Abstract
Accurate prediction of clinical scores (of neuropsychological tests) based on noninvasive structural magnetic resonance imaging (MRI) helps understand the pathological stage of dementia (e.g., Alzheimer’s disease (AD)) and forecast its progression. Existing machine/deep learning approaches typically preselect dementia-sensitive brain locations for MRI feature extraction and model construction, potentially leading to undesired heterogeneity between different stages and degraded prediction performance. Besides, these methods usually rely on prior anatomical knowledge (e.g., brain atlas) and time-consuming nonlinear registration for the preselection of brain locations, thereby ignoring individual-specific structural changes during dementia progression because all subjects share the same preselected brain regions. In this article, we propose a multitask weakly-supervised attention network (MWAN) for the joint regression of multiple clinical scores from baseline MRI scans. Three sequential components are included in MWAN: 1) a backbone fully convolutional network for extracting MRI features; 2) a weakly supervised dementia attention block for automatically identifying subject-specific discriminative brain locations; and 3) an attention-aware multitask regression block for jointly predicting multiple clinical scores. The proposed MWAN is an end-to-end and fully trainable deep learning model in which dementia-aware holistic feature learning and multitask regression model construction are integrated into a unified framework. Our MWAN method was evaluated on two public AD data sets for estimating clinical scores of mini-mental state examination (MMSE), clinical dementia rating sum of boxes (CDRSB), and AD assessment scale cognitive subscale (ADAS-Cog). Quantitative experimental results demonstrate that our method produces superior regression performance compared with state-of-the-art methods. Importantly, qualitative results indicate that the dementia-sensitive brain locations automatically identified by our MWAN method well retain individual specificities and are biologically meaningful.
Keywords: Clinical score prediction, convolutional neural networks (CNNs), dementia, structural magnetic resonance imaging (MRI), weakly supervised localization
I. Introduction
DEMENTIA is an overall term that describes a group of symptoms associated with a decline in memory or other thinking skills that are severe enough to reduce a person’s ability in daily activities. Alzheimer’s disease (AD) accounts for 60% to 80% of dementia cases, characterized by the progressive and irreversible loss of intellectual skills [1]. The prodromal stage of AD is called mild cognitive impairment (MCI), which can be further specified as progressive MCI (pMCI) and stable MCI (sMCI). Patients with pMCI will eventually progress to AD over time, while those with sMCI will remain stable with mild memory and cognitive decline [2]. Accurate disease diagnosis at the early stage of AD is thus of great clinical value, as it is the precondition for timely treatments to delay the disease progression. In clinical practice, various neuropsychological tests, e.g., mini-mental state examination (MMSE) [3], clinical dementia rating sum of boxes (CDRSB) [4], and AD assessment scale cognitive subscale (ADAS-Cog) [5], are commonly used to evaluate the dementia status and identify dementia-related behavioral and mental abnormalities. Considering that clinical scores of these neuropsychological tests are believed to be strongly correlated with the disease status [4], predicting them automatically and accurately is highly desired for comprehensively understanding the stage of dementia pathology and forecasting its future progression.
As a noninvasive imaging modality, structural magnetic resonance imaging (MRI) is sensitive to anatomical changes in the brain and has been routinely used in clinical practice for the assessment and diagnosis of patients with suspected dementia [6]. Using primarily MRI data, diverse computer-aided diagnosis (CAD) methods have also been proposed for automated dementia diagnosis and prognosis [7]–[9]. In contrast to the fact that the majority of these CAD methods were designed to identify category labels (e.g., AD/MCI) [7], relatively fewer of them contributed to clinical score prediction, potentially because regressing continuous variables is a much more challenging task in practice [8].
Many conventional machine learning methods [10]–[14] and, more recently, several deep learning methods [15]–[17] have been developed for MRI-based clinical score prediction. Conventional machine learning methods usually extract handcrafted MRI features (e.g., brain tissue density) for constructing regression models. Differently, most of the existing deep learning methods unify the steps of feature extraction and model construction, and thus, the resulting task-oriented features generally lead to better regression accuracy. As it is challenging to capture subtle disease-related changes in whole-brain MRI scans (especially at the early stage of dementia), existing CAD methods usually need to preselect brain locations that are potentially sensitive to dementia, e.g., according to anatomical prior knowledge. Such a requirement may have a negative influence on automated clinical score prediction, for which the main reason could be twofold. First, predefining brain locations isolated to the subsequent learning stage may lead to suboptimal prediction performance due to potential heterogeneity in two standalone stages. Second, due to the dependence on nonlinear registration, the preselection of brain locations is time-consuming in both the training and test phases. More importantly, the resulting locations may neglect subject-specific structural changes in disease progression because they are strictly consistent across all MRI scans. Even though a recently proposed deep network [18] can identify informative brain regions in a task-oriented manner, it still ignores subject-level specificities in dementia, due to the use of nonlinear registration for the proposal localization shared among all subjects.
In this article, we attempt to develop an end-to-end deep learning framework for automated detection of dementia-related anatomical abnormalities and unified regression of multiple clinical scores. Using directly the whole-brain MRI scan as input, a multi-task weakly-supervised attention network (MWAN) is proposed for this purpose, with the schematic presented in Fig. 1. Specifically, our MWAN model consists of three cascaded blocks: 1) a backbone fully convolutional network (FCN) to extract relatively high-dimensional feature maps; 2) a unique weakly-supervised dementia attention block to localize informative brain regions that indicate subject-specific dementia status; and 3) an attention-aware multitask regression block to jointly predict multiple clinical scores. To evaluate the effectiveness of our proposed method, we jointly predicted the clinical values of MMSE, CDRSB, and ADAS-Cog and conducted cross validation on two public data sets, i.e., Alzheimer’s Disease Neuroimaging Initiative-1 (ADNI-1) and ADNI-2 [19]. The quantitative experimental results demonstrate that our method has superior performance in clinical score regression compared with state-of-the-art methods. Also, qualitative results indicate that the dementia-sensitive brain locations automatically identified by our MWAN method well retain subject-level specificities and are biologically meaningful.
Fig. 1.
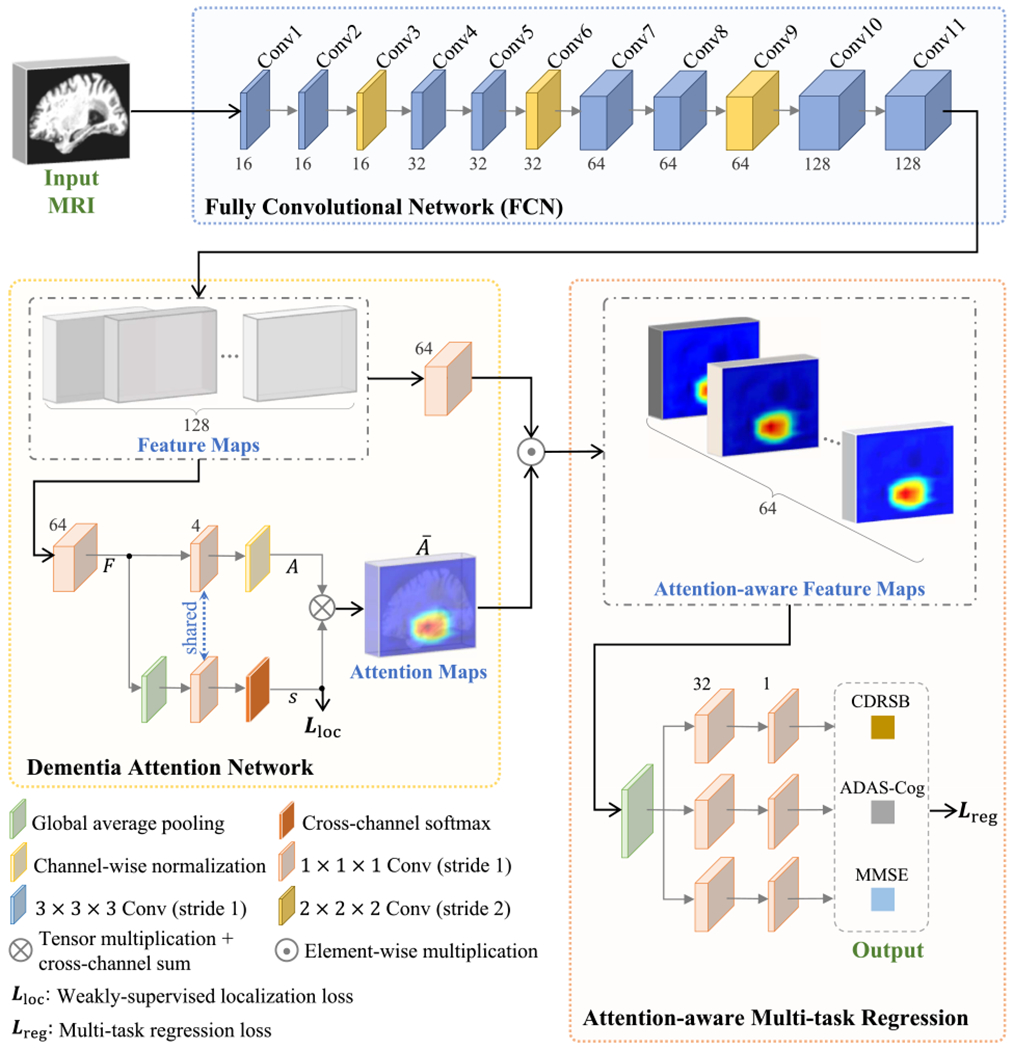
Schematic diagram of the proposed MWAN for joint regression of multiple clinical scores from whole-brain MRI scans. Our MWAN is end-to-end and fully trainable, which consists of a backbone FCN, a weakly-supervised dementia attention block, and an attention-ware multitask regression block. Here, F, A, s, and Ā denote the output of corresponding layers in the dementia attention block. Notably, s is first softmax-normalized for the multiplication with A.
Compared with existing CAD methods for dementia diagnosis/prognosis, the main contributions of the proposed method can be summarized in threefold.
Our MWAN integrates automated discriminative localization, feature extraction, and model construction into a unified framework, which explicitly encourages the homogeneity between different CAD components.
The proposed dementia attention block is task-oriented and fully trainable. Leveraging the comprehensive guidance provided by category labels and multiple clinical scores, it can automatically identify subject-specific dementia-related locations from the whole-brain MR image.
Our MWAN is efficient during inference. After network training, it consumes directly the whole-brain image for end-to-end prediction of multiple clinical scores, without the use of time-consuming nonlinear registration.
