic Review of the Literature in Combination With an Analysis of Experimental Data Collected in the EU-MOODINFLAME Consortium. Front Psychiatry [Internet]. 2019. July 9;10:458. Available from: 10.3389/fpsyt.2019.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol [Internet]. 2016. January;16(1):22–34. Available from: 10.1038/nri.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenblat JD, Kakar R, Berk M, Kessing LV, Vinberg M, Baune BT, et al. Anti-inflammatory agents in the treatment of bipolar depression: a systematic review and meta-analysis. Bipolar Disord [Internet]. 2016. March;18(2):89–101. Available from: 10.1111/bdi.12373 [DOI] [PubMed] [Google Scholar]
- 12.Kappelmann N, Lewis G, Dantzer R, Jones PB, Khandaker GM. Antidepressant activity of anti-cytokine treatment: a systematic review and meta-analysis of clinical trials of chronic inflammatory conditions. Mol Psychiatry [Internet]. 2018. February;23(2):335–43. Available from: 10.1038/mp.2016.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntyre RS, Subramaniapillai M, Lee Y, Pan Z, Carmona NE, Shekotikhina M, et al. Efficacy of Adjunctive Infliximab vs Placebo in the Treatment of Adults With Bipolar I/II Depression: A Randomized Clinical Trial. JAMA Psychiatry [Internet]. 2019. May 8;76(8):783–90. Available from: 10.1001/jamapsychiatry.2019.0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Mansur RB, Brietzke E, Carmona NE, Subramaniapillai M, Pan Z, et al. Efficacy of adjunctive infliximab vs. placebo in the treatment of anhedonia in bipolar I/II depression. Brain Behav Immun [Internet]. 2020. May 4; Available from: 10.1016/j.bbi.2020.04.063 [DOI] [PubMed] [Google Scholar]
- 15.Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF, et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry [Internet]. 2013. January;70(1):31–41. Available from: 10.1001/2013.jamapsychiatry.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson KME, Wasén C, Juzokaite L, Leifsdottir L, Erlandsson MC, Silfverswärd ST, et al. Inflammation in the hippocampus affects IGF1 receptor signaling and contributes to neurological sequelae in rheumatoid arthritis. Proc Natl Acad Sci U S A [Internet]. 2018. December 18;115(51):E12063–72. Available from: 10.1073/pnas.1810553115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinridders A, Cai W, Cappellucci L, Ghazarian A, Collins WR, Vienberg SG, et al. Insulin resistance in brain alters dopamine turnover and causes behavioral disorders. Proc Natl Acad Sci U S A [Internet]. 2015. March 17;112(11):3463–8. Available from: 10.1073/pnas.1500877112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasca C, Dobbin J, Bigio B, Watson K, de Angelis P, Kautz M, et al. Insulin receptor substrate in brain-enriched exosomes in subjects with major depression: on the path of creation of biosignatures of central insulin resistance. Mol Psychiatry [Internet]. 2020. June 15; Available from: 10.1038/s41380-020-0804-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millett CE, Harder J, Locascio JJ, Shanahan M, Santone G, Fichorova RN, et al. TNF-α and its soluble receptors mediate the relationship between prior severe mood episodes and cognitive dysfunction in euthymic bipolar disorder. Brain Behav Immun [Internet]. 2020. April 6; Available from: 10.1016/j.bbi.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haroon E, Welle JR, Woolwine BJ, Goldsmith DR, Baer W, Patel T, et al. Associations among peripheral and central kynurenine pathway metabolites and inflammation in depression. Neuropsychopharmacology [Internet]. 2020. May;45(6):998–1007. Available from: 10.1038/s41386-020-0607-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osimo EF, Pillinger T, Rodriguez IM, Khandaker GM, Pariante CM, Howes OD. Inflammatory markers in depression: A meta-analysis of mean differences and variability in 5,166 patients and 5,083 controls. Brain Behav Immun [Internet]. 2020. July;87:901–9. Available from: 10.1016/j.bbi.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husain M, Roiser JP. Neuroscience of apathy and anhedonia: a transdiagnostic approach. Nat Rev Neurosci [Internet]. 2018. August;19(8):470–84. Available from: 10.1038/s41583-018-0029-9 [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Subramaniapillai M, Brietzke E, Mansur RB, Ho RC, Yim SJ, et al. Anti-cytokine agents for anhedonia: targeting inflammation and the immune system to treat dimensional disturbances in depression. Ther Adv Psychopharmacol [Internet]. 2018. December;8(12):337–48. Available from: 10.1177/2045125318791944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snaith RP, Hamilton M, Morley S, Humayan A, Hargreaves D, Trigwell P. A scale for the assessment of hedonic tone the Snaith-Hamilton Pleasure Scale. Br J Psychiatry [Internet]. 1995. July;167(1):99–103. Available from: 10.1192/bjp.167.1.99 [DOI] [PubMed] [Google Scholar]
- 25.Leventhal AM, Chasson GS, Tapia E, Miller EK, Pettit JW. Measuring hedonic capacity in depression: a psychometric analysis of three anhedonia scales. J Clin Psychol [Internet]. 2006. December;62(12):1545–58. Available from: 10.1002/jclp.20327 [DOI] [PubMed] [Google Scholar]
- 26.Kramer NE, Cosgrove VE, Dunlap K, Subramaniapillai M, McIntyre RS, Suppes T. A clinical model for identifying an inflammatory phenotype in mood disorders. J Psychiatr Res [Internet]. 2019. June;113:148–58. Available from: 10.1016/j.jpsychires.2019.02.005 [DOI] [PubMed] [Google Scholar]
- 27.Mustapic M, Eitan E, Werner JK Jr, Berkowitz ST, Lazaropoulos MP, Tran J, et al. Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front Neurosci [Internet]. 2017. May 22;11:278. Available from: 10.3389/fnins.2017.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins RJ, Mustapic M, Goetzl EJ, Kapogiannis D. Exosomal biomarkers of brain insulin resistance associated with regional atrophy in Alzheimer’s disease. Hum Brain Mapp [Internet]. 2017. April;38(4):1933–40. Available from: 10.1002/hbm.23494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athauda D, Gulyani S, Karnati HK, Li Y, Tweedie D, Mustapic M, et al. Utility of Neuronal-Derived Exosomes to Examine Molecular Mechanisms That Affect Motor Function in Patients With Parkinson Disease: A Secondary Analysis of the Exenatide-PD Trial. JAMA Neurol [Internet]. 2019. April 1;76(4):420–9. Available from: 10.1001/jamaneurol.2018.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sáenz-Cuesta M, Arbelaiz A, Oregi A, Irizar H, Osorio-Querejeta I, Muñoz-Culla M, et al. Methods for extracellular vesicles isolation in a hospital setting. Front Immunol [Internet]. 2015. February 13;6:50. Available from: 10.3389/fimmu.2015.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansur RB, Delgado-Peraza F, Subramaniapillai M, Lee Y, Iacobucci M, Rodrigues N, et al. Extracellular Vesicle Biomarkers Reveal Inhibition of Neuroinflammation by Infliximab in Association with Antidepressant Response in Adults with Bipolar Depression. Cells [Internet]. 2020. April 6;9(4). Available from: 10.3390/cells9040895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breiman L Classification and Regression Trees. Boca Raton: Routledge; 2017. [Google Scholar]
- 33.Bergstra J, Bengio Y. Random Search for Hyper-Parameter Optimization. J Mach Learn Res [Internet]. 2012. [cited 2019 Oct 25];13(Feb):281–305. Available from: http://www.jmlr.org/papers/v13/bergstra12a.html [Google Scholar]
- 34.Shmueli G To Explain or to Predict? Stat Sci [Internet]. 2010. August [cited 2020 May 13];25(3):289–310. Available from: https://projecteuclid.org/euclid.ss/1294167961 [Google Scholar]
- 35.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2020. Available from: https://www.R-project.org/ [Google Scholar]