The remaining part of this article is organized as follows. In Section II, we briefly review related work on MRI-based dementia diagnosis/prognosis and automated discriminative localization in brain MRIs. In Section III, we introduce the studied data and the proposed method. In Section IV, we present the competing methods, experimental setup, and experimental results. In Section V, we discuss the effectiveness of different components of our method, as well as the limitations of our current work and possible future research directions. This article is finally concluded in Section VI.
II. Related Work
A. MRI-Based Dementia Diagnosis/Prognosis
Using primarily structural MRI data, diverse CAD methods have been proposed for category classification or clinical score prediction in dementia. Conventional learning methods usually extract handcrafted MRI features at the voxel level or region level for classification/regression model construction. For example, Stonnington et al. [10] extracted gray matter (GM) density map to construct relevance vector regression (RVR) model [20] for clinical score prediction. Wang et al. [11] extracted regional MRI features based on an adaptive regional clustering method [21] to construct ensemble RVR models for clinical score prediction. Zhang et al. [12] and Zhu et al. [14] parcellated the whole-brain image into multiple regions of interest (ROIs), based on which they further extracted regional features to construct support vector regression models to jointly predict MMSE and ADAS-Cog values, under the assumption that different clinical scores are inherently correlated. Considering that the early stage of dementia may only cause subtle structural changes while the whole-brain images are globally similar, several studies also proposed to extract features at the intermediate scale between voxels and coarse brain regions, i.e., from local image patches, for disease classification/regression. For example, Tong et al. [22] used a set of local MRI patches to construct a multiple-instance learning (MIL) model for AD classification and MCI conversion prediction. Zhang et al. [23] defined anatomical landmarks by groupwise statistical comparison, based on which they further located multiple MRI patches to construct SVM classifiers for AD diagnosis.
More recently, several deep learning methods, e.g., based on convolutional neural networks (CNNs) [24], have been proposed to learn task-oriented MRI features for category classification or clinical score prediction in dementia. For example, Zhang et al. [15] used pretrained CNN models to extract high-level features from predefined ROIs to construct a multitask dictionary learning model for the joint prediction of MMSE and ADAS-Cog scores. Li et al. [25] coarsely extracted the hippocampal regions from MRI scans to construct a residual network [26] for multicategory disease diagnosis. Based on anatomical landmarks [23], Liu et al. [17] extracted a set of MRI patches to construct a deep multitask multichannel learning (DM2L) model to jointly predict multiple clinical scores and identify category class labels, under the motivation that the classification task could provide additional guidance for the regression task. A systematic review of CNN-based AD diagnosis can be found in [27], which includes almost all existing methods for category classification while without attention on clinical score prediction.
Notably, the above-mentioned CAD methods typically rely on the preselection of brain locations in MRIs for classification and/or regression model construction. The reason is that directly analyzing the whole-brain MRI scans cannot be a reliable solution, considering that a volumetric MR scan usually contains millions of voxels and many of them are not relevant to dementia. However, brain locations independently predefined in such a way might not be well coordinated with both the stages of feature learning and learning model construction, thus potentially hampering the diagnostic performance.
B. Discriminative Localization in Brain MRIs
Detection and localization of (potentially) dementia-sensitive regions from the whole-brain MRI scans is a precondition for discriminative feature extraction and classification/regression model construction. Most of the existing CAD methods perform prelocalization empirically according to biological prior knowledge or anatomical brain atlases. For example, Fan et al. [21] proposed an adaptive regional clustering method to separate the whole-brain image as multiple parts in terms of local tissue volumetric measurements, based on which regional volumetric features were further extracted for MRI-based schizophrenia diagnosis [21] and clinical score prediction in dementia [11]. Using nonlinear registration algorithm [28], Zhang et al. [12] warped the anatomical automatic labeling (AAL) atlas onto each MRI scan to define multiple ROIs in terms of macroscopic brain structures, from which brain tissue volumes were quantified as features for joint AD diagnosis and clinical score prediction. Considering that the discriminative capacity of some brain regions (e.g., the hippocampus) have been reported by various clinical studies on dementia, several conventional learning methods [22] and deep learning methods [25] extracted local image patches from these regions for automated dementia diagnosis.
Apart from the utilization of prior knowledge, few methods were proposed to treat the preidentification of discriminative brain locations as a supervised learning task. For example, based on local morphological features, Zhang et al. [23] first performed a statistical group comparison between AD subjects and normal control (NC) subjects to identify brain locations (or anatomical landmarks) that have significant group difference. Then, regarding these brain locations on training data as the ground truth, they trained a shape-constrained random-forest regression model to automatically predict consistent brain locations on unseen subjects for AD diagnosis. Similarly but more precisely, a two-stage deep learning framework was proposed in [29], where a CNN was constructed in the first stage to regress the displacements from local image patches to predefined anatomical landmarks, and then, an FCN was designed in the second stage to jointly predict the coordinates of these anatomical landmarks, under the assumption that different landmarks could provide complementary information to each other. These supervised learning methods greatly improved the efficiency of CAD models in the inference phase, as they generally have no need of time-consuming nonlinear registration step for the localization of predefined brain locations in unseen MRI scans. However, the discriminative capacity of brain locations identified by these supervised learning methods is determined by the ground-truth predefinition in terms of statistical group comparison. In addition, discriminative localization in both those empirical methods (i.e., using biological prior knowledge) and supervised learning methods is isolated to CAD models, which may hamper the learning performance due to potential heterogeneity between different steps.
More recently, Lian et al. [18] proposed a hierarchical fully convolutional network (H-FCN) to perform task-oriented discriminative localization for dementia diagnosis. Using location proposals widely distributed over the whole-brain image as initial input, H-FCN adopts the category label as weakly-supervised guidance to hierarchically identify informative brain locations at both the patch and region levels and then seamlessly learns multiscale imaging features for AD diagnosis and MCI conversion prediction. However, since location proposals are anatomically consistent across all subjects, the multiscale discriminative locations identified by H-FCN ignore potential individual-level specificity of different subjects, e.g., the dementia-induced structural changes may occur at different brain locations or have different severity in certain brain regions for different subjects. Inspired by recent active research in the computer vision community on weakly-supervised discriminative localization [30], [31], in this work, we attempt to design an end-to-end deep learning model for learnable localization of subject-specific structural abnormalities in the whole-brain MRI and seamless prediction of multiple clinical scores.
III. Materials and Method
A. Data sets and Image Preprocessing
Two public data sets downloaded from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) were studied, i.e., ADNI-1 and ADNI-2 consisting of 1396 subjects with baseline structural MRI scans. Considering that several subjects that appear in ADNI-1 also appear in ADNI-2, we removed those subjects from ADNI-2 to ensure that these two data sets are independent. The baseline ADNI-1 and ADNI-2 data sets have 1.5T and 3T T1-weighted MR scans, respectively. The baseline ADNI-1 data set contains 797 subjects, including 226 NCs, 225 sMCI, 165 pMCI, and 181 AD patients. The baseline ADNI-2 data set contains 599 subjects, including 185 NC, 234 sMCI, 37 pMCI, and 143 AD. Three types of neuropsychological test scores were downloaded for ADNI-1 and ADNI-2, including CDRSB, ADAS-Cog, and MMSE. The studied data sets are complete, i.e., each subject has the baseline clinical scores for all neuropsychological tests. The definition of pMCI/sMCI is based on whether MCI would convert to AD within 36 months after the baseline evaluation. The demographic and clinical information of the data sets is summarized in Table I.
TABLE I.
Demographic Information of the Studied Data Sets (i.e., the Baseline ADNI-1 and ADNI-2). The Gender Is Reported as Male/Female (i.e., M/F). The Age (in Years), Education Years, and Clinical Scores (i.e., CDRSB, ADAS-Cog, and MMSE) Are Reported in Terms of Mean ± Standard Deviation
| Dataset | Category | Age | Education Years | Gender (M/F) | Clinical Scores |
||
|---|---|---|---|---|---|---|---|
| CDRSB | ADAS-Cog | MMSE | |||||
| ADNI-1 | NC | 75.85±5.03 | 16.05±2.87 | 118/108 | 0.03±0.12 | 6.22±2.92 | 29.11±1.00 |
| sMCI | 74.87±7.64 | 15.55±3.18 | 151/74 | 1.42±0.78 | 10.35±4.30 | 27.28±1.77 | |
| pMCI | 74.82±6.83 | 15.67±2.85 | 101/64 | 1.85±0.94 | 13.27±4.06 | 26.58±1.71 | |
| AD | 75.30±7.50 | 14.72±3.14 | 94/87 | 4.34±1.61 | 18.58±6.25 | 23.30±1.99 | |
|
| |||||||
| ADNI-2 | NC | 73.47±6.25 | 16.51±2.54 | 88/97 | 0.05±0.23 | 5.78±3.08 | 29.03±1.27 |
| sMCI | 71.66±7.56 | 16.20±2.69 | 123/111 | 1.20±0.78 | 8.22±3.59 | 28.25±1.62 | |
| pMCI | 71.27±7.28 | 16.24±2.67 | 21/16 | 2.24±1.26 | 11.81±4.41 | 26.97±1.66 | |
| AD | 74.24±7.99 | 15.86±2.60 | 85/58 | 4.43±1.75 | 20.73±7.28 | 23.16±2.21 | |
All brain MR images were processed following a standard pipeline, that is, the anterior commissure (AC)-posterior commissure (PC) correction was first performed using the MIPAV software.1 After that, we corrected the intensity of each brain image using the N3 algorithm [32]. The brain skull and dura were stripped by the BET method in the FSL package [33]. The cerebellum was further removed by warping a labeled template to each skull-stripped image. Using the FLIRT method in the FSL package [33], all MR images were linearly aligned onto the Colin27 template [34] to remove global linear difference and also to resample all images for having the same spatial resolution (i.e., 1×1×1 mm3). Finally, we cropped all linearly aligned images to have the same size of 144 × 184 × 152, and these cropped images will be used as the input for our proposed method. It is worth mentioning that, since the MR images of different subjects were linearly aligned and have the same size, they were straightforwardly cropped with the same margin, under the requirement that all brain tissues should be completely preserved without loss of any potentially useful information.
B. Multi-Task Weakly-Supervised Attention Network
Using the whole-brain MRI scan (size: 144 × 184 × 152) as input, we propose an MWAN, an end-to-end and fully trainable deep architecture, to jointly predict multiple clinical scores. In a task-oriented manner, our MWAN can automatically identify subject-specific discriminative locations from the whole-brain image and seamlessly learn high-level MRI feature representations to construct a multitask regression model. As shown in Fig. 1, our MWAN consists of a backbone FCN, a trainable dementia attention block, and an attention-aware multitask regression block.
1). Backbone:
The backbone employs an FCN architecture to generate feature maps (with considerable spatial resolution) for capturing global information from the whole-brain MRI scan. Specifically, in our current implementation, the backbone FCN consists of 11 convolutional (Conv) layers, where eight layers use 3 × 3 × 3 kernels, while the other three layers adopt 2 × 2 × 2 kernels, without additional pooling operations. The stride for these 3 × 3 × 3 kernels is set as 1, whereas the stride for the 2 × 2 × 2 kernels is set as 2. All these kernels are with zero padding, followed by batch normalization (BN) and rectified linear unit (ReLU) activation. The 2 × 2 × 2 Conv layers (i.e., Conv3, Conv6, and Conv9) are placed after 3×3×3 Conv layers (i.e., Conv2, Conv5, and Conv8) to downsample feature maps and increase receptive fields. The numbers of channels for the 11 Conv layers are 16, 16, 16, 32, 32, 32, 64, 64, 64, 128, and 128, respectively. Given an input whole-brain MR image with the size of W × H × L, such a backbone produces a set of 128 features maps (i.e., the output of Conv11) and each with the size of (W/8) × (H/8) × (L/8). It is worth noting that our backbone FCN is designed to be lightweight, especially considering the limited number (e.g., hundreds) of training samples in our task. Also, as a plug-in unit, this fundamental backbone can be easily replaced by any other more advanced FCN architectures (e.g., residual or dense blocks).
2). Dementia Attention Module:
The clinical scores of neuropsychological tests are believed to be intrinsically correlated with category labels in dementia (i.e., NC, sMCI, pMCI, or AD), considering that they point to semantically similar targets from multiple complementary views [8]. The demographic and clinical information presented in Table I also supports this assumption since subjects from different groups have different mean scores in ADNI1 and ADNI2. Notably, based on such assumption, some conventional learning methods [12], [14] and several deep learning methods [17] have been proposed to perform joint category classification and clinical score estimation, by extracting/learning shared MRI features for both tasks. Different from these CAD methods for joint classification and regression, in this article, we leverage primarily the imagewise category labels as weakly-supervised guidance to design a trainable dementia attention module for the automated identification of discriminative brain locations that are strongly relevant to subject-specific dementia status. The resulting dementia attention maps will be further used to assist the construction of a multitask model for the joint regression of multiple clinical scores.
Our dementia attention module is built upon the output of the backbone FCN, which is inspired by [30] and [31] but operates distinctively in another fully trainable way. As shown in Fig. 1, in the proposed dementia attention network, we first employ an 1 × 1 × 1 Conv layer with 64 channels to squeeze the Conv11 feature maps from the backbone. Denote the squeezed feature maps as F = [F1, …, FM], where (m = 1, …, M) is the feature map [size: (W/8) × (H/8) × (L/8)] at the mth channel and M = 64 is the number of channels. A global average pooling (GAP) layer is then applied on F to produce a holistic feature representation capturing the semantic information from the whole-brain image. After that, both the feature representation f and the squeezed feature map F are further mapped by another 1×1×1 Conv layer (without bias) onto the category label space (i.e., with C = 4 units for the Conv layer). Here, the 1 × 1 × 1 Convs for f and F share the same set of learnable weights denoted as w = [w1, …, wC], with (c = 1, …, C), and the corresponding outputs are s = [s1, …, sC] and A = [A1, …, AC], respectively.
Notably, sharing the 1 × 1 × 1 Conv layer between f and F ensures that the resulting attention maps A being dementia-status specific, as A and s are concurrently optimized under the supervision of the respective category label, that is, since implies the individual score for a given subject belonging to the cth category, we can expect that the corresponding actually describes the spatially varying contributions of different brain locations in quantifying subject-specific dementia status related to sc. Specifically, due to the GAP operation, we have f ∝ [∑x,y,z F1(x, y, z), …, ∑x,y,z FM(x, y, z)]T, based on which we can further infer that
| (1) |
where is spatially varying and proportional to the probability of the given subject belonging to the cth category. According to the above relation, we finally quantify the subject-specific dementia attention maps by aggregating s and A, with the form of
| (2) |
where denotes the softmax normalization of classification scores and and are the minimum and maximum elements in Ac, respectively. By jointly training the dementia attention block with other parts of MWAN, the resulting Ā in (2) will highlight discriminative brain regions that are strongly relevant to subject-specific dementia status.
3). Attention-Aware Multitask Regression Module:
Using Ā produced by the dementia attention module as spatial guidance, a multitask regression block is further designed to jointly predict multiple clinical scores, including CDRSB, ADAS-Cog, and MMSE. Specifically, we first adopt an 1 × 1 × 1 Conv layer with M = 64 channels to squeeze the feature maps generated by the backbone FCN (i.e., the output of Conv11). After that, the squeezed feature maps are then elementwisely weighted by Ā for each channel to enhance the influence of features extracted from discriminative brain locations (i.e., with large weights in Ā) and suppress the contribution of features extracted from brain regions that are not relevant to dementia (i.e., with small weights or even zero weight in Ā). The spatially weighted feature maps are then processed by a GAP layer to obtain the attention-aware holistic feature representation (with 64 elements) describing the semantic information of the whole-brain image. Finally, for each clinical score (or regression task), two successive fully connected (FC) layers, with 32 and 1 unit(s), respectively, are applied on the attention-aware feature representation to predict its value.
4). End-to-End Implementation:
Using the whole-brain MRI scan as input, our MWAN model is designed for end-to-end discriminative localization and clinical score regression. The learnable parameters for different modules can be jointly optimized by minimizing a hybrid loss function, denoted as
| (3) |
where α > 0 is a tuning parameter that balances the contributions of two terms and . As shown in Fig. 1, and are the weakly-supervised localization loss and the multitask regression loss, respectively. The parameter α was empirically set as 0.01 in our experiments.
Let be a training set containing N samples, where Xn is the whole-brain MRI scan for the nth subject, yn ∈ {1, …, C} is the corresponding class label, and denotes T types of clinical scores. We assume that Wfcn, Wloc, and Wreg are the learnable parameters for the backbone FCN, the dementia attention module, and the attention-aware multitask regression module, respectively. Let be the softmax-normalized score produced by the dementia attention block. Considering that is a function in terms of Xn given Wfcn and Wloc, i.e., , we thus define as
| (4) |
which is the categorical cross-entropy loss and 1(·) is a binary indicator. On the other hand, let denote the predicted clinical scores yielded by the multitask regression module. As it is a function in terms of Xn given Wfcn, Wloc, and Wreg, i.e., , we can represent as
| (5) |
which is the mean squared error. According to the definition of and in (4) and (5), it is worth noting that both the localization and regression losses are backpropagated to optimize the backbone FCN. In addition, the regression loss is also merged into the dementia attention block to assist its training, implying that the discriminative localization in our proposed method is primarily determined by the category label along with auxiliary guidance provided by complementary views of the clinical scores.
Our MWAN model was implemented using Python based on Keras and Tensorflow backend. The input was the linearly aligned MRI scans (size: 144 × 184 × 152), and our network was constructed for end-to-end regression of T = 3 clinical scores (i.e., CDRSB, ADAS-Cog, and MMSE). At the training stage, four (C = 4) category labels (i.e., NC, sMCI, pMCI, and AD) are used for weakly supervised dementia attention detection. The training set was augmented online by randomly rescaling brain images in a small range and flipping them in the axial plane. The models were trained with the Adam optimizer (learning rate: 0.001; mini-batch size: 2; dropout for Conv layers: 0.5) for 100 epochs in total. Each epoch took around 320 s on the Titan Xp 12 GB graphics card.
IV. Experiments
A. Competing Methods
The clinical score regression performance of our proposed MWAN method was compared with three conventional learning methods using handcrafted features defined at different scales: 1) voxel-based morphometry (VBM) [35]; 2) ROI-based pattern analysis (ROI) [12]; and 3) anatomical landmark-based morphometry (LBM) [23]. Furthermore, it was also compared with three deep learning methods, including a standard CNN trained for single score regression (CNN-S), 5) CNN trained to jointly regress multiple scores (CNN-M), and a state-of-the-art method, i.e., DM2L method [17]. These competing methods are briefly summarized as follows.
VBM: In the VBM-based method, each brain MR image was first aligned onto the AAL template via nonlinear registration [36]. After that, the voxelwise GM density map was extracted as features from each brain image, followed by a feature selection operation based on t-test to select informative features for dimensionality reduction. Finally, using the selected voxelwise features, multiple support vector regressors (SVRs) were trained separately for the regression of each clinical score.
ROI: In line with [12], we segmented each brain MR image into three different tissue types, including GM, white matter (WM), and cerebrospinal fluid (CSF) [37]. On the other hand, we warped the AAL template (with 90 predefined ROIs in cortical and subcortical regions) into the native space of each subject using nonlinearly registration [36], [38], based on which the normalized GM volumes in the 90 ROIs were quantified as features. Finally, using such regional features, we trained an SVR for the regression of each clinical score independently.
LBM: In the LBM method, K = 50 anatomical landmarks [23] were used to locate K 3-D patches (size: 24×24×24) from the whole-brain MR image. From each patch, 100-D morphological features were extracted, and the features for different patches were concatenated as a 100k-D vector. Finally, using these patch-level features, independent SVRs were trained to predict the three clinical scores separately.
CNN-S: In the CNN-S method, the network architecture was designed by attaching a GAP layer and an FC layer onto the backbone FCN shown in Fig. 1. Then, we trained an independent model for the prediction of each clinical score using the mean-squared-error (MSE) loss.
CNN-M: The CNN-M method is similar to CNN-S, except that only one model was trained to predict all clinical scores jointly by attaching parallel FC layers after the GAP layer.
DM2L: The DM2L method constructed a deep learning model on the same set of K local patches that have been used in the LBM method. Notably, the DM2L method utilized the identification of category labels as an auxiliary task to assist the prediction of clinical scores, and multiple clinical scores were jointly predicted, under the assumption that clinical scores and category labels have semantically consistent meaning. Therefore, a multitask multibranch CNN was finally constructed, where each patch was processed by a specific subnetwork to learn patchwise features, and then, all patchwise features were merged to learn higher level feature for joint regression and classification.