- 36.Revelle W psych: Procedures for Psychological, Psychometric, and Personality Research [Internet]. Evanston, Illinois: Northwestern University; 2019. Available from: https://CRAN.R-project.org/package=psych [Google Scholar]
- 37.Hennig C fpc: Flexible Procedures for Clustering [Internet]. 2019. Available from: https://CRAN.R-project.org/package=fpc
- 38.Hennig C Cluster-wise assessment of cluster stability. Comput Stat Data Anal [Internet]. 2007. September 15;52(1):258–71. Available from: http://www.sciencedirect.com/science/article/pii/S0167947306004622 [Google Scholar]
- 39.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 40.Chen G, Bhojani MS, Heaford AC, Chang DC, Laxman B, Thomas DG, et al. Phosphorylated FADD induces NF-kappaB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. Proc Natl Acad Sci U S A [Internet]. 2005. August 30;102(35):12507–12. Available from: 10.1073/pnas.0500397102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature [Internet]. 2005. April 28;434(7037):1138–43. Available from: 10.1038/nature03491 [DOI] [PubMed] [Google Scholar]
- 42.Christian F, Smith EL, Carmody RJ. The Regulation of NF-κB Subunits by Phosphorylation. Cells [Internet]. 2016. March 18;5(1). Available from: 10.3390/cells5010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Block ML, Zecca L, Hong J-S. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci [Internet]. 2007. January;8(1):57–69. Available from: 10.1038/nrn2038 [DOI] [PubMed] [Google Scholar]
- 44.Barger SW, Hörster D, Furukawa K, Goodman Y, Krieglstein J, Mattson MP. Tumor necrosis factors alpha and beta protect neurons against amyloid beta-peptide toxicity: evidence for involvement of a kappa B-binding factor and attenuation of peroxide and Ca2+ accumulation. Proc Natl Acad Sci U S A [Internet]. 1995. September 26;92(20):9328–32. Available from: 10.1073/pnas.92.20.9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenn AM, Henry CJ, Huang Y, Dugan A, Godbout JP. Lipopolysaccharide-induced interleukin (IL)-4 receptor-α expression and corresponding sensitivity to the M2 promoting effects of IL-4 are impaired in microglia of aged mice. Brain Behav Immun [Internet]. 2012. July;26(5):766–77. Available from: 10.1016/j.bbi.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta D, Raison CL, Woolwine BJ, Haroon E, Binder EB, Miller AH, et al. Transcriptional signatures related to glucose and lipid metabolism predict treatment response to the tumor necrosis factor antagonist infliximab in patients with treatment-resistant depression. Brain Behav Immun [Internet]. 2013. July;31:205–15. Available from: 10.1016/j.bbi.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lügering A, Schmidt M, Lügering N, Pauels HG, Domschke W, Kucharzik T. Infliximab induces apoptosis in monocytes from patients with chronic active Crohn’s disease by using a caspase-dependent pathway. Gastroenterology [Internet]. 2001. November;121(5):1145–57. Available from: 10.1053/gast.2001.28702 [DOI] [PubMed] [Google Scholar]
- 48.Chaudhari U, Romano P, Mulcahy LD, Dooley LT, Baker DG, Gottlieb AB. Efficacy and safety of infliximab monotherapy for plaque-type psoriasis: a randomised trial. Lancet [Internet]. 2001. June 9;357(9271):1842–7. Available from: 10.1016/s0140-6736(00)04954-0 [DOI] [PubMed] [Google Scholar]
- 49.Horiuchi T, Mitoma H, Harashima S-I, Tsukamoto H, Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology [Internet]. 2010. July;49(7):1215–28. Available from: 10.1093/rheumatology/keq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitoma H, Horiuchi T, Hatta N, Tsukamoto H, Harashima S-I, Kikuchi Y, et al. Infliximab induces potent anti-inflammatory responses by outside-to-inside signals through transmembrane TNF-alpha. Gastroenterology [Internet]. 2005. February;128(2):376–92. Available from: 10.1053/j.gastro.2004.11.060 [DOI] [PubMed] [Google Scholar]