Apart from clinical score regression, we also visually compared the discriminative localization performance of our proposed dementia attention module with that of a state-of-the-art weakly supervised method, i.e., the class activation map (CAM) method [30]. It is worth noting that, different from our trainable dementia attention module, CAM is an offline method, i.e., it computes the CAM after the network is completely trained. Specifically, using the whole-brain MR image as the input, we first trained the backbone FCN (as shown in Fig. 1) for category classification by minimizing the cross-entropy loss, i.e., (4). After that, the CAM method was called to produce the CAM for each subject based on the weights of the classification layer and the Conv11 feature maps.
B. Experimental Setup
Considering that training and test on independent neuroimaging data sets could relatively more objectively analyze the generalization capacity of a prediction model [27], [39], [40], we performed data setwise twofold cross validation on ADNI-1 and ADNI-2 for the evaluation of the automated regression and discriminative localization performance. Specifically, in the first iteration, we trained the CAD models on ADNI-1, by randomly splitting 15% subjects for validation, and then evaluated the trained model on ADNI-2. In the second iteration, we reversed the training and test sets (i.e., using ADNI-2 for training and ADNI-1 for test) and randomly split 10% subjects from ADNI-2 for validation, considering that the ADNI-2 data set is relatively small. The performance of score regression was quantitatively evaluated by two metrics: 1) the correlation coefficient (CC) and 2) the root mean square error (RMSE) between the ground-truth scores and the clinical scores predicted by each automated method. Let be a set of the ground-truth clinical scores for N test samples, and are the corresponding prediction. The CC is defined as
| (6) |
where z* and denote the mean values for z and , respectively. The RMSE can be written as
| (7) |
C. Results of Clinical Score Regression
The quantitative performance of the five methods on predicting CDRSB, ADAS-Cog, and MMSE scores is summarized in Tables II and III. Note that Table II presents the results obtained by the models trained on ADNI-1 and tested on ADNI-2, and Table III reports the results produced by the models trained on ADNI-2 and tested on ADNI-1. From Tables II and III, we have the following observations.
TABLE II.
Regression Results in Terms of CC and RMSE on ADNI-2 Obtained by Models Trained on ADNI-1
| Method | CDRSB |
ADAS-Cog |
MMSE |
|||
|---|---|---|---|---|---|---|
| CC | RMSE | CC | RMSE | CC | RMSE | |
| VBM | 0.278 | 2.010 | 0.290 | 7.406 | 0.289 | 2.889 |
| ROI | 0.380 | 1.893 | 0.360 | 7.358 | 0.325 | 2.899 |
| LBM | 0.431 | 1.772 | 0.527 | 6.245 | 0.331 | 2.754 |
| CNN-S | 0.373 | 1.777 | 0.511 | 6.671 | 0.447 | 2.591 |
| CNN-M | 0.440 | 1.776 | 0.517 | 6.614 | 0.464 | 2.527 |
| DM2L | 0.533 | 1.666 | 0.565 | 6.200 | 0.567 | 2.373 |
| MWAN (Ours) | 0.621 | 1.503 | 0.648 | 5.701 | 0.613 | 2.244 |
TABLE III.
Regression Results in Terms of CC and RMSE on ADNI-1 Obtained by Models Trained on ADNI-2
| Method | CDRSB |
ADAS-Cog |
MMSE |
|||
|---|---|---|---|---|---|---|
| CC | RMSE | CC | RMSE | CC | RMSE | |
| VBM | 0.197 | 1.851 | 0.146 | 6.382 | 0.208 | 2.685 |
| ROI | 0.190 | 2.024 | 0.205 | 6.507 | 0.211 | 2.710 |
| LBM | 0.417 | 1.922 | 0.512 | 5.835 | 0.435 | 2.664 |
| CNN-S | 0.436 | 1.762 | 0.434 | 6.447 | 0.453 | 2.550 |
| CNN-M | 0.461 | 1.747 | 0.486 | 5.990 | 0.401 | 2.482 |
| DM2L | 0.468 | 1.628 | 0.580 | 5.426 | 0.502 | 2.428 |
| MWAN (Ours) | 0.564 | 1.569 | 0.611 | 5.525 | 0.532 | 2.414 |
First, the deep learning methods (e.g., CNN-M, the state-of-the-art DM2L, and our MWAN) generally led to better regression results than the conventional learning methods (i.e., VBM, ROI, and LBM) on both ADNI-1 and ADNI-2. For example, compared with ROI and LBM for predicting CDRSB on ADNI-2 (Table II), our MWAN resulted in 24.1% and 19.0% improvements in terms of CC, respectively. Compared with VBM for predicting ADAS-Cog and MMSE on ADNI1 (Table III), our MWAN improved the RMSE from 6.382 and 2.685 to 5.525 and 2.414, respectively. These results suggest that learning task-oriented MRI features instead of handcrafted features is beneficial for the task of automated clinical score regression.
Second, CNN-M, the state-of-the-art DM2L, and our MWAN yielded overall better regression performance than CNN-S. For example, compared with CNN-S for predicting MMSE on ADNI-2 (Table II), CNN-M and our MWAN reduced the RMSE from 2.591 to 2.527 and 2.244, respectively. one potential reason could be that jointly predicting multiple clinical scores can bring complementary information for each other, considering that they are inherently associated with the same dementia status.
Besides, our MWAN and the state-of-the-art DM2L methods consistently outperformed the other two fundamental CNN-based methods (i.e., CNN-S and CNN-M) on both data sets. For instance, compared with CNN-M on ADNI-2 (Table II), our MWAN yielded 18.1%, 13.1%, and 14.9% improvements in terms of CC for predicting CDRSB, ADAS-Cog, and MMSE, respectively. It indicates that, without explicit localization of discriminative brain locations using prior-knowledge (in DM2L) or automatically (in our MWAN), developing deep learning models from scratch (i.e., using the whole-brain MRI scans) for dementia diagnosis is challenging in practice. As has been discussed, the main reason could be that the early stage of dementia may only cause subtle structural changes in a whole-brain MRI scan, while millions of voxels exist in each brain MRI. Also, our MWAN method consistently outperformed the state-of-the-art DM2L on both data sets. This implies that performing task-oriented discriminative localization in an end-to-end framework, as we do in MWAN, is desired in the task of clinical score regression.
Finally, we can also observe that, compared with the results on ADNI-2 (Table II), all CAD methods had worse performance on ADNI-1 (Table III). The possible reason is that the models applied in the second case were trained with less (and unbalanced) subjects from ADNI-2. Notably, our MWAN still led to the best results in such a difficult case, implying its robustness in MRI-based clinical score regression.
D. Results of Discriminative Localization
The discriminative localization performance of our MWAN was compared with the offline CAM method [30]. We show several representative dementia attention maps achieved by MWAN and CAM in Fig. 2, where six AD subjects from ADNI-2 are randomly selected and the corresponding models were trained on ADNI-1.
Fig. 2.
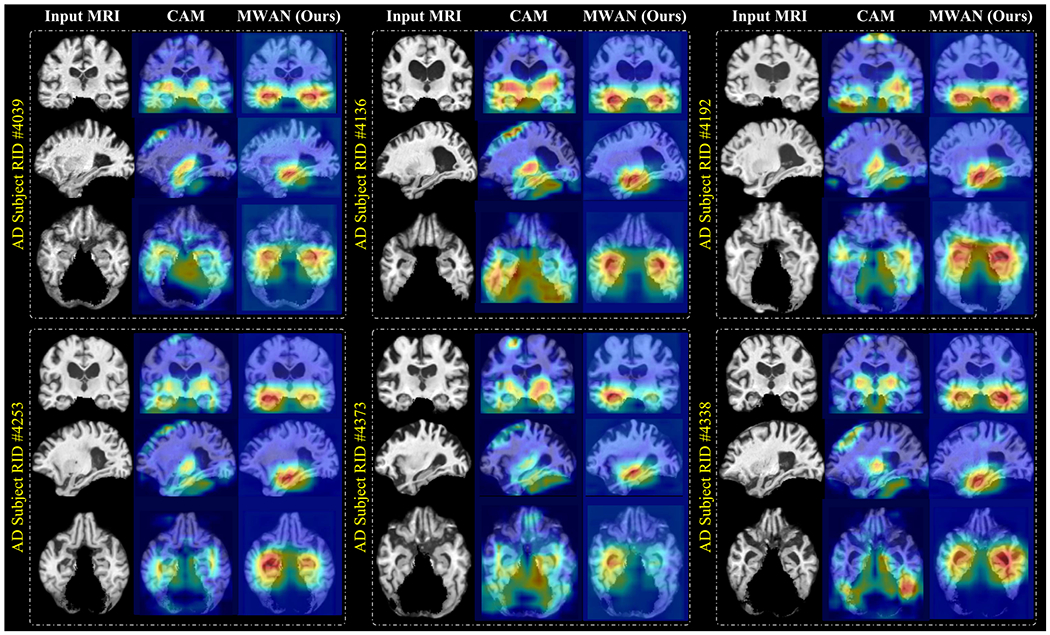
Representative dementia attention maps predicted by the offline CAM method [30] and our end-to-end MWAN method, respectively, for six different AD subjects from ADNI-2, using the models trained on ADNI-1. Red and blue denote high and low discriminative power, respectively.
From Fig. 2, we can see that, compared with the offline CAM method, our end-to-end MWAN can generate biologically more reasonable attention maps, which highlight the subcortical regions that control the memory and decisionmaking in the human brain, while with less emphases on uninformative brain locations, e.g., the boundaries between the brain and (removed) cerebellum. It is also worth mentioning that the highlighted subcortical regions include the hippocampus and amygdala, which is consistent with previous clinical studies that have validated the discriminative capacity of these brain regions for dementia diagnosis. These results suggest that leveraging multiple guidance (i.e., both category labels and clinical scores) to learn the dementia attention maps in a fully trainable framework, such as in our proposed method, can more precisely localize discriminative brain regions for automated clinical score regression in dementia.
V. Discussion
An MWAN is proposed in this work for end-to-end clinical scores prediction from the whole-brain MRI scans. The proposed method has two important components, i.e., a fully trainable dementia attention module and a multitask regression module. In this section, we first perform ablation studies to further analyze the effectiveness of these two modules and the associated hybrid loss function. After that, we discuss the additional role of the attention maps generated by our method in describing subject-specific dementia status, followed by the discussion on the relationship between our dementia attention module and other learnable attention mechanisms in the literature. Finally, we discuss the limitations of our current work and possible future research directions.