- 51.Genovese T, Mazzon E, Crisafulli C, Di Paola R, Muià C, Esposito E, et al. TNF-alpha blockage in a mouse model of SCI: evidence for improved outcome. Shock [Internet]. 2008. January;29(1):32–41. Available from: 10.1097/shk.0b013e318059053a [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene [Internet]. 2006. October 30;25(51):6706–16. Available from: 10.1038/sj.onc.1209933 [DOI] [PubMed] [Google Scholar]
- 53.Lügering A, Lebiedz P, Koch S, Kucharzik T. Apoptosis as a therapeutic tool in IBD? Ann N Y Acad Sci [Internet]. 2006. August;1072:62–77. Available from: 10.1196/annals.1326.013 [DOI] [PubMed] [Google Scholar]
- 54.Nikolaus S, Raedler A, Kühbacker T, Sfikas N, Fölsch UR, Schreiber S. Mechanisms in failure of infliximab for Crohn’s disease. Lancet [Internet]. 2000. October 28;356(9240):1475–9. Available from: 10.1016/s0140-6736(00)02871-3 [DOI] [PubMed] [Google Scholar]
- 55.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science [Internet]. 1996. February 2;271(5249):665–8. Available from: 10.1126/science.271.5249.665 [DOI] [PubMed] [Google Scholar]
- 56.Greene MW, Sakaue H, Wang L, Alessi DR, Roth RA. Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by Serine 312 phosphorylation. J Biol Chem [Internet]. 2003. March 7;278(10):8199–211. Available from: 10.1074/jbc.M209153200 [DOI] [PubMed] [Google Scholar]
- 57.Stagakis I, Bertsias G, Karvounaris S, Kavousanaki M, Virla D, Raptopoulou A, et al. Anti-tumor necrosis factor therapy improves insulin resistance, beta cell function and insulin signaling in active rheumatoid arthritis patients with high insulin resistance. Arthritis Res Ther [Internet]. 2012. June 12;14(3):R141. Available from: 10.1186/ar3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hançer NJ, Qiu W, Cherella C, Li Y, Copps KD, White MF. Insulin and metabolic stress stimulate multisite serine/threonine phosphorylation of insulin receptor substrate 1 and inhibit tyrosine phosphorylation. J Biol Chem [Internet]. 2014. May 2;289(18):12467–84. Available from: 10.1074/jbc.M114.554162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho RCM, Chua AC, Tran BX, Choo CC, Husain SF, Vu GT, et al. Factors Associated with the Risk of Developing Coronary Artery Disease in Medicated Patients with Major Depressive Disorder. Int J Environ Res Public Health [Internet]. 2018. September 21;15(10). Available from: 10.3390/ijerph15102073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstein BI, Carnethon MR, Matthews KA, McIntyre RS, Miller GE, Raghuveer G, et al. Major Depressive Disorder and Bipolar Disorder Predispose Youth to Accelerated Atherosclerosis and Early Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation [Internet]. 2015. September 8;132(10):965–86. Available from: 10.1161/CIR.0000000000000229 [DOI] [PubMed] [Google Scholar]
- 61.Rosenstiel P, Agnholt J, Kelsen J, Medici V, Waetzig GH, Seegert D, et al. Differential modulation of p38 mitogen activated protein kinase and STAT3 signalling pathways by infliximab and etanercept in intestinal T cells from patients with Crohn’s disease. Gut [Internet]. 2005. February;54(2):314–5; author reply 316–6. Available from: https://www.ncbi.nlm.nih.gov/pubmed/15647208 [PMC free article] [PubMed] [Google Scholar]
- 62.Waetzig GH, Seegert D, Rosenstiel P, Nikolaus S, Schreiber S. p38 mitogen-activated protein kinase is activated and linked to TNF-alpha signaling in inflammatory bowel disease. J Immunol [Internet]. 2002. May 15;168(10):5342–51. Available from: 10.4049/jimmunol.168.10.5342 [DOI] [PubMed] [Google Scholar]
- 63.Waetzig GH, Rosenstiel P, Nikolaus S, Seegert D, Schreiber S. Differential p38 mitogen-activated protein kinase target phosphorylation in responders and nonresponders to infliximab. Gastroenterology [Internet]. 2003. August;125(2):633–4; author reply 635–6. Available from: 10.1016/s0016-5085(03)00979-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.