A. Effectiveness of Dementia Attention Module
To further evaluate the influence of the dementia attention module, we performed an ablation study by comparing our MWAN with its variant without discriminative localization, called multitask network (MTN). Based on the same backbone FCN but without automated discriminative localization, MTN jointly predicts multiple clinical scores and also includes category classification as an auxiliary task (as in DM2L and MWAN). The quantitative regression performance of MTN was compared with DM2L and MWAN on both ADNI-1 and ADNI-2, with the results summarized in Fig. 3. From Fig. 3, we can see that both DM2L and MWAN consistently led to better results than MTN for the joint regression of multiple clinical scores on ADNI-1 and ADNI-2. It indicates that discriminative localization is an essential step in developing CAD models for dementia diagnosis. Considering that our MWAN has overall the best performance, these results also suggest the effectiveness of our fully learnable dementia attention module.
Fig. 3.
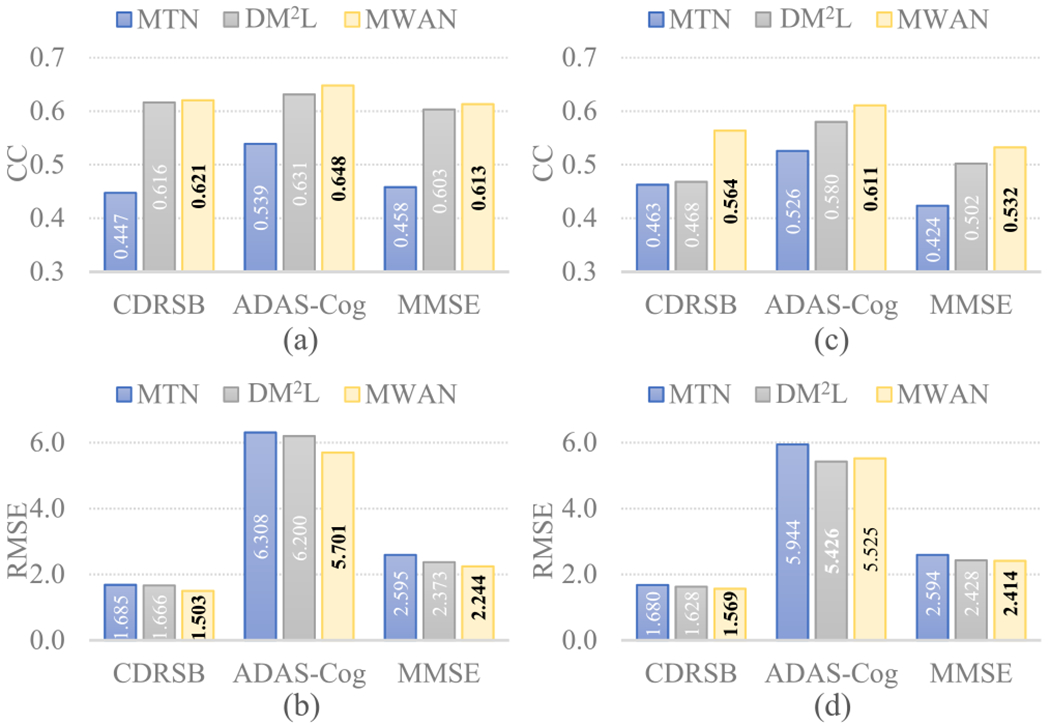
Clinical score regression results (in terms of CC and RMSE) obtained by MTN (a variant of our MWAN without discriminative localization), DM2L, and MWAN. (a) CC and (b) RMSE quantified on ADNI-2, respectively, using the models trained on ADNI-1. (c) CC and (d) RMSE quantified on ADNI-1, respectively, using the models trained on ADNI-2.
To analyze in more detail if the dementia attention module benefits the estimation of clinical scores for patients with varying status, we separated the subjects from ADNI-2 into two groups: 1) AD versus NC and 2) pMCI versus sMCI. On each group, we compared MTN with MWAN. The corresponding results (in terms of CC) are summarized in Table IV, from which we have the following two observations. First, MWAN consistently outperformed MTN in each case, implying that our dementia attention module can capture subject-specific information to improve the regression performance. Second, the results for the AD versus NC group are much better than those for the pMCI versus sMCI group, indicating that accurately estimating clinical scores is challenging but practically meaningful for understanding the early stage of dementia.
TABLE IV.
CC Values for the Subjects From Two Different Groups (i.e., AD versus NC and pMCI versus sMCI). The Results Were Quantified on ADNI-2 Using the Models Trained on ADNI-1
| Group | Method | CDRSB | ADAS-Cog | MMSE |
|---|---|---|---|---|
| AD vs. NC | MTN | 0.523 | 0.571 | 0.537 |
| MWAN | 0.731 | 0.705 | 0.727 | |
|
| ||||
| pMCI vs. sMCI | MTN | 0.258 | 0.421 | 0.330 |
| MWAN | 0.388 | 0.537 | 0.391 | |
B. Effectiveness of Multi-Task Learning
To further evaluate the benefits brought by the multi-task attention-aware regression module, we constructed a variant of our MWAN, called MWAN-S, that predicts each clinical score independently. Specifically, MWAN-S employs a similar architecture with MWAN, except that the attention-aware regression module only contains one branch to predict a single clinical score. On both ADNI-1 and ADNI-2, we compared MWAN-S with MWAN, with the quantitative results in terms of both CC and RMSE summarized in Fig. 4, where Fig. 4(a) and (b) and (c) and (d) corresponds to the results on ADNI-2 (trained on ADNI-1) and ADNI-1 (trained on ADNI-2), respectively. It can be observed that MWAN outperformed MWAN-S on both data sets, and the improvement is more significant in Fig. 4(b), where the task is more challenging because ADNI-2 is relatively smaller and more imbalanced. These results imply that jointly predicting multiple clinical scores, as we do in our multitask attention-aware regression module, could effectively improve the regression performance, as different neuropsychological tests describe the dementia status from complementary views.
Fig. 4.
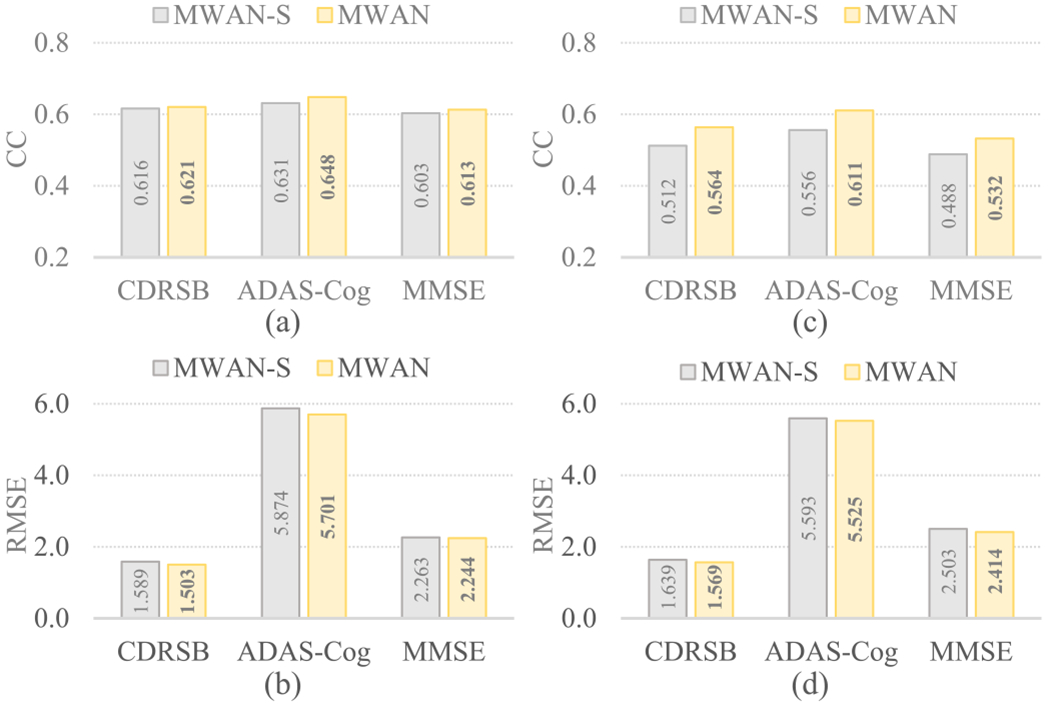
Regression results on (a) and (b) ADNI-2 and (c) and (d) ADNI-1 produced by MWAN-S and MWAN, respectively, where MWAN-S (a variant of our MWAN) predicts each clinical score independently.
C. Training With Hybrid Loss
The experiments in Section IV-D show that the proposed dementia attention module performed well in detecting discriminative brain locations. Besides, the discussion in Section V-A suggests that it can effectively improve the accuracy of the subsequent multitask regression module. As a part of an end-to-end deep network, our dementia attention module was trained concurrently with other network components via minimizing a hybrid loss function (3), that is, it was primarily determined by the category-label supervision, i.e., the weakly-supervised localization loss (4), along with the auxiliary assistance backpropagated from the subsequent multitask regression module, i.e., the regression loss (5). To further check the effectiveness of such a hybrid loss, we implemented the following variants for comparison.
First, to evaluate the primary role of the localization loss (as well as the dementia attention module) in discriminative localization, we implemented two variants: 1) setting α = 0, i.e., using solely to train the network (denoted as w/o ), and 2) moving to the end of the network for joint classification and score regression (denoted as JCR). Based on the models trained on ADNI-1 and applied to ADNI-2, we compared the representative attentions maps produced by these variants in Fig. 5 and summarized the corresponding regression results in Fig. 6. We have two observations from Fig. 5: 1) without the category-label supervision, w/o cannot precisely localize biologically meaningful brain regions (e.g., hippocampus) associated with dementia, and 2) without direct supervision on the dementia attention module, JCR generated anatomically more unconcentrated attention maps than the original hybrid loss. These observations suggest the primary role of for training the dementia attention module. The regression results presented in Fig. 6 also support this argument, as the dementia attention module trained with the original hybrid loss consistently outperformed both w/o and JCR.
Fig. 5.
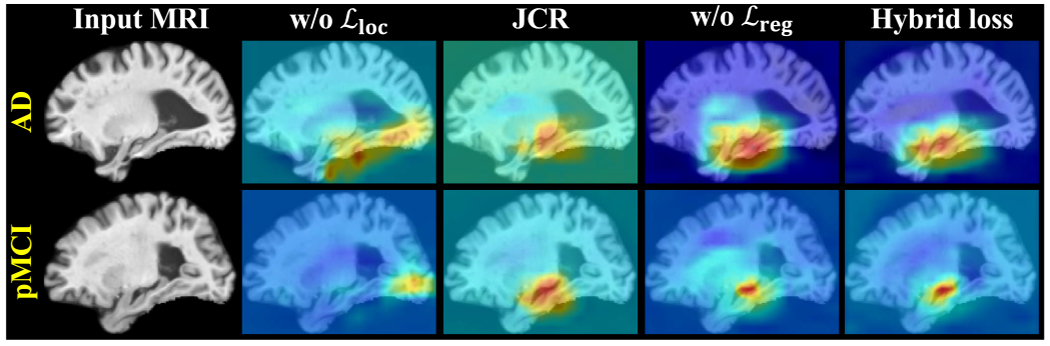
Representative dementia attention maps predicted by using different variants of the hybrid loss for two ADNI-2 subjects with AD and pMCI, respectively, where w/o denotes removing the localization loss, JCR denotes moving the localization loss to the end of MWAN for joint classification and regression, and w/o denotes removing the regression loss.
Fig. 6.
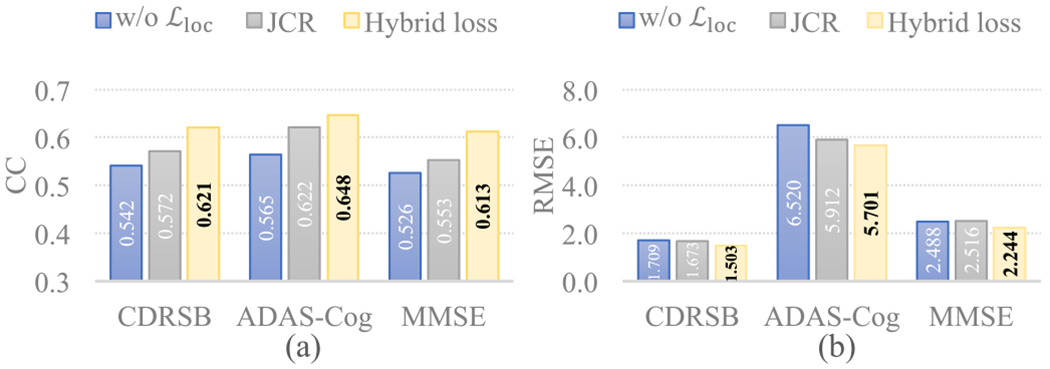
Regression results in terms of (a) CC and (b) RMSE on ADNI-2 obtained by our MWAN trained with different variants of the hybrid loss, where w/o denotes removing the localization loss and JCR denotes moving the localization loss to the end of MWAN for joint classification and regression.
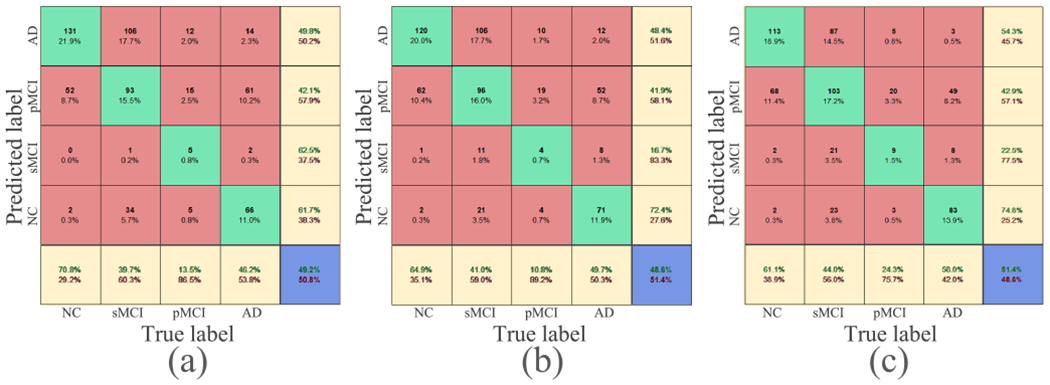
Second, to evaluate whether and the attention-aware multi-task regression module could provide auxiliary guidance to assist the training of the dementia attention module, we implemented another variant of MWAN, for which the regression loss was removed during training (denoted as w/o ). From Fig. 5, we can see that, although more precise than w/o and JCR, the attention maps produced by w/o relatively highlighted more uninformative regions than the original hybrid loss in both the cases of pMCI and AD. As a supplementary, we also compared the classification accuracy (NC versus sMCI versus pMCI versus AD) between the original hybrid loss, w/o , and JCR, with the respective confusion matrices shown in Fig. 7. We can observe from Fig. 7 that, using as an assistance, the dementia attention module could also more precisely differentiate between subjects with different category labels. For example, the overall accuracy for the original hybrid loss is 0.514, which is better than both w/o (0.492) and JCR (0.486). All these results presented in Figs. 5 and 7 show that the multi-task regression module could assist the training of the preceding attention module via backpropagating the loss quantified by .
Fig. 7.
Confusion matrices for multi-class classification (NC versus sMCI versus pMCI versus AD) on ADNI-2 obtained by different variants of the hybrid loss. (a) Removing the regression loss (i.e., w/o ). (b) Moving the localization loss to the end of MWAN for joint classification and regression (i.e., JCR). (c) Original hybrid loss.
D. Dementia Status Revealed by Attention Maps
The dementia attention maps produced by our end-to-end MWAN method could potentially provide additional information to describe subject-specific dementia status. In Fig. 8, we show the MRI scans of four subjects with different category labels (i.e., NC, sMCI, pMCI, and AD) and their corresponding attention maps generated by our MWAN method. It can be observed from Fig. 8 that the attention map for the NC subject nearly highlights all spatial locations with heat values (i.e., red color), indicating that there is no apparent difference between different brain locations in identifying NC subjects, i.e., there are no structural abnormalities in the brain. As the disease progresses from NC to AD, we can see that the heat values (i.e., red color) in the attention maps are gradually decreased (i.e., changed to blue) at most brain locations, while they are eventually accumulated at the hippocampal regions. These results suggest that, by uncovering the potentially gradual atrophic process of the human brain due to dementia, our MWAN method could provide additional information regarding the disease progression based on the generated attention maps. The visual evidence provided by attention maps may facilitate clinicians to fast determine the stage of dementia and the disease progression in clinical practice.
Fig. 8.
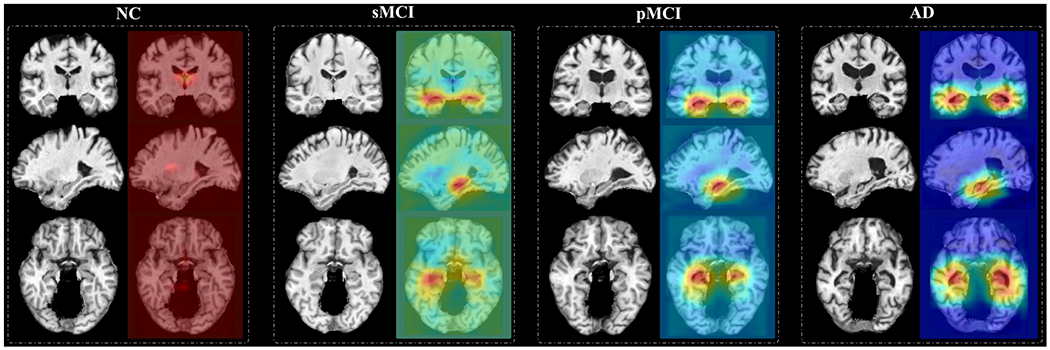
Attention maps produced by our MWAN for four subjects with different category labels (i.e., NC, sMCI, pMCI, and AD), respectively. The attention map for each subject is shown in three views, where red and blue denote high and low discriminative power, respectively.
The subject-specific dementia status revealed by our MWAN method also happens for different subjects from the same clinical category. In Fig. 9, we show the attention maps for 12 patients from the clinical categories of sMCI, pMCI, and AD. It can be seen that, for the subjects from the same category (e.g., the four AD subjects shown in the last row), different brain locations (e.g., the left subcortical region, the right subcortical region, or both) are highlighted by the respective attention maps with spatially different heat values.
Fig. 9.
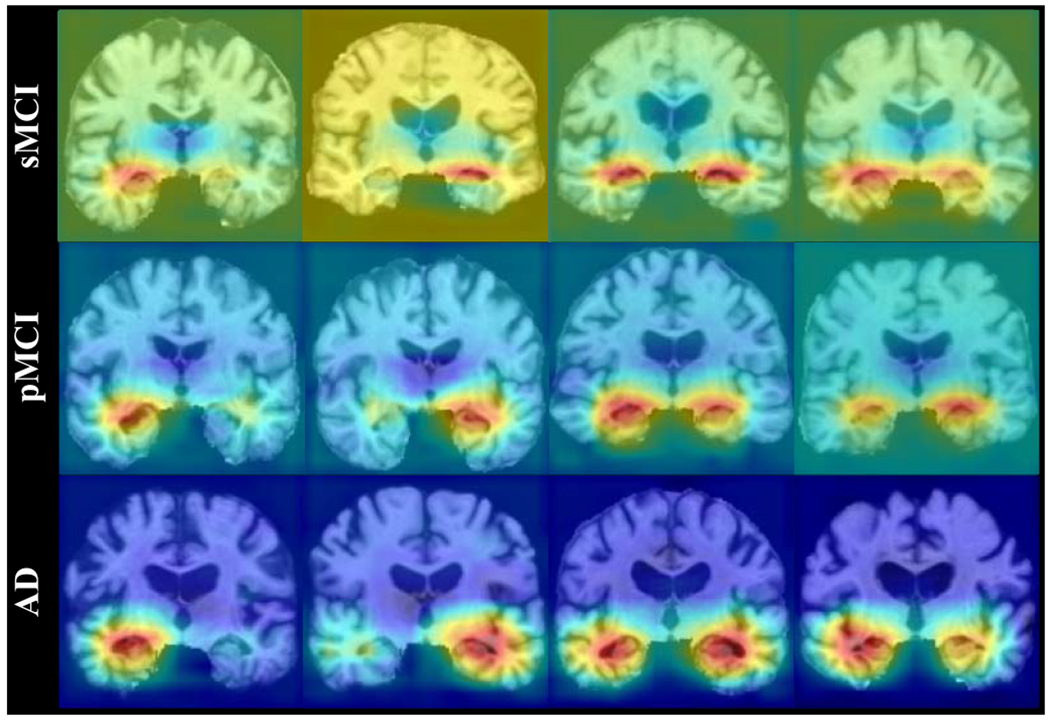
Attention maps produced by our MWAN for 12 different patients from three categories (i.e., AD, pMCI, and sMCI), where each attention map highlights specific brain regions or presents specific weights on the identified brain regions.
E. Comparison With Other Learnable Attention Mechanisms
Learnable attention mechanisms have been actively studied in recent years for dealing with various medical image computing tasks, e.g., segmentation [39]–[42], diagnosis [43], [44], and detection [45], [46]. Existing spatial attention modules were typically designed in a task-driven way. For example, to segment brain tumors from MR images, Zhou et al. [40] proposed a curriculum learning framework, called one-pass multitask network (OM-Net). OM-Net adopts an end-to-end coarse-to-fine structure to form the cascaded spatial attention for the identification of multiple intratumoral classes in one pass, which achieved state-of-the-art segmentation performance on several public data sets. Chen et al. [41] proposed a multitask attention-based semi-supervised learning (MASSL) framework, which leverages both labeled and unlabeled training data to learn a segmentation model. MASSL consists of a supervised segmentation subnetwork and an unsupervised reconstruction subnetwork, where the probability maps produced by the segmentation part serve as soft attentions to ensure the reconstruction of unlabeled training images being semantically meaningful. It is worth noting that these attention mechanisms designed for image segmentation are technically different from our dementia attention module. Specifically, they commonly adopt the ground-truth label maps as the dense supervision to produce soft attention, while our dementia attention module regards the image-level category labels as the weakly-supervised guidance for disease-related discriminative localization. On the other hand, our dementia attention module is also not the same as other learnable attentions for diagnosis/detection [43]–[46], that is, different from these classification frameworks, our MWAN uniquely leverages category labels as auxiliary information to produce subject-specific spatial attention for boosting the regression of intrinsically correlated clinical scores.
To further justify the unique role of the dementia attention module in our MWAN for multi-class clinical score regression, we compared it with another state-of-the-art learnable attention mechanism, i.e., the autofocus module [39]. Specifically, the autofocus module applies multiscale Conv layers (i.e., shared weights but different dilation rates) on an input tensor to produce a set of scale-invariant feature maps. In parallel, it uses an attention branch to generate an attention map for each Conv layer, by which the optimal-scale for each voxel is adaptively selected via the elementwise multiplication between the feature maps and attention maps. Based on the same backbone FCN, we directly replaced our dementia attention module with the autofocus module (with four scales) and constructed a variant of MWAN (denoted as MAFN) for multi-task clinical score regression. Under the same experimental setting, the attention maps produced by MWAN and MAFN for two representative AD subjects from ADNI-2 (using the models trained on ADNI-1) are compared in Fig. 10. Correspondingly, the respective regression results are compared in Fig. 11. Compared with the autofocus module, we can see from Figs. 10 and 11 that our dementia attention module generated obviously more precise attention maps with biologically meaningful highlights, which further result in more precise clinical score regression in terms of both two metrics. The main reason could be twofold. First, our dementia attention module is weakly supervised by the auxiliary category labels that are intrinsically correlated with the clinical scores. Second, different from organ segmentation, our task of MRI-based dementia status estimation has no explicit need of scale-invariant learning formulated in [39], as it principally ignored the fact that the pathological changes induced by dementia could be different across subjects. These results in some sense suggest the unique role of our dementia attention module in discriminative localization for dementia status estimation.
Fig. 10.
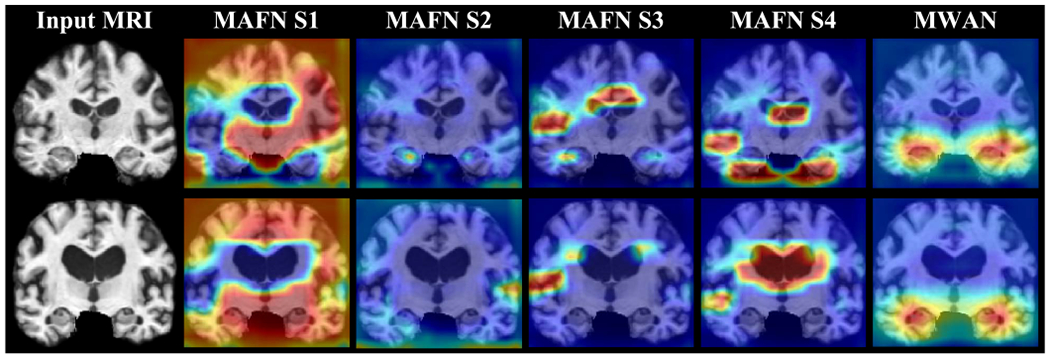
Attention maps produced by MAFN integrating autofocus module [39] and our MWAN, respectively, for two representative AD subjects from ADNI-2. Here, the autofocus module was implemented to have four different scales (i.e., S1–S4).
Fig. 11.
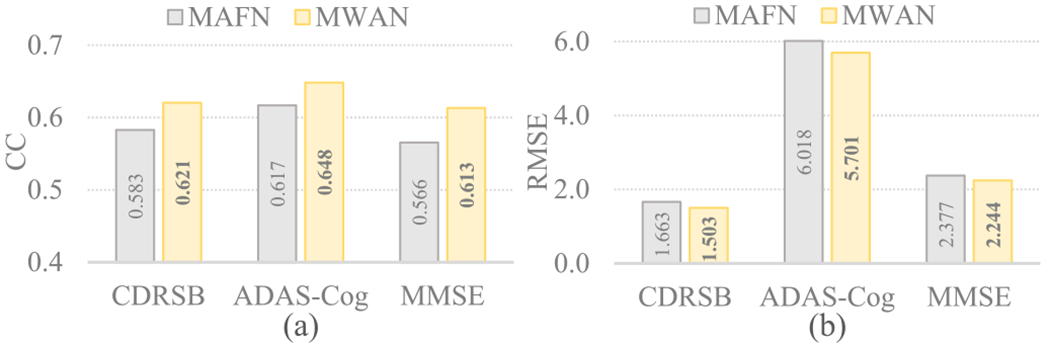
Regression results in terms of CC and RMSE obtained by MAFN integrating autofocus module [39] and our MWAN, respectively, on ADNI-2 (with the models trained on ADNI-1).
F. Limitations and Future Work
Different from previous CAD methods that typically rely on anatomical prior knowledge and time-consuming nonlinear registration to preselect fixed ROIs shared across all subjects, our MWAN is a more efficient method featuring an end-to-end learning architecture. It inputs directly the whole-brain images to automatically localize discriminative brain regions strongly correlated with the patient’s status for the learning of a multi-task regression model. Experimental results and ablation studies presented in Sections IV-C and V-A demonstrate that our MWAN outperforms other competing methods and their variants in the joint estimation of multiple clinical scores. As another practically useful property, our MWAN can produce biologically more informative localization results than methods using other attention mechanisms (see Sections IV-D and V-E). The respective attention maps could potentially provide auxiliary guidance to characterize subject-specific dementia status (see Section V-D). Although our MWAN method achieved good results in end-to-end discriminative localization and multiple clinical score regression, its performance and practical usage could be further improved in the future by carefully dealing with several limitations listed in the following.
First, the dementia attention maps generated by our current work are relatively coarse, considering that the discriminative localization was performed on the final backbone feature maps (size: 1/8 of the input whole-brain image). The coarse attention maps are not sensitive enough to identify very subtle structural changes for the accurate discrimination between different dementia statuses at the prodromal stage of AD (i.e., sMCI versus pMCI). For example, according to the demographic information summarized in Table I, the clinical values of CDRSB and MMSE are largely overlapped between the groups of sMCI and pMCI, resulting in worse regression accuracy than other groups as has been discussed in Table IV. For fine-grained localization in more detail, we could potentially use a top-down strategy (e.g., the feature pyramidal architecture [47]) to hierarchically place our proposed dementia attention module at multiple scales.
Second, while our implementation of MWAN can jointly predict multiple clinical scores, there is no explicit modeling of the correlations between different clinical scores (i.e., different regression tasks). Using the learned correlations between different tasks as auxiliary guidance has been extensively studied in the machine learning and computer vision communities [48], [49], which may also be applied in our task to boost the joint prediction of multiple clinical scores.
Besides, in our current work, only the baseline clinical scores were predicted from the baseline MRI scans. Effectively modeling longitudinal multimodal neuroimaging data (e.g., structural and functional MRI, and positron emission tomography) to automatically predict the progression of clinical scores over time should be a practically meaningful direction.
VI. Conclusion
In this article, we have proposed an MWAN, an end-to-end deep learning method, to automatically identify dementia-sensitive brain locations from the whole-brain MR images for the joint regression of multiple clinical scores. Specifically, based on the feature maps generated by a backbone FCN, our MWAN first adopts a dementia attention module to automatically localize subject-specific discriminative locations from the whole-brain image. After that, a multi-task regression module is then constructed to jointly predict multiple clinical scores. All components of our proposed MWAN can be jointly optimized to ensure their consistency. Experiments on 1,396 subjects from ADNI-1 and ADNI-2 have demonstrated the effectiveness of our proposed method in both the tasks of automated discriminative localization and dementia status estimation.
Acknowledgment
Data used in this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) data set. The investigators within the ADNI did not participate in the analysis or writing of this study. A complete listing of ADNI investigators can be found online.
The work of Mingxia Liu and Li Wang was supported in part by the NIH under Grant AG041721 and Grant MH117943.
Footnotes
Contributor Information
Chunfeng Lian, School of Mathematics and Statistics, Xi’an Jiaotong University, Xi’an 710049, China.
Mingxia Liu, Department of Radiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA; Biomedical Research Imaging Center (BRIC), The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Li Wang, Department of Radiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA; Biomedical Research Imaging Center (BRIC), The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
Dinggang Shen, School of Biomedical Engineering, ShanghaiTech University, Shanghai 201210, China; Department of Research and Development, Shanghai United Imaging Intelligence Co., Ltd., Shanghai 201210, China; Department of Artificial Intelligence, Korea University, Seoul 02841, Republic of Korea.
References
- [1].Jagust W, “Vulnerable neural systems and the borderland of brain aging and neurodegeneration” Neuron, vol. 77, no. 2, pp. 219–234, Jan. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gauthier S et al. , “Mild cognitive impairment,” Lancet, vol. 367, no. 9518, pp. 1262–1270, 2006. [DOI] [PubMed] [Google Scholar]
- [3].Tombaugh TN and McIntyre NJ, “The mini-mental state examination: A comprehensive review,” J. Amer. Geriatrics Soc, vol. 40, no. 9, pp. 922–935, Sep. 1992. [DOI] [PubMed] [Google Scholar]
- [4].O’Bryant SE et al. , “Staging dementia using clinical dementia rating scale sum of boxes scores: A Texas Alzheimer’s research consortium study,” Arch. Neurol, vol. 65, no. 8, pp. 1091–1095, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Graham DP, Cully JA, Snow AL, Massman P, and Doody R, “The Alzheimer’s disease assessment scale-cognitive subscale: Normative data for older adult controls,” Alzheimer Disease Associated Disorders, vol. 18, no. 4, pp. 236–240, 2004. [PubMed] [Google Scholar]
- [6].Frisoni GB, Fox NC, Jack CR Jr., Scheltens P, and Thompson PM, “The clinical use of structural MRI in Alzheimer disease,” Nature Rev. Neurol, vol. 6, no. 2, p. 67, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rathore S, Habes M, Iftikhar MA, Shacklett A, and Davatzikos C, “A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages,” Neuroimage, vol. 155, pp. 530–548, Jul. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sabuncu MR and Konukoglu E, “Clinical prediction from structural brain MRI scans: A large-scale empirical study,” Neuroinformatics, vol. 13, no. 1, pp. 31–46, Jan. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lian C, Liu M, Pan Y, and Shen D, “Attention-guided hybrid network for dementia diagnosis with structural MR images,” IEEE Trans. Cybern., early access, Jul. 28, 2020, doi: 10.1109/TCYB.2020.3005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stonnington CM, Chu C, Klöppel S, Jack CR, Ashburner J, and Frackowiak RSJ, “Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease,” Neuroimage, vol. 51, no. 4, pp. 1405–1413, Jul. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang Y, Fan Y, Bhatt P, and Davatzikos C, “High-dimensional pattern regression using machine learning: From medical images to continuous clinical variables,” Neuroimage, vol. 50, no. 4, pp. 1519–1535, May 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang D and Shen D, “Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease,” Neuroimage, vol. 59, no. 2, pp. 895–907, Jan. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhou J, Liu J, Narayan VA, and Ye J, “Modeling disease progression via multi-task learning,” Neuroimage, vol. 78, pp. 233–248, Sep. 2013. [DOI] [PubMed] [Google Scholar]
- [14].Zhu X, Suk H-I, Wang L, Lee S-W, and Shen D, “A novel relational regularization feature selection method for joint regression and classification in AD diagnosis,” Med. Image Anal, vol. 38, pp. 205–214, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang J, Li Q, Caselli RJ, Ye J, and Wang Y, “Multi-task dictionary learning based convolutional neural network for computer aided diagnosis with longitudinal images,” 2017, arXiv:1709.00042. [Online]. Available: http://arxiv.org/abs/1709.00042
- [16].Lian C, Liu M, Wang L, and Shen D, “End-to-end dementia status prediction from brain mri using multi-task weakly-supervised attention network,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assisted Intervent. Cham, Switzerland: Springer, 2019, pp. 158–167. [PMC free article] [PubMed] [Google Scholar]
- [17].Liu M, Zhang J, Adeli E, and Shen D, “Joint classification and regression via deep multi-task multi-channel learning for Alzheimer’s disease diagnosis,” IEEE Trans. Biomed. Eng, vol. 66, no. 5, pp. 1195–1206, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lian C, Liu M, Zhang J, and Shen D, “Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 42, no. 4, pp. 880–893, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jack CR et al. , “The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods,” J. Magn. Reson. Imag, vol. 27, no. 4, pp. 685–691, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tipping ME, “Sparse Bayesian learning and the relevance vector machine,” J. Mach. Learn. Res, vol. 1, pp. 211–244, Sep. 2001. [Google Scholar]
- [21].Fan Y, Shen D, Gur RC, Gur RE, and Davatzikos C, “COMPARE: Classification of morphological patterns using adaptive regional elements,” IEEE Trans. Med. Imag, vol. 26, no. 1, pp. 93–105, Jan. 2007. [DOI] [PubMed] [Google Scholar]
- [22].Tong T, Wolz R, Gao Q, Guerrero R, Hajnal JV, and Rueckert D, “Multiple instance learning for classification of dementia in brain MRI,” Med. Image Anal, vol. 18, no. 5, pp. 808–818, Jul. 2014. [DOI] [PubMed] [Google Scholar]
- [23].Zhang J, Gao Y, Gao Y, Munsell BC, and Shen D, “Detecting anatomical landmarks for fast Alzheimer’s disease diagnosis,” IEEE Trans. Med. Imag, vol. 35, no. 12, pp. 2524–2533, Dec. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krizhevsky A, Sutskever I, and Hinton GE, “ImageNet classification with deep convolutional neural networks,” in Proc. Adv. Neural Inf. Process. Syst. (NIPS), 2012, pp. 1097–1105. [Google Scholar]
- [25].Li H, Habes M, and Fan Y, “Deep ordinal ranking for multi-category diagnosis of Alzheimer’s disease using hippocampal MRI data,” 2017, arXiv:1709.01599. [Online]. Available: http://arxiv.org/abs/1709.01599
- [26].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 770–778. [Google Scholar]
- [27].Wen J et al. , “Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation,” 2019, arXiv:1904.07773. [Online]. Available: http://arxiv.org/abs/1904.07773 [DOI] [PubMed]
- [28].Shen D and Davatzikos C, “HAMMER: Hierarchical attribute matching mechanism for elastic registration,” IEEE Trans. Med. Imag, vol. 21, no. 11, pp. 1421–1439, Nov. 2002. [DOI] [PubMed] [Google Scholar]
- [29].Zhang J, Liu M, and Shen D, “Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks,” IEEE Trans. Image Process, vol. 26, no. 10, pp. 4753–4764, Oct. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou B, Khosla A, Lapedriza A, Oliva A, and Torralba A, “Learning deep features for discriminative localization,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 2921–2929. [Google Scholar]
- [31].Yang J, She D, Lai Y-K, Rosin PL, and Yang M-H, “Weakly supervised coupled networks for visual sentiment analysis,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., Jun. 2018, pp. 7584–7592. [Google Scholar]
- [32].Sled JG, Zijdenbos AP, and Evans AC, “A nonparametric method for automatic correction of intensity nonuniformity in MRI data,” IEEE Trans. Med. Imag, vol. 17, no. 1, pp. 87–97, Feb. 1998. [DOI] [PubMed] [Google Scholar]
- [33].Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, and Smith SM, “FSL,” NeuroImage, vol. 62, no. 2, pp. 782–790, 2012. [DOI] [PubMed] [Google Scholar]
- [34].Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, and Evans AC, “Enhancement of MR images using registration for signal averaging,” J. Comput. Assist. Tomogr, vol. 22, no. 2, pp. 324–333, Mar. 1998. [DOI] [PubMed] [Google Scholar]
- [35].Ashburner J and Friston KJ, “Voxel-based morphometry: The methods,” NeuroImage, vol. 11, no. 6, pp. 805–821, 2000. [DOI] [PubMed] [Google Scholar]
- [36].Jia H, Wu G, Wang Q, and Shen D, “ABSORB: Atlas building by self-organized registration and bundling,” NeuroImage, vol. 51, no. 3, pp. 1057–1070, Jul. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jia H, Yap P-T, and Shen D, “Iterative multi-atlas-based multi-image segmentation with tree-based registration,” NeuroImage, vol. 59, no. 1, pp. 422–430, Jan. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhan Y, Ou Y, Feldman M, Tomaszeweski J, Davatzikos C, and Shen D, “Registering histologic and MR images of prostate for image-based cancer detection,” Academic Radiol, vol. 14, no. 11, pp. 1367–1381, Nov. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Qin Y et al. , “Autofocus layer for semantic segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assisted Intervent. (MICCAI). Cham, Switzerland: Springer, 2018, pp. 603–611. [Google Scholar]
- [40].Zhou C, Ding C, Wang X, Lu Z, and Tao D, “One-pass multi-task networks with cross-task guided attention for brain tumor segmentation,” 2019, arXiv:1906.01796. [Online]. Available: http://arxiv.org/abs/1906.01796 [DOI] [PubMed]
- [41].Chen S, Bortsova G, Juárez AG-U, van Tulder G, and de Bruijne M, “Multi-task attention-based semi-supervised learning for medical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assisted Intervent. (MICCAI). Cham, Switzerland: Springer, 2019, pp. 457–465. [Google Scholar]
- [42].Nie D, Gao Y, Wang L, and Shen D, “ASDNet: Attention based semi-supervised deep networks for medical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assisted Intervent. (MICCAI). Cham, Switzerland: Springer, 2018, pp. 370–378. [Google Scholar]
- [43].Qaiser T and Rajpoot NM, “Learning where to see: A novel attention model for automated immunohistochemical scoring,” IEEE Trans. Med. Imag, vol. 38, no. 11, pp. 2620–2631, Nov. 2019. [DOI] [PubMed] [Google Scholar]
- [44].Zhang J, Xie Y, Xia Y, and Shen C, “Attention residual learning for skin lesion classification,” IEEE Trans. Med. Imag, vol. 38, no. 9, pp. 2092–2103, Sep. 2019. [DOI] [PubMed] [Google Scholar]
- [45].Schlemper J et al. , “Attention gated networks: Learning to leverage salient regions in medical images,” Med. Image Anal, vol. 53, pp. 197–207, Apr. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xu H, Dong M, Lee M-H, O’Hara N, Asano E, and Jeong J-W, “Objective detection of eloquent axonal pathways to minimize postoperative deficits in pediatric epilepsy surgery using diffusion tractography and convolutional neural networks,” IEEE Trans. Med. Imag, vol. 38, no. 8, pp. 1910–1922, Aug. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, and Belongie S, “Feature pyramid networks for object detection,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 2117–2125. [Google Scholar]
- [48].Zhang Z, Luo P, Loy CC, and Tang X, “Learning deep representation for face alignment with auxiliary attributes,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 38, no. 5, pp. 918–930, May 2016. [DOI] [PubMed] [Google Scholar]
- [49].Liu M, Zhang D, Chen S, and Xue H, “Joint binary classifier learning for ECOC-based multi-class classification,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 38, no. 11, pp. 2335–2341, Nov. 2016. [Google Scholar]


